Abstract
The HDV ribozyme self-cleaves by a chemical mechanism involving general acid-base catalysis to generate a 2′,3′-cyclic phosphate and a 5′-hydroxyl termini. Biochemical studies from several laboratories have implicated C75 as the general acid and hydrated magnesium as the general base. We have previously shown that C75 has a pKa shifted > 2 pH units toward neutrality [Gong, B., Chen, J. H., Chase, E., Chadalavada, D. M., Yajima, R., Golden, B. L., Bevilacqua, P. C., and Carey, P. R. (2007) J. Am. Chem. Soc. 129, 13335–13342.], while in crystal structures, it is well-positioned for proton transfer. However no crystallographic evidence for a hydrated magnesium poised to serve as a general base in the reaction has been observed in high-resolution crystal structures of various reaction states and mutants. Herein, we use solution kinetic experiments and parallel Raman crystallographic studies to examine the effects of pH on rate and Mg2+-binding properties of wild-type and 7-deazaguanosine mutants of the HDV ribozyme. These data suggest that a previously-unobserved hydrated magnesium ion interacts with the N7 of the cleavage site G•U wobble base pair. Integrating this metal ion binding site with the available crystal structures provides a new three-dimensional model for the active site of the ribozyme that accommodates all available biochemical data and appears competent for catalysis. The position of this metal is consistent with a role of a magnesium-bound hydroxide as a general base as dictated by biochemical data.
RNA enzymes, or ribozymes use a variety of catalytic strategies, including general acid-base catalysis and metal-mediated catalysis, to effect biological activity within cells. The hepatitis delta virus (HDV)1 ribozyme is a self-cleaving RNA element integral to the life cycle of the hepatitis delta virus, a human pathogen (1). Similar versions of this ribozyme are encoded within the genomic and antigenomic viral RNAs. Each ribozyme comprises ~85-nucleotides and catalyzes a self-cleavage reaction that separates nascent multimer-copy 1.7 kilobase genomes into single unit-length genomes (Figure 1A). The reverse reaction (ligation) has not been detected for the HDV ribozyme, most likely because it does not contain a significant binding site for the upstream cleavage product (2, 3). The sequences of genomic and antigenomic ribozymes differ slightly but both fold into a similar tertiary structure and catalyze strand scission through the same mechanism (2). A similar ribozyme has recently been discovered within mammalian transcriptomes (4).
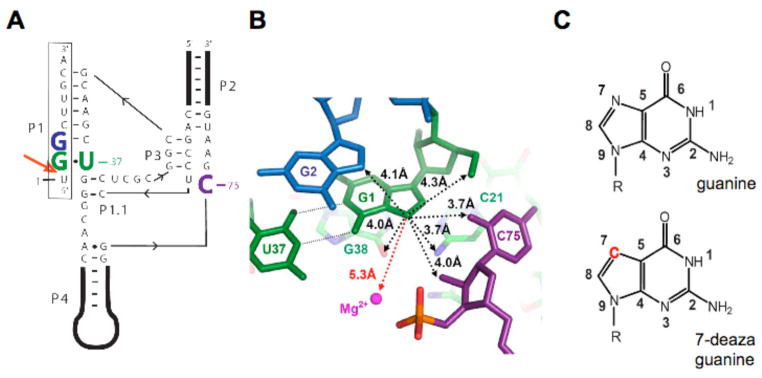
Figure 1.
Overview of the genomic HDV ribozyme structure. (A) Secondary structure of the genomic HDV ribozyme used in this study. The 9-nt substrate strand (boxed) is introduced in trans. The solid line with embedded arrow shows the 5′ to 3′ direction of chain connectivity in the ribozyme strand. The cleavage site between U(−1) and G1 is indicated with a heavy red arrow. Important nucleotides are colored as follows: C75 is purple, the cleavage site G•U wobble pair is green and G2 is blue. (B) Atoms surrounding the G1·U37 wobble pair in post-cleaved HDV ribozyme (PDB code1cx0)(5). Nucleotides are colored as in panel a. Non-hydrogen atoms within 4.5 Å to the N7 of G1 are shown by black dotted lines. The crystallographic metal ion found closest to G1 is 5.3 Å away and highlighted by a red dashed line. (c) The chemical structures of guanosine and 7-deazaguanosine. Modification in 7-deazaguanine relative to guanine is highlighted in red.
Crystal structures have been solved for the genomic ribozyme both after (5) and prior (6, 7) to self-cleavage. The post cleavage structure reveals C75 positioned for proton transfer as a general acid in the cleavage reaction. In contrast, the pre-cleavage structure does not show C75 in position to be either a general acid or a general base. In the pre-cleavage structure, a hydrated magnesium ion is positioned to act as the general acid and C75 was modeled into a position where it could serve as a general base. High resolution structures of the pre-cleaved ribozyme may not reflect the catalytically active form of the ribozyme because they were solved either with an inactivating C75U mutation or in the absence of magnesium cofactor. In the post-cleavage structure, no magnesium ions were found at the active site. We are interested in the positioning of the catalytic Mg2+ ion within the active site of the pre-cleaved ribozyme in the presence of an unmodified C75. None of the magnesium ions observed in the available crystal structures to date have the characteristics expected for this catalytic Mg2+ ion.
Inspection of the active site of the HDV ribozyme post-cleavage structure reveals a potential metal binding site within the major goove of the cleavage site base-pair. The leaving group nucleotide is a guanosine (G1) base-paired to a uridine within the ribozyme to generate a G•U wobble. The genomic HDV, antigenomic HDV, and human ribozymes all contain a G•U pair, or rarely a different purine-pyrimidine pair, at the cleavage site. The major groove of G•U wobble pairs is known to provide binding sites for divalent metal ions and their hexammine analogs (8–10). In addition, the HDV cleavage site wobble pair is surrounding by phosphate groups, including the scissile phosphate and G76 which could potentially provide inner- or outer sphere ligands to a Mg2+ ion.
The role of the cleavage site G•U wobble has been previously examined in some detail (see Results). Substituting the wobble pair with most base combinations results in significant defects (11, 12). Purine–pyrimidine combinations, however, are well tolerated and have similar metal ion- and pH-dependencies for reactivity (13). While the wobble geometry is not necessary for efficient reaction, a purine at the cleavage site position 1 in either Watson-Crick or wobble geometry is required. These findings indicate that the purine at the cleavage site plays an important structural role in the ribozyme reaction (13) and could even participate in binding a metal ion. Notably, in the available crystal structures, there are no ligands binding in the major groove of the cleavage site purine (Figure 1B) (5, 6).
To probe the role of the cleavage site G•U wobble base pair of the HDV ribozyme further, we have characterized the wild-type (WT) ribozyme and ribozymes with 7-deazaguanosine (7DG) substitutions (Figure 1C) using pH- and metal-dependent solution kinetic experiments and Raman crystallography. It was found that introduction of 7DG at the cleavage site guanosine (G1) was detrimental to cleavage. This observation strongly supports involvement of the N7 of G1 in the reaction mechanism despite structural analyses showing no interactions with this atom. Characterization of the pH-dependence and Mg2+-binding kinetic properties of the WT and modified ribozymes suggests that the 7DG modification destabilizes binding of the catalytic metal ion. Raman crystallography on the WT ribozyme indicates that protonation of C75 results in decoordination of a guanosine N7 and loss of an inner-sphere bound magnesium ion. Substitution of G1 with 7DG localized the source of the Raman guanosine signal to the cleavage site guanosine and strongly suggests that the magnesium ion directly coordinates the N7 of G1.
