Abstract
Acute lung injury (ALI) is a common and frequently devastating illness characterized by acute hypoxemic respiratory failure, profound inflammation, and flooding of the alveoli. Despite recent advances in ALI care, the morbidity and mortality of ALI continues to be unacceptably high. ALI-inciting events (e.g., sepsis, trauma, aspiration, pneumonia) are quite common, yet only a fraction of patients develop the syndrome. This heterogeneity of patients presenting with ALI has sparked interest in identifying the role of genetic factors that contribute to ALI susceptibility and prognosis. Recent advances in high-throughput sequencing and expression technologies now provide the tools to perform large-scale genomic analyses in complex disorders such as ALI; gene expression profiling and pathway analysis provide further insight into previously described molecular pathways involved in the syndrome. In this article, we describe the use of genomewide association studies, ortholog in silico techniques, utility of consomic rat methods, and candidate gene approaches using expression profiling and pathway analyses. These methods have confirmed suspected ALI candidate genes (e.g., IL-6 and MIF), but more impressively have identified novel genes (e.g., GADD45α and PBEF) not previously suspected in ALI. The analysis of the molecular pathways (e.g., the cytoskeleton in vascular barrier regulation) has identified additional genes contributing to the development and severity of ALI (e.g., MLCK), thereby providing therapeutic targets in this devastating illness.
Keywords: acute lung injury, genetics, ventilator-induced lung injury
GENETICS OF LUNG INJURY AND REPAIR
Acute lung injury (ALI) describes a syndrome characterized by an acute onset of hypoxemic respiratory failure and bilateral radiographic infiltrates in the absence of elevated left atrial pressures (1). ALI, and its more severe form, acute respiratory distress syndrome (ARDS), occur in an estimated 190,000 cases in the United States every year and continue to carry an unacceptable mortality rate of 30 to 40%, despite recent advances in treatment of the critically ill (2). Aggressive exploration into pathogenesis of ALI occurring in conditions as varied as sepsis, trauma, aspiration, or pneumonia (3) has highlighted the critical role of severe inflammation and alveolar flooding by protein-rich fluid due to increased vascular permeability, resulting in the profound physiologic impairment characteristic of the syndrome. However, despite recent increased systems biology–like understanding of ALI (genes to populations), there remains a need to explain the heterogeneity of patients with ALI and to improve therapeutic options, which are currently severely limited in this critically ill group of patients.
In prior work, we focused our efforts on two aspects of ALI pathobiology: understanding the genetic factors involved in determining ALI risk/severity and identifying novel biomarkers to allow selection of susceptible populations and to drive new therapies. The molecular basis of ALI, however, has proven difficult to study due to the following factors: (1) the diversity in stimuli capable of evoking lung injury, (2) the heterogeneous nature of the syndrome itself, (3) the presence of varied comorbid illnesses, (4) potential incomplete gene penetrance, and (5) complex gene–environment interactions. Traditionally, the approach to explain the molecular basis of complex disorders such as ALI involved a “gene by gene” approach focusing on a putative single gene/gene product important in disease pathogenesis. More recently, a global gene approach has involved linkage studies using DNA from afflicted families to query the genome using known genetic “markers” that span the genome to identify “genomic hot spots” associated with a given phenotype. This has led to successes in identifying ALI genes, such as components of inflammatory pathways (e.g., tumor necrosis factor [TNF]-β and IL-10) (4, 5), genes encoding key alveolar products (i.e., surfactant protein B) (6–8), and genes involved in the coagulation pathways (9). However, linkage studies of this type are labor intensive and expensive and difficult to conduct in ALI as there is rarely a family history of lung injury, the disease occurs sporadically, and the effects of genetic polymorphisms on the susceptibility and severity of ALI are likely modest at best.
In this report, we describe a variety of approaches to identify genes and gene variations responsible for ALI in the era following mapping of the human genome. The advent of high-throughput gene sequencing and expression technologies, and complete genome sequencing of model organisms, now provide the tools to perform large-scale analyses of the genome in complex disorders such as ALI. Whole genome scans, in silico approaches, utilization of consomic rats, and a candidate gene approach involving expression profiling and pathway analysis are proving exceptionally useful in identifying novel candidate genes and genetic variations.
DISCOVERING ALI SUSCEPTIBILITY GENES: GENOMEWIDE ASSOCIATION APPROACHES
The concept that genetic factors may be involved in the development of ALI was suggested by the possible association of a previously known insertion/deletion polymorphism (D) in the angiotensin converting enzyme (ACE) gene with worsened mortality among patients with ARDS (DD genotype with fourfold increase in mortality) (10). This landmark association study set the stage for additional studies and approaches to more firmly establish a genetic basis of ALI and to identify ALI candidate genes. Despite the availability of linkage techniques, given that there is no known family inheritance of ALI susceptibility, no linkage studies in ALI have ever been performed. However, high-throughput whole genome scanning technology has become a more powerful tool than linkage studies, particularly in detecting disease susceptibility genes with modest effects. The International HapMap Project (11), which identified blocks of single nucleotide polymorphisms (SNPs) linked to each other, has allowed selection of the most informative SNPs for further disease association studies (12). Currently, the most common high-throughput SNP platforms, the Illumina platform and the Affymetrix SNP chip, involve high-throughput assessment of nearly a million SNPs spanning the genome (i.e., genomewide association studies [GWAS]). Both GWAS platforms are effective and have been successfully used in diverse disorders such as age-related macular degeneration (13), inflammatory bowel disease (14), and type 2 diabetes (15). Although this approach has yet to be used in either ALI or sepsis, the application of GWAS to these complex lung diseases is undoubtedly imminent.
