Abstract
The entire epithelium of the lung is generated from a small pool of undifferentiated progenitor cells. At least during the early stages of development these reside in the distal tips of the embryonic lung. They respond to multiple signals from the surrounding mesenchyme and play a critical role as morphogenetic organizing centers. In addition, they proliferate rapidly and give rise to daughter cells that differentiate into all the specialized epithelial cells types of the newborn lung. Despite the importance of the progenitor cells, we still know relatively little about the mechanisms controlling their proliferation, morphogenesis, and developmental fate. Here, we discuss new data on the potential role of microRNAs in co-coordinately regulating multiple signaling pathways in embryonic progenitor cells. In particular, our recent transgenic experiments suggest that microRNAs encoded by the miR-17-92 cluster positively promote proliferation of epithelial progenitor cells and inhibit their differentiation. We speculate on how this information might be exploited therapeutically in the long term.
Keywords: miR-17-92, transgenic mouse, Sox9, NMyc, cancer
The newborn lung contains at least six epithelial cell types—basal cells; ciliated, secretory, and neuroendocrine cells; and type I and type II alveolar cells—organized in the correct proportion and spatial pattern. All of these cell types arise from undifferentiated epithelial progenitor cells in the embryonic lung (Reference 1 and our own unpublished observations). In addition, their generation involves a delicate balance between the opposing forces of proliferation and differentiation. Rapid proliferation of the progenitors, in association with the outgrowth and branching of the epithelial tubes, is essential for producing a lung that is large enough and has a sufficient number of alveoli to sustain life. When undifferentiated cells, or their descendants, leave the progenitor pool, they give rise first to lineage-committed precursor cells with a slower rate of proliferation and eventually to fully differentiated cells. This progression is responsible for generating the diversity of cell types needed for normal lung function. A major challenge is to understand not only the basic properties of progenitor cells but also how the balance between their proliferation and differentiation is regulated. For example, it would be clinically advantageous to know how to maintain this balance in the lungs of premature babies and to avoid therapies that drive differentiation at the expense of retaining a large enough pool of progenitor cells to complete alveolar formation. It is also important to know when in embryonic development the cells that are responsible for repairing the epithelium of the adult lung after injury are laid down. This information will enable us to better understand how prematurity or adverse events such as infection during the fetal and early neonatal period can affect the cellular composition and repair capacity of the adult lung. Armed with this knowledge, we can begin to explore therapeutic treatments to prevent long-term respiratory problems.
EPITHELIAL PROGENITOR CELLS OF THE EMBRYONIC LUNG
There is considerable evidence that, at least during the pseudoglandular stage of lung development, the distal tips of the branching tubules contain a population of undifferentiated multipotent epithelial progenitor cells (Figure 1). Among the questions our lab is addressing, using the mouse as a model system, are the following:
Are the progenitor cells a homogeneous population of multipotent cells, each having an equal potential of giving rise to progeny that can differentiate into any of the specialized cell types, or are there several different subpopulations of progenitors, each with more restricted developmental fates?
How is the proliferation and undifferentiated state of the progenitor cells maintained?
What mechanisms control the diversification of the progenitor cell population left behind by the outgrowth of the distal tips?
Here, we focus on recent experiments that provide evidence that microRNAs (miRNAs) regulate the proliferation and undifferentiated phenotype of lung epithelial progenitor cells.
Figure 1.
Epithelial progenitor cells of the embryonic lung. (A) Schematic representation of epithelial progenitor cells during early lung development. During the pseudoglandular stage (∼E10.5-16 in the mouse), the lung contains a system of actively branching epithelial tubes. Cells at the distal tip are morphologically unspecialized and are characterized by the expression of a number of genes encoding signaling proteins, transcription factors, and components of downstream signaling pathways. These include Sox9, NMyc, Id2 (Idb2), Pea3, Erm, Bmp4 (see References 25 and 31). There are numerous reciprocal interactions (red arrows) between the epithelium and the surrounding mesenchyme and vasculature that involve multiple secreted signaling factors, including sonic hedgehog, Fgfs, and Wnts (25–27). As the tips grow out and branch, they leave behind cells in the proximal stalks that give rise to differentiated cell types. Markers for these phenotypes first appear around E14.5 in the most proximal regions (32), and the process of differentiation proceeds from proximal to distal. Just before birth, the cells in the distal tips give rise to the precursors of the terminal alveoli containing type I and type II cells. It is not known how the switch in fate of the daughter cells—from proximal airway cells to alveolar cells—is regulated. (B) The distal progenitor cells rapidly transit the cell cycle so that after a 1-hour pulse of bromodeoxyuridine (BrdU) in utero at E12.5, about 80% of the distal tip cells are labeled (arrow) (29). Scale bar = 50 μm. (C) Whole mount in situ hybridization of E11.5 mouse lung shows that distal epithelial progenitor cells (arrow) preferentially express the transcription factor, Id2 (also known as Idb2). (D) Immunohistochemistry of section of E14.5 mouse lung shows that distal epithelial cells preferentially express Sox9 (arrow). In the adult lung, Sox9 expression is absent (see also Reference 33). Scale bar = 60 μm.