These data suggest the cleavage site G•U wobble pair is involved in coordinating a partially hydrated Mg2+ ion that has many of the biochemical properties associated with the catalytic magnesium ion of the HDV ribozyme. A model for the active site of the HDV ribozyme is proposed that is consistent with the available biochemical data for the WT HDV ribozyme.
MATERIALS AND METHODS
Sample preparation
The HDV ribozyme sequence was based on studies of fast-folding ribozyme presented previously (14–16). In all experiments, two strands of RNA were used. The larger strand spanned nucleotides 9 to 70, contained most of the active site, and is formally considered the ribozyme, or enzyme, strand. This RNA (Figure 1A) was made by T7 transcription and purified by a 6% polyacylamide, 7 M urea denaturing gel. The smaller oligomer was either an all-RNA, cleavable substrate, or a modified, uncleavable inhibitor. The full sequence of the ribozyme is provided in Figure S1 and the sequence and modification of the oligomers are listed below.
Substrates:
WT: 5′UGGCUUGCA3′
G1(7DG): 5′U(7DG)GCUUGCA3′
G2(7DG): 5′UG(7DG)CUUGCA3′
G1G2(7DG): 5′U(7DG)(7DG)CUUGCA3′
Inhibitors:
2′-OCH3: 5′(mU) GGCUUGCA3′
G1(7DG)_2′-OCH3: 5′(mU) (7DG)GCUUGCA3′
All oligonucleotides were purchased from Dharmacon except those containing 7-deazaguanosine, which were purchased from the W.M. Keck Facility, Yale. After deprotection and desalting using standard protocols provided by the companies, the 7-deazaguanosine containing oligonucleotides were further purified by TLC (PTLC Glass Plates Si 60, F254 20 x 20 cm, 0.5 mm thickness, from EMD Chemicals Inc.) using solvent system 1-propanol/ammonium hydroxide/water (55/35/10). The chromatogram was run until the solvent reached 0.75 x the height of the plate. The plate was air dried and bands were visualized by UV-shadowing. The full-length product was excised with a razor blade and extracted with 2 mL water three times. The collected supernatant containing the oligonucleotides was recovered by centrifugation. After desalting, the eluted fractions were dried using a speed-vac concentrator. The substrate strands that were used to study the ribozyme kinetics were 5-end radiolabeled by T4 polynucleotide kinase and [γ-32P] ATP (3000Ci/mmol).
Ribozyme kinetics and data fitting
The cleavage reactions were carried out under single turnover conditions with the ribozyme in 250-fold excess over substrate strand. The ribozyme 32P–labeled substrate strands were mixed in 25 mM Tris-HCl, pH 7.0 or sodium acetate, pH 5.0, heated at 90 °C for 2 minutes, and cooled to room temperature for 10 minutes. The mixture was pre-warmed at 37 °C in the absence of magnesium ion for 2 minutes and an aliquot was withdrawn as zero time point. The cleavage reaction was initiated by adding MgCl2 solution. At appropriate time points, aliquots of the reaction were removed and quenched with equal volume of formamide buffer (95% formamide, 50–200 mM EDTA). Samples were analyzed on an 18% acrylamide, 7 M urea denaturing gel. The radioactivity signal was recorded on a PhosphorImager (Molecular Dynamics) and the fraction of RNA cleaved was quantified using ImageQuant (Molecular Dynamics). Plots of the fraction reacted (f) versus time were fit using KaleidaGraph (Synergy software) to the single exponential equation:
| eq (1) |
where f is the fraction of ribozyme cleaved at time t, A is the fraction of ribozyme cleaved at completion, A+B is the burst fraction and kobs is the observed first-order rate constant. The values of kmax and Kd,Mg2+ were determined from a fit of kobs and MgCl2 concentration to equation (2):
| eq (2) |
where Kd,Mg2+ is the apparent dissociation constant, kmax is the maximal observed rate constant, and αMg is the Hill coefficient.
Crystal preparation
Equimolar concentrations of the ribozyme strand and the inhibitor strands, which contained a 2′-OCH3 at U-1, were mixed in final concentration of 5 mg/mL in 5 mM Tris-HCl buffer pH 7.5. The mixture was heated at 90 °C for 1 min and cooled to room temperature. MgCl2 was added to a final concentration of 10 mM and the mixture was incubated at 50 °C for 10 min and at room temperature 10 min. The cooled sample was then mixed with an equal volume of precipitant (50 mM, pH 5.0 sodium acetate, 1 mM spermine, 28–33 % MPD), and crystallized using the hanging-drop vapor-diffusion method. Cubic crystals with dimensions of 0.2 mm * 0.2 mm * 0.2 mm could be obtained in 1–3 weeks. These crystals diffract X-rays to a resolution of ~ 5 Å (unpublished data). The crystals were held in a stabilization solution containing 20 mM MgCl2, 2 mM spermine, 50% MPD with sodium cacodylate buffers pH 5.0–5.4.
Raman spectroscopy
The key experimental Raman data were recorded for a single crystal of HDV using a Raman microscope system (Kaiser Optical Systems) as described (17). In brief, a single HDV crystal in a hanging drop, containing 20 mM MgCl2 in 50 mM acetate (pH 5.0) or cacodylate (pH 7.5) buffers, was excited with 100 mW of 647-nm laser irradiation. The 180° back-scattered Raman light from the crystal was collected via the microscope objective with spectral acquisition times of 100 s. The Raman spectra of HDV crystal with buffer and buffer alone were obtained at pH 5.0 and 7.5, respectively. The computer aided subtraction of [HDV crystal + buffer] minus [buffer] using GRAMS/32 software (ThermoGalactic, Inc., Salem, NH) gave the spectra of [HDV crystal] at pH 5.0 and pH 7.5, as well as the Raman difference spectrum of [pH 7.5] minus [pH 5.0].
RESULTS
Cleavage site base pair considerations
Previous characterization of the HDV ribozyme implicated the cleavage site G•U wobble in a structural role but did not exclude its involvement in binding metal ions. The cleavage site base pair is a G•U in 61 out of 76 genomic HDV isolates and a G-C in the other 15, while it can be a G•U, G-C, or A–U in the mammalian version (4, 13, 18). In the antigenomic HDV ribozyme, it is exclusively G•U (269 out of 269 isolates). Thus, in nature, the cleavage site base pair is exclusively in a 5′ purine-3′ pyrimidine orientation. We recently reported pH- and Mg2+-dependent kinetics for genomic HDV ribozymes with G•U, G-C, A-U, and A+•C base pairs at the cleavage site (13). These four variants showed remarkably similar behavior, with only the A+•C ribozyme deviating and only at high pH. The behavior observed with the A+•C wobble pair is expected because formation of this base pair requires protonation at the N1 of A (pKa ~ 6.5) (19, 20). Our recent study indicted that the cleavage site base pair plays a structural role in the ribozyme (13). Because the four purine-pyrimidine base pairs functioned similarly regardless of the identity of the purine, if a metal ion does bind to this base pair it likely does so through the N7 of the purine at 1 position since this is the only common molecular feature.