DISCOVERING ALI SUSCEPTIBILITY GENES: IN SILICO, GENOMIC, AND CONSOMIC APPROACHES
One method we have successfully used to identify ALI candidate genes is an in silico ortholog gene approach. The thematic underpinning of this approach is the hypothesis that patients with ALI and preclinical animal models of ALI would exhibit commonality in expression of evolutionarily conserved genes across species. Using profiling results from more than 50 Affymetrix microarray chips obtained from ventilator-associated ALI models (VALI) (human, rat, mouse, dog), we identified 3,077 genes whose expression was altered across all four species in response to ventilator-associated mechanical stress (16, 17). Filtering these results for unidirectional change in gene expression (with >1.3-fold change) refined the list to 69 genes, reflecting specific ALI-associated gene categories (modules/ontologies): coagulation, inflammation, chemotaxis/cell motility, and immune response. This approach identified multiple genes already recognized as highly likely to play a role in ALI (IL-6, aquaporin 1 [AQP-1], plasminogen activator inhibitor type 1 [PAI-1]) (12, 13, 22, 31), as well as several novel genes not previously known to be mechanistically involved in ALI (17) (Table 1).
TABLE 1.
EXPRESSION PROFILING FOR CANDIDATE GENE SELECTION IN VENTILATOR-ASSOCIATED ACUTE LUNG INJURY: IN SILICO APPROACH
| Gene* | Gene Symbol | Fold Change (ventilation vs. control) |
|---|---|---|
| ALI related | ||
| Interleukin 1β | IL-1β | 1.53 |
| Interleukin 6 | IL-6 | 1.84 |
| Tissue factor/thromboplastin | F3 | 1.52 |
| Plasminogen activator inhibitor type 1 | PAI-1 | 1.47 |
| Cyclooxygenase II | COX2 | 1.79 |
| Interleukin 13 | IL-13 | 1.30 |
| Aquaporin 1 | AQP-1 | −1.30 |
| Plasminogen activator, urokinase receptor | PLAUR | 1.47 |
| FGA, fibrinogen alpha; | FGA | 1.30 |
| CCAAT/enhancer-binding protein | C/EBP | 1.40 |
| Interleukin 1 receptor antagonist | CCL2 | 2.00 |
| Adrenomedullin receptor | ADMR | −1.35 |
| VALI candidates | ||
| Chemokine (C-X-C motif) receptor 4 | CXCR4 | 1.62 |
| Gap junction protein, alpha 1 (connexin 43) | GJA-1 | 1.33 |
| Interleukin 1 receptor, type II | IL1R2 | 1.88 |
| Growth arrest DNA damage-inducible, alpha | GADD45α | 1.71 |
| B-cell translocation gene 1 | BTG-1 | 1.38 |
| Trefoil factor 2 (spasmolytic protein 1) | TFF-2 | −1.32 |
Definition of abbreviations: ALI = acute lung injury; VALI = ventilator-associated acute lung injury.
Candidate genes achieved more than 1.30-fold change in expression from rat, murine, and canine models and human samples with ALI or VALI compared with controls (P < 0.05).
One novel gene surviving this ortholog gene-filtering approach after mechanical stress is the growth arrest and DNA damage-inducible 45α (GADD45α) gene (17), a member of an evolutionarily conserved gene family whose expression increases after genotoxic or environmental stress (18, 19). GADD45α, a small, 21-kD, predominantly nuclear protein, induces G2/M cell cycle arrest via direct inhibition of Cdc2–cyclinB1 complex, induces G1/S cell cycle arrest through cyclin-dependent kinase inhibitor p21, and interacts with proliferating cell nuclear antigen (PCNA), a nuclear protein that plays a central role in DNA repair (20). In addition, GADD45α induces apoptosis in most cells but may also promote survival in hematopoetic cells likely due to DNA repair functions (19). GADD45α maintains genomic stability in a p53-responsive manner (21) and acts as a negative regulator of T-cell proliferation (22).
Despite its multiple known functions, the role of GADD45α in ALI, endothelial/epithelial barrier dysfunction, or repair of injured lung is unknown (17). We and others have demonstrated that GADD45α expression is increased in an LPS/mechanical ventilation murine model (23) or by hyperoxic exposure (24, 25). The GADD45α gene contains upward of 25 validated SNPs (National Center for Biotechnology Information [NCBI] SNP database) whose role in risk of development, severity, or recovery from ALI is completely unknown (18). We recently validated the potential importance of increased GADD45a expression in ALI, demonstrating that GADD45a knockout mice experience increased injury after LPS challenge (26) and are currently pursuing further characterization of the role of GADD45a and its variants in VALI.
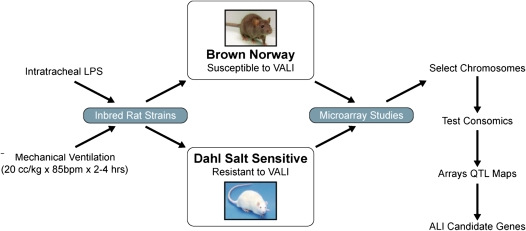
Complementing the in silico approach described above, we recently used a consomic rat approach to identify novel ALI gene candidates (27). Two strains of inbred rodents were determined to have differing susceptibility to VALI (20 cc/kg, 4 h): the VALI-sensitive Brown Norway (BN) rat strain and the VALI-resistant Dahl salt-sensitive (SS) strain. Using microarray analysis and a bioinformatics-intense candidate gene approach, we identified 245 differentially expressed potential VALI genes with ontologies such as transcriptional regulation, chemotaxis, and inflammation (27). Because chromosomes 2, 13, 16, and 17 were found to contain the highest density of VALI-response genes, consomic SS rats containing substituted BN chromosome 13 were exposed to VALI mechanical stress resulting in conversion of the resistant SS rat to VALI sensitivity (27). We are currently exploring genes residing on chromosome 13 as potential ALI-associated candidates involved in inflammation and blood coagulation, including the chemokine receptor 4 (CXCR4) gene previously shown to be associated with ALI (27–29) (Figure 1).