miRNAs AND LUNG DEVELOPMENT
miRNAs are small, single-stranded, noncoding RNAs (∼22 bp in length) that negatively regulate messenger RNA translation and stability at a post-transcriptional level (2–7). There are currently about 533 miRNAs encoded in the human genome and 442 in the mouse (http://microrna.sanger.ac.uk/); most of the genes are conserved between species, but some are specific for humans versus mice. They are transcribed as large primary RNAs, which then enter a processing pathway (8, 9). First, they are cut into smaller precursors by Drosha, an RNAse-III enzyme that is part of a large protein complex with Dgcr8 (DiGeorge syndrome critical region gene 8). After transport to the cytoplasm, another enzyme, Dicer, converts the precursors into mature miRNAs that are recruited into the RNA-induced silencing complex (RISC). Finally, this complex interferes with the translation and stability of target messenger RNAs, in general by imperfect base-pairing to target sequences in the 3′ untranslated region (4). Individual miRNAs may target multiple mRNAs. Conversely, individual mRNAs may contain sequences complementary to multiple miRNA family members (10–12).
The first evidence that miRNAs play a role in lung development was the abnormal phenotype of embryonic lungs in which the gene encoding the enzyme Dicer was deleted specifically in the endoderm of the primary lung buds (13). The lungs showed a dramatic reduction in branching morphogenesis, beginning at E12.5, and there is extensive apoptosis. Interpretation of this result is complicated by the fact that Dicer processes precursors not only of miRNAs but also of short interfering RNAs (siRNAs). These are essential for maintaining normal heterochromatin structure (14, 15), so that defects in organ development due to absence of Dicer cannot to attributed to failure to process miRNAs alone.
A number of publications have reported the expression of miRNAs in the mouse or rat lung (16–18). However, there were no studies on the function of specific miRNAs in lung development. To address this problem, we first identified the miRNA expression profile at different stages of development (19). This was performed in three ways. First, total lung RNA isolated from several stages was hybridized to custom-built arrays containing 198 mouse miRNA probes (20, 21). Second, small RNAs were cloned from E11.5 and E17.5 lung total RNA, and 1,668 and 1,941 clones sequenced, respectively. This provided a more quantitative measure of the different miRNAs present at these two stages, and the most abundant miRNAs are shown in Tables 1 and 2. The location of some of these miRNAs was then analyzed by whole mount and section in situ hybridization using commercially generated short “locked nucleic acid” probes. Initial analysis of these data revealed several interesting features. First, some of the miRNAs most highly expressed in the early embryonic lung (E11.5) are also elevated in small cell lung cancer tumors (22) and adenocarcinomas (23). Among the miRNAs that have high levels in both the embryonic lung and lung cancer are members of the miR-17-92 cluster (see below). Second, the converse is also true; a number of the miRNAs more highly expressed in the adult compared with embryonic lung are expressed at lower levels in tumors compared with normal lung. Among the latter category are members of the let-7 family. This was one of the first miRNA families to be described, and among the known targets are mRNAs for Ras proteins (24). Ras lies downstream of receptor tyrosine kinase signaling, including the fibroblast growth factor (FGF) receptors. FGF signaling is known to be important for lung branching morphogenesis and epithelial cell proliferation (see References 25–27). It therefore makes sense that let-7 members would be more highly expressed in the normal adult or neonatal lung when growth is reduced compared with early embryonic stages.
Among the miRNAs most highly expressed in early pseudoglandular stage are members of the miR-17-92 cluster (Tables 1 and 2). In situ hybridization shows that at least three members, miR-17-5p, miR-20a, and miR-19b, are present in both the epithelium and mesenchyme of the embryonic lung and confirms that the levels decline as development proceeds. The miR-17-92 cluster is encoded by a single locus on chromosome 14 in mice and chromosome 13 in humans. It is transcribed as a single RNA that is then processed into six mature members. However, this processing does not seem to be equally efficient for all members. For example, mature miR-19a has very low abundance compared with other members. There is recent evidence for regulation of miRNA processing during mouse embryonic development at the level of Drosha but the role of miRNA processing in lung development has not yet been addressed (28).