Ribozyme design
The ribozyme strand used in this study was designed to crystallize readily for use in Raman crystallography experiments. The sequence is based on a fast-folding variant with less propensity to misfold than the native sequence (15, 16). The substrate was provided to the ribozyme in trans and had a single nucleotide upstream of the cleavage site (Figure 1A). To facilitate crystallization, the P4 helix was shortened and capped with a GAAA tetraloop as previously described (17). As shown in the present study, these modifications do not affect the catalytic activity of the ribozyme in any detectable manner. In RNAs used in crystallization experiments, a methoxyuridine was substituted for U(−1) of the substrate to inhibit the reaction and prevent cleavage.
The N7 of the cleavage site wobble pair is required for maximal reactivity
The sp2 nitrogen has stronger electronegativity than carbon and is inclined to interact with metal ions (21). Therefore the nitrogen-to-carbon substitution provides a potential means to disrupt binding of metal ions and assess the contributions of the metal ions to activity (22). This strategy was employed herein to probe the existence of a metal ion binding site within the major groove of the cleavage site base pair in the HDV ribozyme. At pH 7.0, the substrate containing a 7DG modification at residue G1 [abbreviated as G1(7DG)] exhibited a 6.3-fold decrease in kmax compared to WT substrate (Figure 2A). Kinetic parameters are compiled in Table 1. This result suggests that the N7 of G1 contributes to catalysis in some manner. This finding was surprising because there are no RNA atoms or metal ions within 3.3 Å of the N7 of G1 in any of the available crystal structures (5, 6).
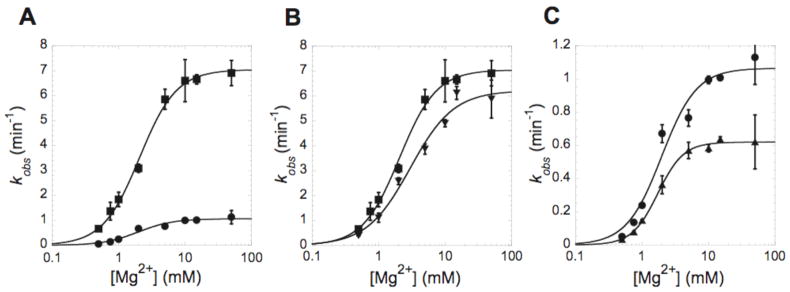
Figure 2.
7-deazaguanosine at the cleavage site reduces ribozyme activity. Plots of kobs with varying concentrations of MgCl2. (A) Comparison of wild type (WT) (■) and G1(7DG) substrates (●). (B) WT (■) and G2(7DG) (▼) substrates. (C) G1(7DG) (●) and G1G2(7DG) substrates (▼). The cleavage reactions were performed at 37 °C in 25 mM Tris buffer pH 7.0. Each point is the average of three independent experiments and the standard deviation is shown as error bars. Data were fit to equation 2 described in methods. The resulting kinetic parameters are provided in Table 1.
Table 1.
Single turnover kinetics parametersa
| kmax (min−1) | Kd,Mg2+ (mM) | αMg | ||||
|---|---|---|---|---|---|---|
| Substrates | pH 5.0 | pH 7.0 | pH 5.0 | pH 7.0b | pH 5.0 | pH 7.0 |
| WT | 2.0 ± 0.2 | 7.0 ± 0.2 | 30 ± 8 | 2.1 ± 0.1 | 0.78 ± 0.08 | 1.6 ± 0.1 |
| G1(7DG) | 1.2 ± 0.1 | 1.1 ± 0.1 | 60 ± 22 | 1.9 ± 0.3 | 0.78 ± 0.09 | 1.7 ± 0.4 |
| G2(7DG) | NDc | 6.2 ± 0.4 | ND | 3.0 ± 0.6 | ND | 1.3 ± 0.3 |
| G1G2(7DG) | ND | 0.62 ± 0.01 | ND | 1.7± 0.1 | ND | 2.3 ± 0.2 |
The values and standard errors were obtained by non-linear least-squares fitting in KaleidaGraph (Synergy software).
Kd,Mg2+ should be treated as apparent Kd,Mg2+ when the Hill coefficients are greater than 1.
Not determined.
To test whether the reduced rate of reaction is specific to the N7 of G1, two control substrates, G2(7DG) and G1G2(7DG), were studied. Substitution of G2 with 7DG gave a kmax that is quite close to that of the WT substrate (Figure 2B). The double substitution with 7DG at G1 and G2, on the other hand, exacerbated the effect of the G1(7DG) substitution by another factor of ~2 (Figure 2C). These results suggest that in the WT ribozyme, the N7 of G1 plays an important role in the reaction.
We then studied the magnesium dependence of the ribozyme reaction and compared the apparent Kd (Kd,Mg2+) and Hill values (αMg) for Mg2+ binding to the WT and three variant ribozymes. At pH 7.0, the 7DG modifications had little effect on the magnesium binding parameters. All of the Kd,Mg2+ and αMg values were similar to each other, 1.7–3.0 mM and 1.3–2.3, respectively (Table 1). These values are also similar to those obtained previously with a one-piece self-cleaving RNA construct (13, 23), which confirms that the variations in the sequence and the use of a trans-acting form of the HDV ribozyme do not affect activity.
The values of kmax and Kd,Mg2+ determined for G2(7DG) and G1G2(7DG) suggest that when the N7 of G1 is absent, the metal binding site is shifted or distorted to involve the nearby N7 of G2. Thus in the G1(7DG) modified ribozyme, the magnesium ion is likely bound in a suboptimal position that results in the observed reduction in kmax but without a measurable effect on Kd,Mg2+ (see Discussion). The G1G2(7DG) modified ribozyme further alters the Mg2+-binding pocket, which results in a somewhat more pronounced defect in kmax.
The Mg2+ at G1 senses the protonation state of C75
According to the post-cleavage crystal structure (5), C75 is well positioned to be a general acid in the HDV ribozyme cleavage reaction, and experimental studies show that it possesses a pKa near neutrality (17, 23, 24). If the metal ion is indeed located in the active site, the electrostatic repulsion from the nearby protonated C75 is expected to weaken its binding. Anticooperative binding between the proton on C75 and the catalytic metal ion has been observed by multiple methods (17, 23, 25). To further characterize the effect that the G1(7DG) modification had on metal binding, the magnesium dependence of the rate constant was examined at pH 5.0 where C75 is expected to be almost entirely protonated.
When the pH is lowered to 5.0, the WT substrate exhibits a ~3.5-fold lower kmax (Figure 3). In addition, αMg drops from ~1.6 to 0.8, a trend that is consistent with previous reports (13). In contrast, when the G1(7DG) substrate is used, the kmax is observed to be unchanged upon lowering the pH from 7.0 to 5.0. (Figure 3). In the WT ribozyme, the overall observed rate-pH profile has been ascribed to the involvement of a general acid (C75) with a pKa of near neutrality, along with a magnesium-bound hydroxide general base with a pKa of > 9. For WT, the increase in rate with pH from 5 to 7 is ascribed largely to deprotonation of the magnesium-bound water. Thus, the lack of pH effect on kmax is consistent with lesser (or no) involvement of the catalytic Mg2+ ion in the G1(7DG) modified ribozyme (23, 26, 27).