Figure 1.
Schematic algorithm identifying acute lung injury candidate genes via the consomic rat approach. ALI = acute lung injury; VALI = ventilator-associated acute lung injury. QTL = quantitative trait loci.
DISCOVERING ALI SUSCEPTIBILITY GENES: CANDIDATE GENE APPROACH–ENABLED GENETIC STUDIES
Similar to the GADD45 studies described above, we have used a “candidate gene approach,” which extends extensive expression profiling across preclinical ALI models and identifies ALI gene candidates to determine allelic frequencies of gene polymorphisms (SNPs) that may confer ALI risk or severity of disease. Cited below are several examples of genes identified via this approach with hypothesized significant mechanistic roles in lung injury, inflammation, or repair in the setting of ALI and VALI.
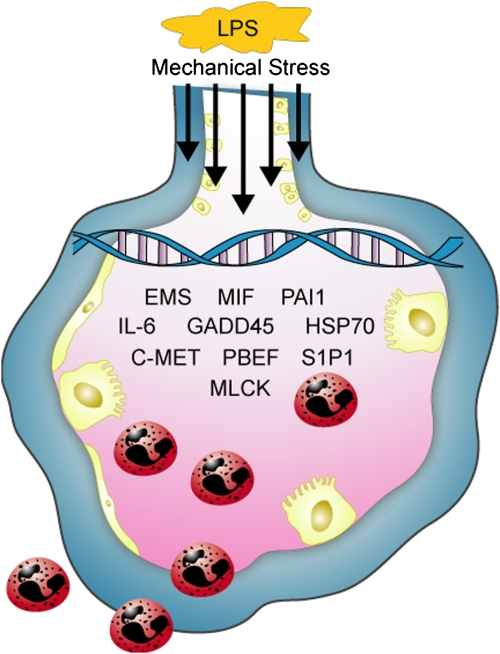
ALI displays significant variation in areas of maximal involvement of the inflammatory process with the basilar and dependent areas of the lung being the most deranged. We hypothesized that anatomic variation may reflect extremes of mechanical stress that occur with mechanical ventilation (30–32). Using a canine model of unilateral saline-induced lung injury replicating the regional tissue variation observed in the clinical syndrome (confirmed by computed tomography scanning) (30, 31), we employed a candidate gene approach with site-specific gene expression profiling to discover novel genetic contributors to VALI (33). Tissues from the dependent areas of the injured lung displayed significantly unique gene expression patterns when compared with nondependent tissues, again reflecting gene ontologies such as protein degradation, protein synthesis, blood coagulation, and inflammation (33) (Figure 2).
Figure 2.
Genomic schema depicting pathobiological events in acute lung injury. Mechanical stress and inflammatory stimuli (e.g., LPS) activate both alveolar epithelium and endothelium, resulting in gene activation facilitating inflammatory pathways, leukocyte diapedesis, and increased vascular permeability. C-MET = mesenchymal–epithelial transition factor; EMS = cortactin; GADD45 = growth arrest and DNA damage-inducible 45α; HSP70 = heat shock protein 70; MIF = macrophage migration inhibitory factor; MLCK = myosin light chain kinase; PAI1 = plasminogen activator inhibitor type 1; PBEF = pre–B-cell colony–enhancing factor; S1P1 = shingosine 1 phosphate receptor.
Specific genes that were markedly altered in our preclinical models of ALI included profound up-regulation of inflammatory cytokines that drive the extent of inflammatory lung injury or antiinflammatory cytokines that may potentially serve as therapeutic targets in ALI. IL-6, a pleiotropic proinflammatory cytokine that activates B and T cells and plays a significant role in the acute phase response, is a well-known ALI biomarker with increased levels associated with worsened outcome in both animals and humans with ARDS, sepsis, and trauma (34, 35). We identified markedly increased IL-6 levels in each ALI/VALI model (mouse, rat, dog, humans with ALI) (17, 36), and preliminarily evaluated IL-6 genetic variants that may affect production and participate in ALI (36). Prior reported studies of IL-6 SNPs in ALI evaluated the commonly studied G/C IL-6 promoter SNP at position −174 associated with reduced circulating levels of IL-6 (37). The C allele and CC genotype frequencies were reduced in intensive care unit (ICU) nonsurvivors, but failed to identify an association with ALI susceptibility (38). A similar study evaluated several IL-6 gene polymorphisms in a cohort of critically ill patients with sepsis inflammatory response syndrome (SIRS) failed to find an association between any single IL-6 SNP and ALI (including the −174 G/C SNP), but did link increased mortality and organ failure in sepsis patients with a three-SNP haplotype (39). We recently reported genotyping results assessing eight IL-6 SNPs from subjects of European descent with sepsis and ALI as well as healthy control subjects and identified the presence of an ALI-protective haplotype (40). This three-allele haplotype includes the −174 G/C SNP and therefore conflicts with the study by Sutherland and colleagues (39), which found this SNP to be within a detrimental haplotype. Together, these studies indicate that the influence of IL-6 polymorphisms in ALI is complex, stimulus specific, and requires further exploration.