TABLE 1.
RELATIVE ABUNDANCE OF MICRORNAS IN THE EMBRYONIC LUNG: THE 20 MOST ABUNDANT MICRORNA FAMILIES IN E11.5 LUNG
| Family Name | Family Members in Mouse | E11.5 Clones | Relative Abundance (%) |
|---|---|---|---|
| miR-17 | miR-17-5p/106a/106b/18/20a/20b/93 | 175 | 10.5 |
| miR-125 | miR-125a/125b | 91 | 5.5 |
| miR-130 | miR-130a/130b/301a/301b | 70 | 4.2 |
| miR-199 | miR-199a/199b | 64 | 3.8 |
| miR-15 | miR-15a/15b/16 | 58 | 3.5 |
| miR-8 | miR-200a/200b/200c/141/429 | 47 | 2.8 |
| miR-27 | miR-27a/27b | 44 | 2.6 |
| miR-199a* | miR-199a* | 41 | 2.5 |
| miR-25 | miR-25/92 | 40 | 2.4 |
| let-7 | let-7a/b/c/d/e/f/g/i/miR-98 | 38 | 2.3 |
| miR-21 | miR-21 | 38 | 2.3 |
| miR-136 | miR-136 | 32 | 1.9 |
| miR-99 | miR-99a/99b/100 | 22 | 1.3 |
| miR-214 | miR-214 | 20 | 1.2 |
| miR-19 | miR-19a/19b | 20 | 1.2 |
| miR-23 | miR-23a/23b | 19 | 1.1 |
| miR-30 | miR-30a/30b/30c/30d/30e | 18 | 1.1 |
| miR-126-3p | miR-126-3p | 17 | 1.0 |
| miR-351 | miR-351 | 17 | 1.0 |
| miR-26 | miR-26a/26b | 17 | 1.0 |
A total of 1,668 and 1,941 clones was obtained from E11.5 and E17.5 total lung RNA, respectively; 69.1% of E11.5 clones and 89.2% of E17.5 clones are known microRNAs (miRNAs). The definition of miRNA families was derived from http://microrna.sanger.ac.uk.
TABLE 2.
RELATIVE ABUNDANCE OF MICRORNAS IN THE EMBRYONIC LUNG: THE 20 MOST ABUNDANT MICRORNA FAMILIES IN E17.5 LUNG
| Family Name | Family Members in Mouse | E17.5 Clones | Relative Abundance (%) |
|---|---|---|---|
| let-7 | let-7a/b/c/d/e/f/g/i/miR-98 | 237 | 12.2 |
| miR-15 | miR-15a/15b/16 | 134 | 6.9 |
| miR-17 | miR-17-5p/106a/106b/18/20a/20b/93 | 96 | 4.9 |
| miR-125 | miR-125a/125b | 86 | 4.4 |
| miR-21 | miR-21 | 69 | 3.6 |
| miR-126-3p | miR-126-3p | 63 | 3.2 |
| miR-130 | miR-130a/130b/301a/301b | 61 | 3.1 |
| miR-8 | miR-200a/200b/200c/141/429 | 59 | 3.0 |
| miR-351 | miR-351 | 58 | 3.0 |
| miR-30 | miR-30a/30b/30c/30d/30e | 55 | 2.8 |
| miR-199 | miR-199a/199b | 53 | 2.7 |
| miR-449 | miR-449 | 51 | 2.6 |
| miR-126-5p | miR-126-5p | 48 | 2.5 |
| miR-322 | miR-322 | 40 | 2.1 |
| miR-199a* | miR-199a* | 39 | 2.0 |
| miR-142-3p | miR-142-3p | 36 | 1.9 |
| miR-27 | miR-27a/27b | 34 | 1.8 |
| miR-25 | miR-25/92 | 31 | 1.6 |
| miR-99 | miR-99a/99b/100 | 30 | 1.5 |
| miR-136 | miR-136 | 28 | 1.4 |
A total of 1,668 and 1,941 clones were obtained from E11.5 and E17.5, respectively' 69.1% of E11.5 clones and 89.2% of E17.5 clones are known microRNAs (miRNAs). The definition of miRNA families was derived from http://microrna.sanger.ac.uk.