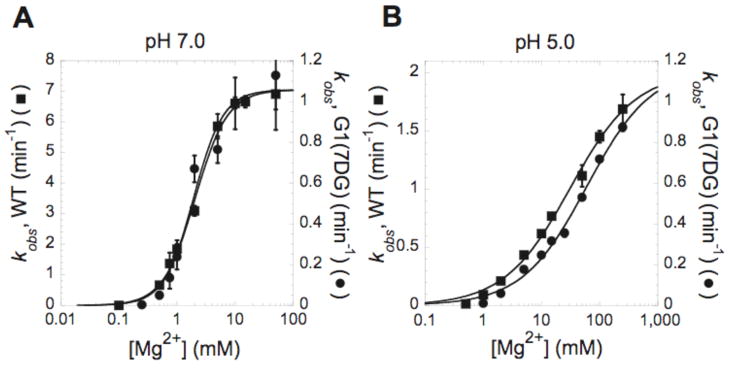
Figure 3.
Effect of 7-deazaguanosine on Mg2+ affinity is apparent at pH 5.0. Plots of kobs as a function of MgCl2 at (A) pH 7.0 and (B) pH 5.0. The cleavage reaction was performed at 37 °C in 25 mM Tris-Cl pH 7.0 or 25 mM sodium acetate pH 5.0. In each plot, the scale for kobs measured with the WT substrate (■) is shown on left Y axis, and the scale for kobs measured with the G1(7DG) (●) is shown on the right Y axis. Each point is the average of at least two independent experiments and the standard deviation is shown as error bars. Data were fit to equation 2. The resulting kinetic parameters are provided in Table 1.
An effect of the 7DG modification on the Kd,Mg2+ is observable when the pH is lowered to 5.0. In both the WT and the G1(7DG) variants, an increase in Kd,Mg2+ is seen at pH 5.0, which is attributed to binding of the proton at C75 interfering with binding of the positively-charged magnesium ion. While the kinetic data for the WT and G1(7DG) modified ribozyme are superposable at pH 7, at pH 5, the Mg2+ requirement to reach partial saturation of kobs for the G1(7DG) variant is shifted higher in every measurement made. Apparently, protonation of C75 exacerbates the effect of the G1(7DG) modification on binding of the active site magnesium ion. These data suggest that the magnesium binding site within the active site of the G1(7DG) modified ribozyme is measurably different (or less tightly bound) than that in the WT ribozyme.
The active site Mg2+ is detectable by Raman difference spectroscopy
We have previously used Raman crystallography to measure the pKa of C75 (17). Consistent with kinetics studies (23), this value is shifted from 4.2, the value expected for an unperturbed cytosine base (21), to 6.4 within the active site of the HDV ribozyme (at 2 mM Mg2+). In addition, proton binding to C75 is anticooperative with magnesium ion binding (17, 23). In this prior study, we used crystals of the HDV ribozyme in which the catalytic 2′-OH on the U-1 base had been modified to -H, -OCH3 and -F to inhibit the reaction (17). These modifications all returned similar pKa values suggesting these changes do not significantly perturb the active site of the HDV ribozyme. Importantly, these constructs allow the ribozyme to be crystallized with both the scissile phosphate and the catalytic nucleobase C75. The strong correlation between the pKa values measured in the crystal and in solution suggests that the WT ribozyme is in a catalytically relevant conformation within these crystals. Using the same RNA crystals, we show here that Raman difference spectroscopy collected as a function of pH reveals details of the active site Mg2+ within the HDV ribozyme that support and extend the kinetic data presented above.
The positive and negative features in the Raman pH-difference spectra in Figure 4 represent the changes in the parent Raman spectrum upon changing pH. Positive peaks are from HDV groups that are present at pH 7.5 but absent at pH 5.0 and negative peaks vice versa. The assignments are listed in Table S1. The details of the feature identifications and assignments in the HDV difference spectra (Figure 4) can be found in the Supporting Information. Most features could be assigned based on earlier assignments of reference compounds (28, 29) and HDV ribozyme crystals (17, 30). The peak at 1528 cm−1 in Figure 4A was previously assigned to a neutral cytosine ring mode possessing a major contribution from N3-C4 stretching (17). We also demonstrated that one cytosine base, identified as C75, was protonated when the pH is reduced from 7.5 to 5.0. Thus, we attribute the cytosine-based changes in Figure 4A to C75 in the active site.
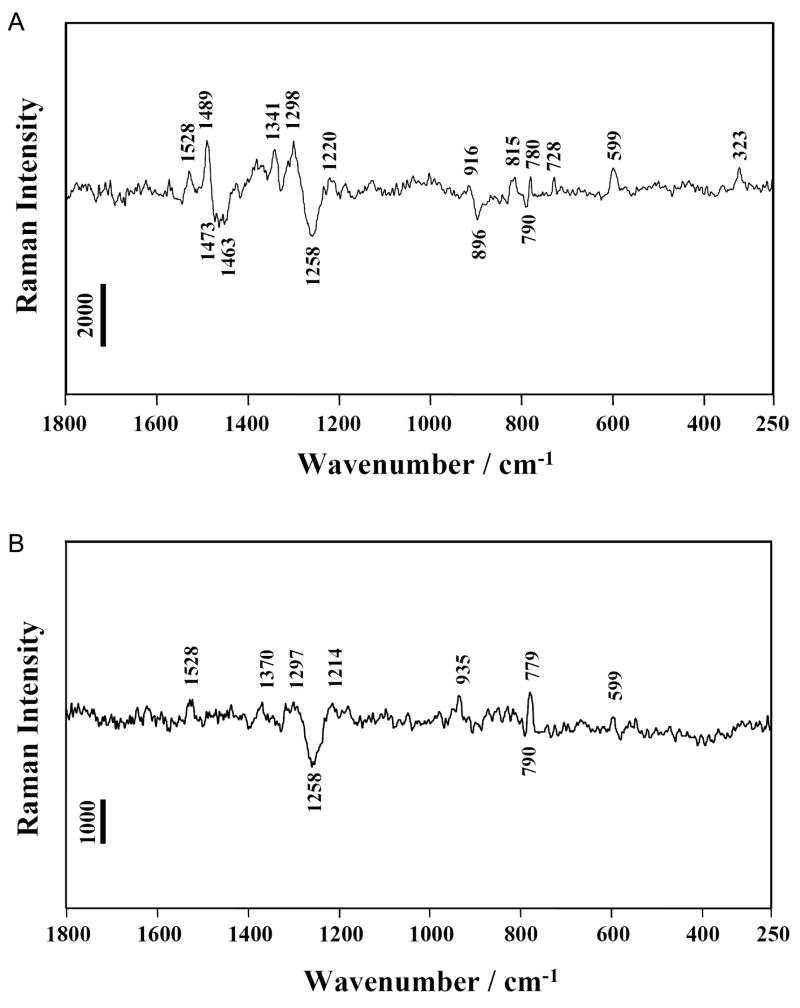
Figure 4.