Another recognized ALI candidate gene and biomarker is macrophage migration inhibitory factor (MIF) (41), a proinflammatory cytokine initially discovered as a soluble product of activated T cells. MIF is produced by many cell types, including monocytes/macrophages, pituitary cells, vascular endothelium, and respiratory epithelium (42, 43), and may regulate cytokine balance between the immune system and inflammation through an ability to override the immunosuppressive effects of glucocorticoids and to up-regulate TNF-α expression (42–44). Several studies (41, 43), including our own (45), have shown increased MIF gene expression and protein levels in both the serum and bronchoalveolar lavage (BAL) fluid of patients with ARDS as well as enhanced MIF production in alveolar endothelium in patients with ARDS when compared with other critically ill patients. Interestingly, pretreatment of mice with anti-MIF antibodies before exposure to LPS attenuates the lung injury (43), suggesting a direct role in sepsis pathobiology. Recently, we used a sepsis-induced ALI DNA cohort composed of patients of African and European descent to examine eight MIF polymorphisms, including the most studied gene promoter G/C SNP at position −173. Similar to IL-6, individual MIF SNPs were not significantly associated with either ALI or sepsis; however, several MIF haplotypes located in the 3′ region of the gene displayed strong association with ALI and sepsis, conferring both protection as well as susceptibility to ALI, in populations of European and African descent (45). These results provide further evidence for MIF as an ALI biomarker and therapeutic target with a role in ALI pathogenesis.
Another example underscoring the power of the candidate gene approach in selecting ALI genes was the identification of pre–B-cell colony–enhancing factor (PBEF), a cytokine involved in B-cell maturation whose expression was induced in amniotic membranes during gestation and by proinflammatory mediators (46, 47). Expression profiling in preclinical ALI animal models and BAL fluid obtained from patients with ALI demonstrated marked increases in PBEF expression with biochemical evidence indicating PBEF as a novel biomarker in ALI (48). Supporting the human and animal data, PBEF appears to have a role in vascular barrier regulation via Ca2+-dependent cytoskeletal rearrangement as well as in regulation of neutrophil apoptosis, two essential features of the inflammatory response (49, 50).
Our mechanistic studies were accompanied by exploration of PBEF genetic variants via an association analysis of sepsis-induced ALI cohort. In this study, 11 SNPs were identified by directly sequencing the PBEF gene in 36 subjects (12 healthy controls, 12 with sepsis, and 12 with ALI) followed by genotyping of two PBEF promoter SNPs (−1543 C/T and −1001 T/G) due to their proximity to mapped transcription factor binding sites (48). The G-allele of the −1001 T/G SNP increased promoter activity and was found to have higher frequency in patients with sepsis-mediated ALI. In contrast, the T variant of the −1543 C/T SNP resulted in a twofold decrease in promoter activity and conferred ALI protection. The −1543C/−1001G haplotype displayed the strongest association with ALI risk (threefold increase) as well as increased risk of sepsis. In contrast, the −1543T/−1001T haplotype conferred ALI protection (48).
There is increasing appreciation that several requirements need to be met before a specific polymorphism (SNP or insertion/deletion) can be said to be truly associated with the expression of a particular pathologic phenotype. We believe that requirements to be considered in the assessment of specific polymorphisms in human diseases include the presence of linkage between the chromosomal site where the gene resides and the speculated phenotype. PBEF resides on chromosome 7q22, a genomic site associated with a number of inflammatory disorders, such as inflammatory bowel disease (51), multiple sclerosis (52), and asthma (53). In the absence of ALI family studies, linkage studies are difficult, rendering this criterion as inapplicable to ALI and therefore should be waived as a criterion for PBEF. A second considered criterion is successful completion of an association study that confirms linkage of the putative disease-associated polymorphism and the phenotype, a requirement fulfilled by our initial report (48) in which we determined significant association between PBEF promoter SNPs and ALI susceptibility. A third, potentially important, criterion is replication of the genotype–phenotype association in a fundamentally distinct population with the affected phenotype. Bajwa and colleagues recently reported significant association between the above mentioned PBEF SNPs −1543 C/T and −1001 T/G and ALI susceptibility in a comparable but separate ALI population (54), thereby validating our earlier findings. Furthermore, the gene GC haplotype at −1001G was associated with increased ICU mortality and the TT haplotype at −1543T was associated with fewer ventilator days and decreased ICU mortality (54). Two additional criteria to be entertained (fourth and fifth) are the finding of the putative polymorphism as a somatic mutation and the determination of the functional consequences of the polymorphism. We are unaware of somatic mutations in PBEF; however, our prior reports (48, 50) describe the functional consequences of these two PBEF SNPs on promoter activity. Thus, although the precise pathogenic mechanisms by which PBEF affects ALI require further investigation, PBEF represents an outstanding example of the candidate gene approach in identifying viable candidate genes and functional polymorphisms discovered that affect ALI susceptibility.
DISCOVERING ALI SUSCEPTIBILITY GENES: ANALYSIS OF BIOLOGICAL PATHWAYS
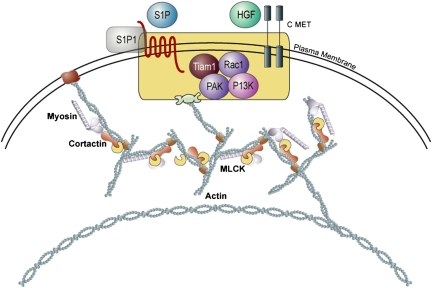
Enabled by sophisticated bioinformatics methodologies and increasing knowledge of the molecular and cellular mechanisms of lung injury, candidate genes are also identified via analysis of cellular pathways involved in ALI pathogenesis. As noted earlier, a marked increase in lung vascular permeability is a hallmark of ALI and reflects the loss of integrity of both endothelial and alveolar epithelial cellular barriers. Barrier integrity is governed to a major extent by cytoskeletal elements that provide the scaffolding for cell architecture as well as the capacity to undergo dynamic changes in cell shape. Genes encoding key cytoskeletal elements, such as cortactin and myosin light chain kinase (MLCK), as well as receptors for barrier-regulatory agonists, such as sphingosine-1 phosphate and hepatocyte growth factor, are a fertile area of exploration for candidate SNPs likely to further understanding of the workings of the vascular endothelial barrier as well as associated signaling pathways (55–59) (Figure 3).