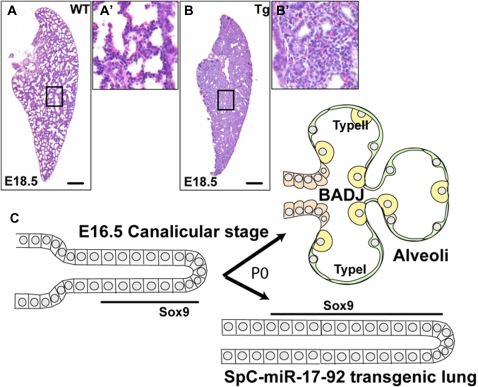
To explore the role of the miR-17-92 cluster in lung development, we generated transgenic embryos in which the whole cluster is expressed at high levels in the endoderm using the 6-kB regulatory fragment (“promoter”) of the mouse surfactant protein C (SftpC) gene. This drives transgene expression in the distal epithelium from soon after the primary buds appear and remains on in the distal progenitor cells through development. Levels gradually increase and then reach very high levels in the differentiated type II alveolar cells. All transgenic Sftpc–miR-17-92 lungs in which the transgene is expressed are very abnormal and no pups survive beyond a few days (19). The abnormal phenotype at E18.5 is illustrated in Figure 2. Terminal sacs, which represent the future alveoli, are completely absent and instead the lung is full of tubes lined by cuboidal epithelium. The cuboidal cells are highly proliferative, as judged by incorporation of bromodeoxyuridine (BrdU) after a 1-hour in utero pulse, and express high levels of Sox9 and NMyc, two markers of distal progenitor cells. By contrast, the expression of markers of differentiated Clara and ciliated cells is reduced.
Figure 2.
Transgenic overexpression of the miR-17-92 cluster in the epithelium of the embryonic mouse lung promotes expansion of the pool of progenitor cells and inhibits their differentiation. (A) Section of wild-type (nontransgenic) E18.5 lung stained with hematoxylin and eosin to show normal structure with multiple terminal sacs that will give rise to the alveoli. The black box indicates typical area, which is shown at higher magnification in A′. Scale bar = 200 μm. (B) Section of transgenic Sftpc–miR-17-92 lung at E18.5 showing the persistence of tubes lined with cuboidal epithelial cells; B′ shows area boxed in black at higher magnification. The cuboidal epithelial cells express Sox9 and NMyc (19). (C) Schematic representation of the effect of miR-17-92 overexpression. At E16.5, the terminal tubes of the lung at the canalicular stage still express Sox9 (see Figure 1D). In the wild-type lung, the terminal tubes give rise to the alveolar cells. These include type I (green) and type II (yellow) epithelial cells, whereas the more proximal cells differentiate into Clara and ciliated cells (orange). BADJ = bronchioalveolar junction; P0 = Postnatal Day 0 (birth). In Sftpc–miR-17-92 transgenic lungs, there is persistence of the progenitor cells expressing Sox9 and absence of differentiated alveolar cells.
The phenotype of SftpC–miR-17-92 transgenic lungs is very similar to that seen when NMyc is overexpressed using the same promoter (29). The results suggest that miR-17-92 family members normally function to promote the proliferation of progenitor cells and that they have to be down-regulated in the progeny, either at the transcriptional or processing stage (or both) in order for the cells to differentiate. Overexpression of miR-17-92 enhances the proliferation and self-renewal of the embryonic progenitor cells and inhibits or delays their differentiation (Figure 2).
The precise mechanism by which overexpression of the miR-17-92 cluster promotes the expansion of the embryonic distal epithelial progenitor pool and the identity of the downstream targets for the different family members are currently under investigation. We have already shown that the cell-cycle regulator Rbl2 is one potential target (19), and it is likely that many more will be found. This is based on the assumption that miRNAs co-coordinately regulate multiple mRNAs within one physiologically relevant program.
miRNAs AS POTENTIAL THERAPEUTIC AGENTS
In the future, we need to know whether all miR-17-92 family members are required to promote the proliferation of the progenitor cells, or only one, or a few. Although up-regulation of miR-17-92 members is seen in some lung cancers (22), it is likely that, in the short term, and in untransformed cells, the effect of overexpression is reversible, and this needs to be tested in vivo. By analogy, transient overexpression of the transcription factor Oct4 in transgenic mouse embryos leads to massive epithelial hyperplasia. However, this resolves after the Oct4 expression is switched off, and the expanded pools of tissue-specific progenitor cells apparently differentiate normally (30). We therefore plan to test whether up-regulation of miR-17-92 under the control of an inducible promoter can expand the pool of lung progenitor/stem cells in a reversible way. If so, it might be possible, in the future, to therapeutically target transient expression of one or a few miR-17-92 family members to the injured adult lung, or even to the lungs of severely premature babies, in such a way as to promote repair or the continued proliferation of endogenous progenitor or stem cells.