Raman difference spectra. (A) [HDV pH 7.5] minus [HDV pH 5.0] (B) [HDV(G1(7DG)) pH 7.5] minus [HDV(G1(7DG)) pH 5.0]. The HDV ribozyme crystal was surrounded by cacodylate buffer pH 7.5 or acetate buffer pH 5.0 containing 20 mM Mg2+. Experimental details were described in previous Raman studies(17). Peak assignments are provided in Table S1. Vertical bars represent photon events.
In addition to features associated with protonation of C75, a band at 323 cm−1 corresponding to a hydrated Mg2+ ion with at least one non-water ligand is observed (Figure 4A). This ion is referred to as an inner-sphere Mg2+ hydrate. Several lines of evidence support the conclusion that this feature is due to a single inner-sphere Mg2+ hydrate bound within the active site. Raman analysis of Mg2+ binding to HDV crystals (at 20 mM Mg2+) (30) and analysis of the X-ray data (5) both demonstrated the presence of approximately 5 inner-sphere Mg2+ hydrates per HDV molecule. These give rise to a standardized Raman intensity of ~21,000 units at the signature 323 cm−1 position for inner-sphere Mg2+ hydrate. The present data (Figure 4B) have a standardized intensity of about 3,400 units at the 323 cm−1 peak. The ratio 3,400/21,000 strongly supports the finding that the 323 cm−1 band in Figure 4A emanates from a single inner-sphere Mg2+ hydrate per HDV molecule. This conclusion finds further support from our observation that introduction of a single 7DG modification is sufficient to disrupt the inner-sphere Mg2+ hydrate detectable in this assay (next section, Figure 4B).
Taken together, it can be concluded that upon going from pH 7.5 to 5.0 there is one inner-sphere Mg2+ hydrate per HDV lost as a single cytosine ring (C75) protonates. The anticooperative binding between this Mg2+ and the proton on C75 is consistent with our kinetics results in solution that support competition between the proton bound to C75 and the active site Mg2+ (23, 24). Therefore, it is likely that this inner-sphere Mg2+ hydrate detected by the Raman difference spectroscopy is the active site Mg2+.
Structural characterization of the active site Mg2+ and its binding site by Raman difference spectroscopy
When the pH is shifted from 7.5 to 5.0 and the catalytic Mg2+ is ejected, the Raman difference spectrum, [pH 7.5] minus [pH 5.0], is expected to reveal differences that correspond to the metal’s ligands. Often, phosphate oxygens provide ligands to Mg2+ and, indeed, we recently showed that inner-sphere interactions between Mg2+ and PO2− groups are detectable and quantifiable in HDV ribozyme crystals (30). The characteristic marker on Raman difference spectroscopy for interaction of PO2− with Mg2+ was the very clear intensity decrease and upshift (to 1117 cm−1) of the 1100 cm−1 peak (E.L. Christian, V. Anderson and M.E. Harris, personal communication and reference (30)). In Figure 4A, the striking absence of a differential feature near 1100 cm−1 upon the pH change indicates that the active site Mg2+ does not form inner-sphere interactions with PO2− groups of the HDV ribozyme. Thus, although the Raman marker of Mg(H2O)x2+ (x ≤ 5) at 323 cm−1 in Figure 4A unambiguously specified that the active site Mg2+-hydrate forms inner-sphere interactions with HDV groups, these do not arise from PO2−.
A potential RNA ligand for interacting with Mg(H2O)x2+ is the N7 of a guanine. This feature is evidenced in the Raman spectra by the differential peak near 1489 cm−1. This spectral region contains ring modes of purine, with a strong N7-C8 stretching component (31, 32). Due to the fact that guanine makes an intense contribution in this region as compared to adenine, nucleic acid peak perturbations in the 1485 cm−1 region have been considered as an indicator of coordination of electrophilic agents (e.g. metal ions) at the N7 position of guanine (32). Thus, the differential peak observed near 1489 cm−1 with its trough near 1473 cm−1 (Figure 4A) suggests that the active site Mg2+ binds inner-sphere to N7 of a guanine. Because this Mg2+ ion is close enough to C75 to sense its protonation state, the guanine N7 observed in the Raman spectra must also be close in space to C75. These results are not explained by the HDV crystal structures (5, 6). There are no Mg2+ions that are both in proximity to C75 and directly coordinate the N7 of a guanosine base. A limited number of candidates exist for a guanine within the HDV active site that could provide a ligand to the Mg2+ ion observed spectroscopically, and G1 is the most likely nucleotide.
To probe the role of the N7 of G1 in coordinating the active site magnesium, we prepared and crystallized the G1(7DG) ribozyme variant and recorded Raman spectra of these crystals. The Raman difference spectrum of [G1(7DG) pH 7.5] minus [G1(7DG) pH 5.0] is shown in Figure 4B. The most significant observation is the absence of guanine N7 Raman marker around 1485 cm−1, which supports the N7 of G1 making a direct inner-sphere contact with Mg2+ in WT crystals. In addition, this difference spectrum lacks the Raman marker of Mg(H2O)x2+ at 323 cm−1. Simultaneous loss of the guanine N7 and inner-sphere Mg2+ hydrate Raman markers indicates that protonation of C75 is correlated with both a change in inner-sphere magnesium ion binding and change in coordinate of the N7 of G1. These data suggest that the G1(7DG) ribozyme either does not bind a metal in the active site, perhaps instead binding the metal in an alternate pocket in a way that is less sensitive to changes in the protonation state of C75, or it binds the active site magnesium as a fully-hydrated Mg(H2O)62+ ion where it will likely be undetectable by Raman difference spectroscopy.
A model for the active site of the HDV ribozyme
A model of the catalytic metal ion in the active site was made by superposing the pre-cleavage crystal structure of the HDV ribozyme (PDB code 1sj3) (6) with the post-cleavage crystal structure (PDB code 1cx0) (5) (Figure 5). The coordinates of the scissile phosphate and U-1 were modeled according to the coordinates of the pre-cleavage structure, while the rest of the structure was of the post-cleavage structure. Although the exact position of the U(−1) nucleotide is not precisely defined by this process, there are very few conformations in which the −1 nucleotide can be accommodated within the active site, and none of the possible variations affect the subsequent positioning of the Mg2+ at the active site. The active site Mg2+ ion was modeled on the basis of the ion observed to have an inner-sphere interaction with the N7 of G163 of the high resolution P4–P6 ΔC209 RNA crystal structure (33) (PDB code 1hr2, chain B). HDV G1 was superposed with G163 to provide the position of the associated hydrated Mg2+ relative to the G•U wobble pair. As presented in the Discussion, this model accommodates the kinetic and spectroscopic data presented herein, as well as earlier biochemical data on the HDV ribozyme, which together support C75 as the general acid and Mg2+-bound hydroxide as the general base for cleavage.
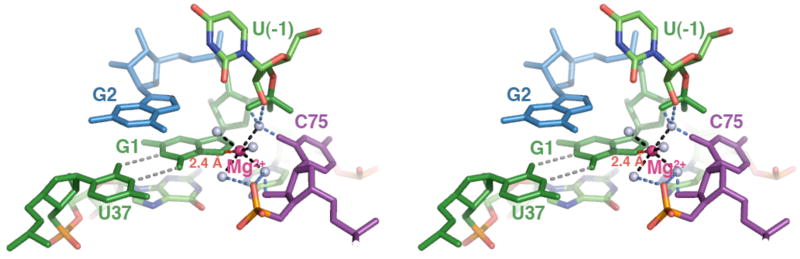
Figure 5.