Figure 3.
Cytoskeletal regulation of endothelial cell barrier function. Depicted are the signaling pathways involved after cell surface growth factor receptor ligation (S1P and HGF), which lead to cytoskeletal rearrangement driven by components such as actin, myosin, cortactin and MLCK. The enhanced formation of cortical actin rings results in stabilization of cell shape, and an increase in vascular barrier function. C-MET = mesenchymal–epithelial transition factor; HGF = hepatocyte growth factor; MLCK = myosin light chain kinase; PAK = p21/cdc42/rac1-activated kinases; PI3K = phosphoinositide-3 kinase; rac1 = rac GTPase 1; S1P = sphingosine-1 phosphate; tiam1 = T-cell lymphoma invasion and metastasis 1.
The gene for human MLCK (MYLK) encodes three proteins, including both nonmuscle and smooth muscle MLCK isoforms and telokin. We have shown the nonmuscle MLCK isoform is centrally involved in the cytoskeleton rearrangement regulating vascular barrier function and permeability, angiogenesis, endothelial cell apoptosis, and leukocyte diapedesis, suggesting a possible mechanistic role for MLCK in the elaboration of ALI (60–63). Nonmuscle MLCK-deficient mice (which retain the smooth muscle MLCK isoform) are less susceptible to LPS-induced ALI (64), whereas MLCK-overexpressing transgenic mice demonstrate increased vascular permeability (65), indicating that nonmuscle MLCK expression is a major determinant of vascular leak in the acutely inflamed murine lung. In addition, MLCK inhibition prevents the increased lung permeability produced by thrombin (60, 61), LPS (64, 66), and mechanical stress (67).
To further evaluate the contribution of MYLK genetic variations to ALI, we directly sequenced MYLK (32 exons and 2 kb of 5′ untranslated region) in 36 subjects (European- and African-Americans with sepsis, sepsis-associated ALI, healthy controls) and identified over 50 SNPs (58). SNP and haplotype association analysis demonstrated strong association in sepsis and sepsis-induced ALI (58) and a haplotype present only in African-American subjects that may highlight potential genetic contributions to observed differences between European- and African-American patients with ALI/ARDS (68). The propensity for MYLK SNPs to influence lung inflammatory disease was validated recently in studies in which we described the contribution of MYLK SNPs to trauma-induced ALI (69) and to severe asthma susceptibility in two distinct African descent populations (70, 71).
CONCLUSIONS
Although the genetic and pathologic mechanisms of ALI are not yet fully understood, the advent of high-throughput molecular technologies and availability of extensive bioinformatics data has accelerated the discovery and validation of ALI candidate genes (e.g., MIF and IL-6). Identification of evolutionarily conserved genes that have altered expression in models of ALI/VALI has further yielded novel candidates, such as GADD45α, a gene with a promising but as yet undefined role in ALI. Regional lung gene expression differences yielded the discovery of PBEF, a strong ALI candidate as not only a biomarker but as a potential target for therapy. Finally, it is highly likely that greater understanding of the role of specific genes/proteins in pathways regulating ALI pathogenesis and permeability (e.g., MLCK) will generate prime targets for therapy in this devastating illness. Together, these novel genomic and genetic approaches may prove exceptionally useful in ushering in the era of personalized medicine for the critically ill.
Supported by NIH grants 2P01HL058064 (J.G.M.G.) and 5T32HL007605-23 (J. Solway).
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Bernard GR, Artigas A, Brigham KL, Carlet J, Falke K, Hudson L, Lamy M, Legall JR, Morris A, Spragg R. The American-European consensus conference on ARDS: definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am J Respir Crit Care Med 1994;149:818–824. [DOI] [PubMed] [Google Scholar]
- 2.Rubenfeld GD, Caldwell E, Peabody E, Weaver J, Martin DP, Neff M, Stern EJ, Hudson LD. Incidence and outcomes of acute lung injury. N Engl J Med 2005;353:1685–1693. [DOI] [PubMed] [Google Scholar]
- 3.Ware LB, Matthay MA. The acute respiratory distress syndrome. N Engl J Med 2000;342:1334–1349. [DOI] [PubMed] [Google Scholar]
- 4.Gong MN, Zhou W, Williams PL, Thompson BT, Pothier L, Boyce P, Christiani DC. −308GA and TNFb polymorphisms in acute respiratory distress syndrome. Eur Respir J 2005;26:382–389. [DOI] [PubMed] [Google Scholar]
- 5.Gong MN, Thompson BT, Williams PL, Zhou W, Wang MZ, Pothier L, Christiani DC. Interleukin-10 polymorphism in position -1082 and acute respiratory distress syndrome. Eur Respir J 2006;27:674–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin Z, Pearson C, Chinchilli V, Pietschmann SM, Luo J, Pison U, Floros J. Polymorphisms of human SP-A, SP-B, and SP-D genes: association of SP-B Thr 131lle with ARDS. Clin Genet 2000;58:181–191. [DOI] [PubMed] [Google Scholar]
- 7.Quasney MW, Waterer GW, Dahmer MK, Kron GK, Zhang Q, Kessler LA, Wunderink RG. Association between surfactant protein B + 1580 polymorphism and the risk of respiratory failure in adults with community-acquired pneumonia. Crit Care Med 2004;32:1115–1119. [DOI] [PubMed] [Google Scholar]
- 8.Gong MN, Wei Z, Xu LL, Miller DP, Thompson BT, Christiani DC. Polymorphism in the surfactant protein-B gene, gender, and the risk of direct pulmonary injury and ards. Chest 2004;125:203–211. [DOI] [PubMed] [Google Scholar]
- 9.Sapru A, Wiemels JL, Witte JS, Ware LB, Matthay MA. Acute lung injury and the coagulation pathway: potential role of gene polymorphisms in the protein C and fibrinolytic pathways. Intensive Care Med 2006;32:1293–1303. [DOI] [PubMed] [Google Scholar]
- 10.Marshall RP, Webb S, Bellingan GJ, Montgomery HE, Chaudhari B, McAnulty RJ, Humphries SE, Hill MR, Laurent GJ. Angiotensin converting enzyme insertion/deletion polymorphism is associated with susceptibility and outcome in acute respiratory distress syndrome. Am J Respir Crit Care Med 2002;166:646–650. [DOI] [PubMed] [Google Scholar]
- 11.International HapMap Project. Available from: http://www.hapmap.org/(accessed Sept. 1, 2001).