Acknowledgments
The authors thank J. Michael Thomson and Scott M. Hammond of the Department of Cell and Developmental Biology, Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, North Carolina, for their very generous support and help with microarray analysis.
Supported by NIH grant HL071303 (to B.L.M.H.).
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Perl AK, Wert SE, Nagy A, Lobe CG, Whitsett JA. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proc Natl Acad Sci USA 2002;99:10482–10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bushati N, Cohen SM. MicroRNA functions. Annu Rev Cell Dev Biol 2007;23:175–205. [DOI] [PubMed] [Google Scholar]
- 3.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science 2003;299:1540. [DOI] [PubMed] [Google Scholar]
- 4.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 2006;20:515–524. [DOI] [PubMed] [Google Scholar]
- 5.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature Rev 2004;5:522–531. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci 2007;32:189–197. [DOI] [PubMed] [Google Scholar]
- 7.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell 2006;11:441–450. [DOI] [PubMed] [Google Scholar]
- 8.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 2005;6:376–385. [DOI] [PubMed] [Google Scholar]
- 9.Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell 2004;16:861–865. [DOI] [PubMed] [Google Scholar]
- 10.Rajewsky N. MicroRNA target predictions in animals. Nat Genet 2006;38:S8–S13. [DOI] [PubMed] [Google Scholar]
- 11.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell 2006;126:1203–1217. [DOI] [PubMed] [Google Scholar]
- 12.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 2007;27:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci USA 2006;103:2208–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 2005;19:489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol 2004;6:784–791. [DOI] [PubMed] [Google Scholar]
- 16.Williams AE, Perry MM, Moschos SA, Lindsay MA. MicroRNA expression in the aging mouse lung. BMC Genomics 2007;8:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moschos SA, Williams AE, Perry MM, Birrell MA, Belvisi MG, Lindsay MA. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genomics 2007;8:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Weng T, Gou D, Chen Z, Chintagari NR, Liu L. Identification of rat lung-specific microRNAs by micoRNA microarray: valuable discoveries for the facilitation of lung research. BMC Genomics 2007;8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA mir-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol 2007;310:442–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature 2005;435:828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods 2004;1:47–53. [DOI] [PubMed] [Google Scholar]
- 22.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microrna cluster, mir-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res 2005;65:9628–9632. [DOI] [PubMed] [Google Scholar]
- 23.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006;9:189–198. [DOI] [PubMed] [Google Scholar]
- 24.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. Ras is regulated by the let-7 microRNA family. Cell 2005;120:635–647. [DOI] [PubMed] [Google Scholar]
- 25.Cardoso WV, Lu J. Regulation of early lung morphogenesis: questions, facts and controversies. Development 2006;133:1611–1624. [DOI] [PubMed] [Google Scholar]
- 26.White AC, Xu J, Yin Y, Smith C, Schmid G, Ornitz DM. Fgf9 and Shh signaling coordinate lung growth and development through regulation of distinct mesenchymal domains. Development 2006;133:1507–1517. [DOI] [PubMed] [Google Scholar]
- 27.Ramasamy SK, Mailleux AA, Gupte VV, Mata F, Sala FG, Veltmaat JM, Del Moral PM, De Langhe S, Parsa S, Kelly LK, et al. Fgf10 dosage is critical for the amplification of epithelial cell progenitors and for the formation of multiple mesenchymal lineages during lung development. Dev Biol 2007;307:237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 2006;20:2202–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okubo T, Knoepfler PS, Eisenman RN, Hogan BL. Nmyc plays an essential role during lung development as a dosage-sensitive regulator of progenitor cell proliferation and differentiation. Development 2005;132:1363–1374. [DOI] [PubMed] [Google Scholar]
- 30.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 2005;121:465–477. [DOI] [PubMed] [Google Scholar]
- 31.Maeda Y, Dave V, Whitsett JA. Transcriptional control of lung morphogenesis. Physiol Rev 2007;87:219–244. [DOI] [PubMed] [Google Scholar]
- 32.Rawlins EL, Ostrowski LE, Randell SH, Hogan BL. Lung development and repair: contribution of the ciliated lineage. Proc Natl Acad Sci USA 2007;104:410–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perl AK, Kist R, Shan Z, Scherer G, Whitsett JA. Normal lung development and function after sox9 inactivation in the respiratory epithelium. Genesis 2005;41:23–32. [DOI] [PubMed] [Google Scholar]