The proposed model with hydrated metal ion close to the GU pair.
Nucleotides are colored as in Figure 1A,B. C75 in purple, the cleavage site G•U wobble in green, and G2 in blue. The N7 of G1 interacts with a hydrated Mg2+ ion (magenta sphere) by inner-sphere coordination (shown as a red dashed line). The distance criteria used for inner-sphere (1.9–2.5Å) and outer-sphere (2.5–3.3Å) are based on the studies of Juneau and Cech(33). 2. Five water molecules (light blue sphere) surround the active site Mg2+ and complete the inner-sphere shell (1.9–2.1 Å, shown in black dash lines). The exact positions of the waters in this model are not known, however the water coordinates as drawn to provide a model of the expected radius of the hydration shell. The possible interaction between these coordinated waters and RNA functional groups are denoted by blue dotted lines. Other experimental criteria used to build the model are described in the text.
DISCUSSION
RNA performs many critical functions in biology including gene regulation and catalysis. Its wide range of cellular functions are possible because it both contains decodable genetic information and has the ability to fold into complex three-dimensional structures. Perhaps due in part to its binary role in the cell, RNA lacks the chemical diversity of protein catalysts. This renders RNA prone to misfolding, and therefore high-resolution crystal structures need to be reconciled with biochemical data. For example, in the case of a small lead-dependent ribozyme, a structural model based on biochemical data (34) is more consistent with recent functional probing of the active site with conformationally restricted nucleotides (35) than experimental structures determined by NMR (36) and X-ray crystallography (37, 38). For the hammerhead ribozyme, a crystal structure of a construct with extended regions that engage in a tertiary structure (39) is consistent with extensive biochemical data (40–43), but these solution experiments could not be reconciled with previous structures that used a minimal hammerhead sequence (44, 45).
Similar structure-function disparities exist in the HDV ribozyme. Work from the Been lab implicates a metal ion at the cleavage site. In particular, switching the scissile phosphate from the native 3′-5′ linkage to 2′-5′ or changing the identity of the (−1) nucleotide alters the metal ion specificity of the reaction (46, 47). Solution kinetic experiments implicate a magnesium-bound water as the general base in the reaction (23, 27, 48). However a metal ion that would explain these results is not observed in the available crystal structures. Moreover, in the crystal structure of the HDV ribozyme in the pre-cleavage state, the catalytic nucleobase at position 75 is not in position to perform general acid or general base catalysis (5, 6). In an effort to develop a functionally relevant model of the HDV ribozyme active site, we have acquired additional solution and Raman crystallographic data on WT and active site mutants and combined these with existing functional and structural data to arrive at a new structural model of this ribozyme in a state that is catalytically competent.
Consequences of 7DG substitution support a metal ion at the active site
Previous biochemical studies support a general acid role for C75. Briefly, the catalytically inactive C75U mutants are rescuable by imidazole. This result supports a mechanism in which C75 could be either a general acid (49) or a general base. However, pH-rate profiles in the presence and absence of Mg2+ are inverted (23, 27), and C75U mutants are rescuable by a very good leaving group (48). These data provide strong support that C75 serves as a general acid catalyst, in agreement with the conformation of C75 observed in the crystal structure of the HDV ribozyme post cleavage (5).
Biochemical data are consistent with a multichannel model for catalysis, in which a hydrated magnesium hydroxide acts as the general base under physiological ion conditions but water acts as the general base at neutral pH in the absence of divalent metal ions (25, 27, 50). Rates in the presence and absence of a catalytic metal ion differ by only ~25-fold, consistent with a relatively minor contribution of the catalytic magnesium to rate enhancement.
In our present study, the G1(7DG) change resulted in a kmax decrease of ~7-fold, rather than the full 25-fold observed in the absence of magnesium. One possibility is that the catalytic metal ion is still positioned within the active site of the G1(7DG) variant, albeit suboptimally, allowing it to make only a partial contribution to catalysis. Consistent with a suboptimal positioning of the catalytic ion, there is loss of the Raman spectroscopic signature of the metal at the active site. Also consistent with partial, rather than no, contribution of the metal ion in G1(7DG), the G1G2(7DG) showed a larger (11-fold) decrease in kmax, which is within 2-fold of the total contribution of the metal ion and lower pH increased the Mg2+-requirement of G1(7DG) relative to wild-type. This suggests that the catalytic metal ion stays loosely associated with the active site in G1(7DG), and is almost completely ejected from the active site in the double mutant or by lowering pH. Lack of any discernable kinetic or binding effect in single mutant G2(7DG) supports G2 not having a role in metal retention in the WT background.
Effects of 7DG mutants on Kd,Mg2+ are, for the most part, subtle. This may be at least partially attributable to the nature of the metal binding site. The catalytic metal ion appears to make a single inner-sphere contact directly to the HDV ribozyme, at the N7 of G1. Other interactions with the ribozyme are likely outer-sphere contacts, mediated by magnesium-bound water molecules. Disruption of the inner-sphere ligand may result in a metal binding site that is not measurably destabilized at pH 7.0, but that results in metal ion orientations that are non-ideal for general base catalysis. Thus, effects are observed on kmax but not on Kd,Mg2+. A 2-fold increase in Kd,Mg2+ for G1(7DG) variant relative to WT HDV ribozyme is observable, but only at low pH, a condition that disfavors metal binding. Overall, both the biochemical and Raman data on the WT and single and double 7DG mutants support a metal ion interacting with the N7 of G1 at the cleavage site.
It is possible that the effects of 7DG incorporation at the cleavage site are the result of destabilization of the P1 helix. However, 7DG substitutions have a relatively minor effect on RNA helix stability. For example, Burkard and Turner (51) showed that this substitution destabilizes an RNA helix with a GG base pair by an entropy-corrected value of only 0 to 0.6 kcal/mol in free energy, with the lesser effect for mismatches near the end of a helix. Thermodynamic studies on DNA hairpins led to a penalty of just 0.34 kcal/mol for 7DG substitution into a Watson-Crick base pair just one base from end of the helix (52). Moreover, the active site of the HDV ribozyme is relatively tolerant at the cleavage site base pair, as evidence by recent work showing that the HDV ribozyme is equally active with GU, GC, AU, and protonated AC pairs at the cleavage site base pair (13). Based on the minor thermodynamic penalty of the 7-deazaguanosine substitution, especially near the end of helices, and the tolerance of the active site with respect to the cleavage site base pair, it is unlikely that 7DG substitution is destabilizing the active site.