- 12.Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nat Genet 2001;29:229–232. [DOI] [PubMed] [Google Scholar]
- 13.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, et al. Complement factor H polymorphism in age-related macular degeneration. Science 2005;308:385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. A genome-wide association study identifies IL23r as an inflammatory bowel disease gene. Science 2006;314:1461–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007;445:881–885. [DOI] [PubMed] [Google Scholar]
- 16.Grigoryev DN, Finigan JH, Hassoun P, Garcia JG. Science review: searching for gene candidates in acute lung injury. Crit Care 2004;8:440–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grigoryev DN, Ma SF, Irizarry RA, Ye SQ, Quackenbush J, Garcia JG. Orthologous gene-expression profiling in multi-species models: search for candidate genes. Genome Biol 2004;5:R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lal A, Gorospe M. Egad, more forms of gene regulation: The GADD45a story. Cell Cycle 2006;5:1422–1425. [DOI] [PubMed] [Google Scholar]
- 19.Liebermann DA, Hoffman B. GADD45 in the response of hematopoietic cells to genotoxic stress. Blood Cells Mol Dis 2007;39:344–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin S, Antinore MJ, Lung FD, Dong X, Zhao H, Fan F, Colchagie AB, Blanck P, Roller PP, Fornace AJ Jr, et al. The GADD45 inhibition of CDC2 kinase correlates with GADD45-mediated growth suppression. J Biol Chem 2000;275:16602–16608. [DOI] [PubMed] [Google Scholar]
- 21.Hollander MC, Philburn RT, Patterson AD, Wyatt MA, Fornace AJ Jr. Genomic instability in GADD45a-/- cells is coupled with S-phase checkpoint defects. Cell Cycle 2005;4:704–709. [DOI] [PubMed] [Google Scholar]
- 22.Salvador JM, Hollander MC, Nguyen AT, Kopp JB, Barisoni L, Moore JK, Ashwell JD, Fornace AJ Jr. Mice lacking the p53-effector gene gadd45a develop a lupus-like syndrome. Immunity 2002;16:499–508. [DOI] [PubMed] [Google Scholar]
- 23.Altemeier WA, Matute-Bello G, Gharib SA, Glenny RW, Martin TR, Liles WC. Modulation of lipopolysaccharide-induced gene transcription and promotion of lung injury by mechanical ventilation. J Immunol 2005;175:3369–3376. [DOI] [PubMed] [Google Scholar]
- 24.O'Reilly MA, Staversky RJ, Watkins RH, Maniscalco WM, Keng PC. p53-independent induction of gadd45 and gadd153 in mouse lungs exposed to hyperoxia. Am J Physiol 2000;278:L552–L559. [DOI] [PubMed] [Google Scholar]
- 25.Roper JM, Gehen SC, Staversky RJ, Hollander MC, Fornace AJ Jr, O'Reilly MA. Loss of GADD45a does not modify the pulmonary response to oxidative stress. Am J Physiol 2005;288:L663–L671. [DOI] [PubMed] [Google Scholar]
- 26.Meyer NJ, Moreno-Vinasco L, Husain AN, Garcia JGN. Loss of gadd45a imparts a phenotype of more severe endotoxin-induced inflammatory lung injury in mice. Proc Am Thorac Soc (in press)
- 27.Nonas SA, Moreno-Vinasco LM, Ma SF, Jacobson JR, Desai AA, Dudek SM, Flores C, Hassoun PM, Sam L, Ye SQ, et al. Use of consomic rats for genomic insights into ventilator-associated lung injury. Am J Physiol Lung Cell Mol Physiol 2007;293:L292–L302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomperts BN, Belperio JA, Rao PN, Randell SH, Fishbein MC, Burdick MD, Strieter RM. Circulating progenitor epithelial cells traffic via CXCR4/CXCL12 in response to airway injury. J Immunol 2006;176:1916–1927. [DOI] [PubMed] [Google Scholar]
- 29.Xu J, Mora A, Shim H, Stecenko A, Brigham KL, Rojas M. Role of the SDF-1/CXCR4 axis in the pathogenesis of lung injury and fibrosis. Am J Respir Cell Mol Biol 2007;37:291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gattinoni L, Bombino M, Pelosi P, Lissoni A, Pesenti A, Fumagalli R, Tagliabue M. Lung structure and function in different stages of severe adult respiratory distress syndrome. JAMA 1994;271:1772–1779. [PubMed] [Google Scholar]
- 31.Gattinoni L, Caironi P, Pelosi P, Goodman LR. What has computed tomography taught us about the acute respiratory distress syndrome? Am J Respir Crit Care Med 2001;164:1701–1711. [DOI] [PubMed] [Google Scholar]
- 32.Rouby JJ, Puybasset L, Nieszkowska A, Lu Q. Acute respiratory distress syndrome: lessons from computed tomography of the whole lung. Crit Care Med 2003;31(4, Suppl):S285–S295. [DOI] [PubMed] [Google Scholar]
- 33.Simon BA, Easley RB, Grigoryev DN, Ma SF, Ye SQ, Lavoie T, Tuder RM, Garcia JG. Microarray analysis of regional cellular responses to local mechanical stress in acute lung injury. Am J Physiol 2006;291:L851–L861. [DOI] [PubMed] [Google Scholar]