Support for the active site model
In the proposed model (Figure 5), the catalytic metal ion is bound in the major groove of the P1 helix and is directly coordinated to the N7 of G1. A wide variety of biochemical data presented here and elsewhere support this interaction. First, kinetic characterization of ribozyme variants have demonstrated that either Watson-Crick or wobble geometry can be accommodated at the cleavage site, but position 1 must be a purine (13). This suggests that the N7, but neither the keto oxygen nor the Watson-Crick face of G1, is important for catalysis. Second, we have now demonstrated biochemically that the N7 of position 1 is required for optimal activity of the HDV ribozyme. Third, a ribozyme with a A+•C wobble pair at the cleavage site exhibits behavior consistent with close approach of the active site Mg2+ to nucleotide 1: the pKa of the A+•C wobble pair decreases as the Mg2+ concentration increases, suggesting binding of the proton to N1 is destabilized by the close approach of the positively charged Mg2+ (13). This is consistent with a binding site for the catalytic metal ion in close proximity to the N1 of nucleotide at position 1. Fourth, Raman difference spectroscopy indicates that C75 protonation is coupled to the N7 of a guanine base through an inner-sphere metal-mediated interaction, and the G1(7DG) modification pinpoints this N7 as that of G1.
If, as previously proposed, a magnesium-bound water serves as a catalytic base in the ribozyme reaction, then the magnesium ion must be close in space to 2′-hydroxyl of U(−1), which is accommodated in the model. In addition, C75, which has been shown biochemically to serve as the general acid in the reaction (23, 27, 48), is positioned next to the leaving group, the 5′-bridging oxygen of U(−1), in the model.
Anticooperative interaction of the proton on C75 and the active site Mg2+, which has been demonstrated by solution kinetic experiments and by Raman crystallography (17, 23, 25) is accommodated by the model. In our model, the active site magnesium is found 5.2 Å from the N3 of C75. Thus, in our proposed model, the Mg(H2O)52+ ion is juxtaposed with functional groups known to be close in space to the catalytic metal ion.
Several structural studies support this model. The HDV ribozyme has been co-crystallized with thallium ions, allowing the position of the monovalent ions within the ribozyme to be unambiguously identified by anomalous scattering (7). In these maps, a thallium ion is observed to bind in a position quite similar to the location of the Mg(H2O)52+ ion in the model presented here. Molecular dynamics simulations have also implicated this site in monovalent ion binding (53). In addition, the imino proton signals in the NMR spectra of the HDV ribozyme have been assigned (54). In this study, the region near the cleavage site wobble pair exhibited the greatest change in chemical shift in response to magnesium ion, suggesting a metal binding site is close in space.
Potential ligands to the catalytic Mg(H2O)52+ ion
Inspection of the ribozyme active site suggests other potential ligands to the catalytic [Mg(H2O)5]2+ ion (Figure 5). The pro-RP oxygens of the scissile phosphate and the phosphate of G76 are strong candidates as outer-sphere ligands. Because these atoms are not directly coordinated to the magnesium ion, their roles in ion binding are not readily testable experimentally. Substitution of these atoms with sulfur is not necessarily expected to have a profound deleterious effect nor have such modifications ever been shown to be rescuable with thiophilic metal ions (J. A. Piccirilli, personal communication). Nonetheless modifications of these pro-RP oxygen atoms do, in fact, interfere with catalysis (55–57). Substitution of the pro-Rp oxygen at the scissile phosphate results in a substrate that only cleaves to 20% completion (57), and substitution at G76 inhibits catalysis (56).
The O2 keto oxygen of C75 represents another potential outer-sphere ligand to the catalytic metal ion. Several studies have reported effects of base substitutions at C75, and all of the functional modifications at this position retain the keto oxygen (48, 58). While the C75A mutant lacks a keto oxygen and can function as the general acid in the HDV ribozyme reaction, its activity is down 300-fold relative to the WT sequence, suggesting that this mutant lacks a key feature(s) relative to the C75, WT sequence (23, 49). Last, the 2′-hydroxyl of C75 is also a potential ligand. Nishikawa et al characterized a ribozyme with a deoxyribose modification at C75 and found that removal of the 2′-hydroxyl of C75 decreased the reaction rate over 2 orders of magnitude, which was the most disruptive of all the modifications tested in the study (59).
Conclusions
We have generated a catalytically competent model for the active site of the HDV ribozyme in a state just prior to cleavage, which positions C75 to act as the general acid and a hydrated magnesium ion to act as the general base. It accommodates all prior biochemical data on this ribozyme, as well as 7DG mutant biochemical and spectroscopic data reported here.
One appealing feature of this model that it is consistent with data from kinetics studies and Raman spectroscopy is its ability to explain repulsion between a protonated C75 and a cationic divalent ion. This interaction should be relieved in the transition state as the proton transfers, especially given Brønsted coefficients for general acid and base catalysis near 0.5 (25, 60). Thus the repulsion between protonated C75 and the magnesium ion may drive the reaction through the catalytic strategy of ground state destabilization (61). Observation that binding of the magnesium ion is saturable in the presence of a protonated C75 is consistent with the reaction requiring binding of these like-charged species (24).
The present crystal structures of the HDV ribozyme are not consistent with microscopic reversibility. In particular, the crystal structure of the cleaved ribozyme suggests that C75 serves as the general acid for cleavage, while the pre-cleaved structures suggests it serves as the general base for cleavage (5, 6). Our model resolves this discrepancy by providing a picture of the pre-cleaved state in which C75 serves as the general acid in the cleavage reaction. Although the ligation reaction of the HDV ribozyme is not observed experimentally (2, 3), presumably due to low affinity of the ribozyme for the upstream product, our model predicts that the crystal structure of the cleaved ribozyme is catalytically relevant, as C75 is well positioned to serve as the general base in a ligation reaction. Thus we provide a picture of the HDV active site that is compatible with the principle of microscopic reversibility.
Lastly, we do not assert that the precise details of the active site are correct; this will have to await additional crystal structures. Nonetheless, the active site model presented herein is congruent with a wealth of biochemical, spectroscopic, and structural data and should help guide mechanistic analysis of this catalytic RNA.
Supplementary Material
The basis for assignments in the HDV Raman difference spectrum
Quantum mechanical calculations of pentahydrate Mg2+ bound inner sphere to N7 of a guanine
Table S1: Positions (cm−1) and assignment of major Raman difference bands observed in Raman difference spectra of [HDV pH 7.5] minus [HDV pH 5.0] and [CMP pH 6.0] minus [CMP pH 3.0]
Figure S1: Sequence of the HDV ribozyme used in this study
Figure S2: Raman difference spectrum of [CMP pH 6.0] minus [CMP pH 3.0]
Figure S3: Model used in quantum mechanical calculations of a pentahydrate Mg2+ bound inner sphere to N7 of a guanine.
This material is available free of charge via the Internet at hppt://pubs.acs.org.
Acknowledgments
We thank Durga Chadalavada, Elaine Chase, and Rieko Yajima for assistance, Eric Christian and Mike Harris for stimulating intellectual contributions to this project, Mark Hermodson, Paul Huber and Joe Kappock for critical reading of the manuscript.
Footnotes
This project was supported by NSF Grant MCB-0527102 to PCB and NIH Grant GM-54072 to PRC, the Purdue University Department of Biochemistry and the Purdue University Cancer Center.
Abbreviations. HDV, hepatitis delta virus; 7DG, 7-deazaguanosine; G1(7DG), 7-deazaguanosine at position 1 of the HDV ribozyme; G2(7DG), 7-deazaguanosine at position 2 of the HDV ribozyme; G1G2(7DG), 7-deazaguanosine at positions 1 and 2 of the HDV ribozyme; C75, the cytosine residue at position 75 of the genomic HDV ribozyme; C75U, the cytosine-to-uracil mutation at position 75 of the genomic HDV ribozyme; Kd,Mg2+, apparent dissociation constant for Mg2+; αMg, Hill number for Mg2+ binding.