- 34.Holub M, Lawrence DA. Influence of endotoxin-induced acute lung injury on pulmonary innate and adaptive immunity. APMIS 2003;111:571–580. [DOI] [PubMed] [Google Scholar]
- 35.Park WY, Goodman RB, Steinberg KP, Ruzinski JT, Radella F II, Park DR, Pugin J, Skerrett SJ, Hudson LD, Martin TR. Cytokine balance in the lungs of patients with acute respiratory distress syndrome. Am J Respir Crit Care Med 2001;164:1896–1903. [DOI] [PubMed] [Google Scholar]
- 36.Nonas SA, Finigan JH, Gao L, Garcia JG. Functional genomic insights into acute lung injury: role of ventilators and mechanical stress. Proc Am Thorac Soc 2005;2:188–194. [DOI] [PubMed] [Google Scholar]
- 37.Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, Humphries S, Woo P. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest 1998;102:1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marshall RP, Webb S, Hill MR, Humphries SE, Laurent GJ. Genetic polymorphisms associated with susceptibility and outcome in ARDS. Chest 2002;121(3, Suppl):68S–69S. [DOI] [PubMed] [Google Scholar]
- 39.Sutherland AM, Walley KR, Manocha S, Russell JA. The association of interleukin 6 haplotype clades with mortality in critically ill adults. Arch Intern Med 2005;165:75–82. [DOI] [PubMed] [Google Scholar]
- 40.Flores CMS, Maresso K, Wade M, Villar J, Garcia JGN. An IL6 gene-wide haplotype is associated with susceptibility to acute lung injury. Proc Am Thorac Soc (In press) [DOI] [PubMed]
- 41.Donnelly SC, Haslett C, Reid PT, Grant IS, Wallace WA, Metz CN, Bruce LJ, Bucala R. Regulatory role for macrophage migration inhibitory factor in acute respiratory distress syndrome. Nat Med 1997;3:320–323. [DOI] [PubMed] [Google Scholar]
- 42.Baugh JA, Bucala R. Macrophage migration inhibitory factor. Crit Care Med 2002;30(1, Supp):S27–S35. [PubMed] [Google Scholar]
- 43.Lai KN, Leung JC, Metz CN, Lai FM, Bucala R, Lan HY. Role for macrophage migration inhibitory factor in acute respiratory distress syndrome. J Pathol 2003;199:496–508. [DOI] [PubMed] [Google Scholar]
- 44.Calandra T, Bernhagen J, Metz CN, Spiegel LA, Bacher M, Donnelly T, Cerami A, Bucala R. MIF as a glucocorticoid-induced modulator of cytokine production. Nature 1995;377:68–71. [DOI] [PubMed] [Google Scholar]
- 45.Gao L, Flores C, Fan-Ma S, Miller EJ, Moitra J, Moreno L, Wadgaonkar R, Simon B, Brower R, Sevransky J, et al. Macrophage migration inhibitory factor in acute lung injury: expression, biomarker, and associations. Transl Res 2007;150:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nemeth E, Tashima LS, Yu Z, Bryant-Greenwood GD. Fetal membrane distention: I. Differentially expressed genes regulated by acute distention in amniotic epithelial (wish) cells. Am J Obstet Gynecol 2000;182:50–59. [DOI] [PubMed] [Google Scholar]
- 47.Ognjanovic S, Bao S, Yamamoto SY, Garibay-Tupas J, Samal B, Bryant-Greenwood GD. Genomic organization of the gene coding for human pre-B-cell colony enhancing factor and expression in human fetal membranes. J Mol Endocrinol 2001;26:107–117. [DOI] [PubMed] [Google Scholar]
- 48.Ye SQ, Simon BA, Maloney JP, Zambelli-Weiner A, Gao L, Grant A, Easley RB, McVerry BJ, Tuder RM, Standiford T, et al. Pre–B-cell colony–enhancing factor as a potential novel biomarker in acute lung injury. Am J Respir Crit Care Med 2005;171:361–370. [DOI] [PubMed] [Google Scholar]
- 49.Jia SH, Li Y, Parodo J, Kapus A, Fan L, Rotstein OD, Marshall JC. Pre-B cell colony-enhancing factor inhibits neutrophil apoptosis in experimental inflammation and clinical sepsis. J Clin Invest 2004;113:1318–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye SQ, Zhang LQ, Adyshev D, Usatyuk PV, Garcia AN, Lavoie TL, Verin AD, Natarajan V, Garcia JG. Pre-B-cell-colony-enhancing factor is critically involved in thrombin-induced lung endothelial cell barrier dysregulation. Microvasc Res 2005;70:142–151. [DOI] [PubMed] [Google Scholar]
- 51.Satsangi J, Parkes M, Louis E, Hashimoto L, Kato N, Welsh K, Terwilliger JD, Lathrop GM, Bell JI, Jewell DP. Two stage genome-wide search in inflammatory bowel disease provides evidence for susceptibility loci on chromosomes 3, 7 and 12. Nat Genet 1996;14:199–202. [DOI] [PubMed] [Google Scholar]
- 52.Vandenbroeck K, Fiten P, Heggarty S, Goris A, Cocco E, Hawkins SA, Graham CA, Marrosu MG, Opdenakker G. Chromosome 7q21-22 and multiple sclerosis: evidence for a genetic susceptibility effect in vicinity to the protachykinin-1 gene. J Neuroimmunol 2002;125:141–148. [DOI] [PubMed] [Google Scholar]
- 53.Koppelman GH, Stine OC, Xu J, Howard TD, Zheng SL, Kauffman HF, Bleecker ER, Meyers DA, Postma DS. Genome-wide search for atopy susceptibility genes in Dutch families with asthma. J Allergy Clin Immunol 2002;109:498–506. [DOI] [PubMed] [Google Scholar]
- 54.Bajwa EK, Yu CL, Gong MN, Thompson BT, Christiani DC. Pre-B-cell colony-enhancing factor gene polymorphisms and risk of acute respiratory distress syndrome. Crit Care Med 2007;35:1290–1295. [DOI] [PubMed] [Google Scholar]
- 55.Dudek SM, Birukov KG, Zhan X, Garcia JG. Novel interaction of cortactin with endothelial cell myosin light chain kinase. Biochem Biophys Res Commun 2002;298:511–519. [DOI] [PubMed] [Google Scholar]
- 56.Dudek SM, Jacobson JR, Chiang ET, Birukov KG, Wang P, Zhan X, Garcia JG. Pulmonary endothelial cell barrier enhancement by sphingosine 1-phosphate: roles for cortactin and myosin light chain kinase. J Biol Chem 2004;279:24692–24700. [DOI] [PubMed] [Google Scholar]
- 57.Finigan JH, Dudek SM, Singleton PA, Chiang ET, Jacobson JR, Camp SM, Ye SQ, Garcia JG. Activated protein C mediates novel lung endothelial barrier enhancement: role of sphingosine 1-phosphate receptor transactivation. J Biol Chem 2005;280:17286–17293. [DOI] [PubMed] [Google Scholar]
- 58.Gao L, Grant A, Halder I, Brower R, Sevransky J, Maloney JP, Moss M, Shanholtz C, Yates CR, Meduri GU, et al. Novel polymorphisms in the myosin light chain kinase gene confer risk for acute lung injury. Am J Respir Cell Mol Biol 2006;34:487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singleton PA, Dudek SM, Ma SF, Garcia JG. Transactivation of sphingosine 1-phosphate receptors is essential for vascular barrier regulation: novel role for hyaluronan and CD44 receptor family. J Biol Chem 2006;281:34381–34393. [DOI] [PubMed] [Google Scholar]
- 60.Dudek SM, Garcia JG. Cytoskeletal regulation of pulmonary vascular permeability. J Appl Physiol 2001;91:1487–1500. [DOI] [PubMed] [Google Scholar]
- 61.Garcia JG, Davis HW, Patterson CE. Regulation of endothelial cell gap formation and barrier dysfunction: role of myosin light chain phosphorylation. J Cell Physiol 1995;163:510–522. [DOI] [PubMed] [Google Scholar]
- 62.Garcia JG, Verin AD, Herenyiova M, English D. Adherent neutrophils activate endothelial myosin light chain kinase: role in transendothelial migration. J Appl Physiol 1998;84:1817–1821. [DOI] [PubMed] [Google Scholar]
- 63.Petrache I, Verin AD, Crow MT, Birukova A, Liu F, Garcia JG. Differential effect of MLC kinase in TNF-alpha-induced endothelial cell apoptosis and barrier dysfunction. Am J Physiol 2001;280:L1168–L1178. [DOI] [PubMed] [Google Scholar]
- 64.Wainwright MS, Rossi J, Schavocky J, Crawford S, Steinhorn D, Velentza AV, Zasadzki M, Shirinsky V, Jia Y, Haiech J, et al. Protein kinase involved in lung injury susceptibility: evidence from enzyme isoform genetic knockout and in vivo inhibitor treatment. Proc Natl Acad Sci USA 2003;100:6233–6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moitra J, Evenoski C, Wadgaoncar R, Turner J, Ma SF, Garcia JGN. A mouse transgenic over-expressing non-muscle myosin light chain kinase in endothelium increases susceptibility to inflammatory lung injury: role of sexual dimorphism and age. Transl Res 2008;151:141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eutamene H, Theodorou V, Schmidlin F, Tondereau V, Garcia-Villar R, Salvador-Cartier C, Chovet M, Bertrand C, Bueno L. LPS-induced lung inflammation is linked to increased epithelial permeability: role of mlck. Eur Respir J 2005;25:789–796. [DOI] [PubMed] [Google Scholar]
- 67.Parker JC. Inhibitors of myosin light chain kinase and phosphodiesterase reduce ventilator-induced lung injury. J Appl Physiol 2000;89:2241–2248. [DOI] [PubMed] [Google Scholar]
- 68.Moss M, Mannino DM. Race and gender differences in acute respiratory distress syndrome deaths in the united states: an analysis of multiple-cause mortality data (1979–1996). Crit Care Med 2002;30:1679–1685. [DOI] [PubMed] [Google Scholar]
- 69.Christie JD, Aplenc R, Li M, Lanken PN, Fuchs B, Albelda S, Flores C, Garcia JGN. Variation in the MYLK gene is associated with development of acute lung injury following major trauma. Crit Care Med (In press) [DOI] [PubMed]
- 70.Flores C, Ma SF, Maresso K, Ober C, Garcia JG. A variant of the myosin light chain kinase gene is associated with severe asthma in African Americans. Genet Epidemiol 2007;31:296–305. [DOI] [PubMed] [Google Scholar]
- 71.Gao L, Grant AV, Rafaels N, Stockton-Porter M, Watkins T, Gao P, Chi P, Munoz M, Watson H, Dunston G, et al. Polymorphisms in the myosin light chain kinase gene that confer risk of severe sepsis are associated with a lower risk of asthma. J Allergy Clin Immunol 2007;119:1111–1118. [DOI] [PubMed] [Google Scholar]