References
- 1.Lai MM. The molecular biology of hepatitis delta virus. Annu Rev Biochem. 1995;64:259–286. doi: 10.1146/annurev.bi.64.070195.001355. [DOI] [PubMed] [Google Scholar]
- 2.Been MD, Wickham GS. Self-cleaving ribozymes of hepatitis delta virus RNA. Eur J Biochem. 1997;247:741–753. doi: 10.1111/j.1432-1033.1997.00741.x. [DOI] [PubMed] [Google Scholar]
- 3.Wadkins TS, Been MD. Ribozyme activity in the genomic and antigenomic RNA strands of hepatitis delta virus. Cell Mol Life Sci. 2002;59:112–125. doi: 10.1007/s00018-002-8409-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–1792. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- 5.Ferre-D’Amare AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 6.Ke A, Zhou K, Ding F, Cate JH, Doudna JA. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 7.Ke A, Ding F, Batchelor JD, Doudna JA. Structural roles of monovalent cations in the HDV ribozyme. Structure. 2007;15:281–287. doi: 10.1016/j.str.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Cate JH, Doudna JA. Metal-binding sites in the major groove of a large ribozyme domain. Structure. 1996;4:1221–1229. doi: 10.1016/s0969-2126(96)00129-3. [DOI] [PubMed] [Google Scholar]
- 9.Konforti BB, Abramovitz DL, Duarte CM, Karpeisky A, Beigelman L, Pyle AM. Ribozyme catalysis from the major groove of group II intron domain 5. Mol Cell. 1998;1:433–441. doi: 10.1016/s1097-2765(00)80043-x. [DOI] [PubMed] [Google Scholar]
- 10.Masquida B, Sauter C, Westhof E. A sulfate pocket formed by three G•U pairs in the 0.97 A resolution X-ray structure of a nonameric RNA. RNA. 1999;5:1384–1395. doi: 10.1017/s1355838299991173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu HN, Lee JY, Huang HW, Huang YS, Hsueh TG. Mutagenesis analysis of a hepatitis delta virus genomic ribozyme. Nucleic Acids Res. 1993;21:4193–4199. doi: 10.1093/nar/21.18.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perrotta AT, Been MD. Core sequences and a cleavage site wobble pair required for HDV antigenomic ribozyme self-cleavage. Nucleic Acids Res. 1996;24:1314–1321. doi: 10.1093/nar/24.7.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerrone-Szakal AL, Chadalavada DM, Golden BL, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: The cleavage site base pair plays a structural role in facilitating catalysis. J Am Chem Soc. 2008;130:14504–14520. doi: 10.1261/rna.1140308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chadalavada DM, Knudsen SM, Nakano S, Bevilacqua PC. A role for upstream RNA structure in facilitating the catalytic fold of the genomic hepatitis delta virus ribozyme. J Mol Biol. 2000;301:349–367. doi: 10.1006/jmbi.2000.3953. [DOI] [PubMed] [Google Scholar]
- 15.Chadalavada DM, Senchak SE, Bevilacqua PC. The folding pathway of the genomic hepatitis delta virus ribozyme is dominated by slow folding of the pseudoknots. J Mol Biol. 2002;317:559–575. doi: 10.1006/jmbi.2002.5434. [DOI] [PubMed] [Google Scholar]
- 16.Brown TS, Chadalavada DM, Bevilacqua PC. Design of a highly reactive HDV ribozyme sequence uncovers facilitation of RNA folding by alternative pairings and physiological ionic strength. J Mol Biol. 2004;341:695–712. doi: 10.1016/j.jmb.2004.05.071. [DOI] [PubMed] [Google Scholar]
- 17.Gong B, Chen JH, Chase E, Chadalavada DM, Yajima R, Golden BL, Bevilacqua PC, Carey PR. Direct measurement of a pK(a) near neutrality for the catalytic cytosine in the genomic HDV ribozyme using Raman crystallography. J Am Chem Soc. 2007;129:13335–13342. doi: 10.1021/ja0743893. [DOI] [PubMed] [Google Scholar]
- 18.Chadalavada DM, Cerrone-Szakal AL, Bevilacqua PC. Wild-type is the optimal sequence of the HDV ribozyme under cotranscriptional conditions. RNA. 2007;13:2189–2201. doi: 10.1261/rna.778107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai Z, Tinoco I., Jr Solution structure of loop A from the hairpin ribozyme from tobacco ringspot virus satellite. Biochemistry. 1996;35:6026–6036. doi: 10.1021/bi952985g. [DOI] [PubMed] [Google Scholar]
- 20.Legault P, Pardi A. Unusual Dynamics and pKa Shift at the Active Site of a Lead-Dependent Ribozyme. J Am Chem Soc. 1997;119:6621–6628. [Google Scholar]
- 21.Saenger W. Principles of nucleic acid structure. Springer-Verlag; New York: 1984. [Google Scholar]
- 22.Nakamatsu Y, Warashina M, Kuwabara T, Tanaka Y, Yoshinari K, Taira K. Significant activity of a modified ribozyme with N7-deazaguanine at g10.1: the double-metal-ion mechanism of catalysis in reactions catalysed by hammerhead ribozymes. Genes Cells. 2000;5:603–612. doi: 10.1046/j.1365-2443.2000.00352.x. [DOI] [PubMed] [Google Scholar]
- 23.Nakano S, Chadalavada DM, Bevilacqua PC. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- 24.Nakano S, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: a mutant of the C41 motif provides insight into the positioning and thermodynamic linkage of metal ions and protons. Biochemistry. 2007;46:3001–3012. doi: 10.1021/bi061732s. [DOI] [PubMed] [Google Scholar]
- 25.Nakano S, Proctor DJ, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: assessing the catalytic and structural contributions of divalent metal ions within a multichannel reaction mechanism. Biochemistry. 2001;40:12022–12038. doi: 10.1021/bi011253n. [DOI] [PubMed] [Google Scholar]
- 26.Bevilacqua PC. Mechanistic considerations for general acid-base catalysis by RNA: revisiting the mechanism of the hairpin ribozyme. Biochemistry. 2003;42:2259–2265. doi: 10.1021/bi027273m. [DOI] [PubMed] [Google Scholar]
- 27.Cerrone-Szakal AL, Siegfried NA, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: solvent isotope effects and proton inventories in the absence of divalent metal ions support C75 as the general acid. J Am Chem Soc. 2008;130:14504–14520. doi: 10.1021/ja801816k. [DOI] [PubMed] [Google Scholar]
- 28.Carey PR. Biochemical Applications of Raman and Resonance Raman Spectroscopies. Academic Press; New York: 1982. [Google Scholar]
- 29.Thomas GJ, Jr, Tsuboi M. Raman Spectroscopy of Nucleic Acids and Their Complexes. Advances in Biophysical Chemistry. 1993;3:1–70. [Google Scholar]
- 30.Gong B, Chen Y, Christian EL, Chen JH, Chase E, Chadalavada DM, Yajima R, Golden BL, Bevilacqua PC, Carey PR. Detection of innersphere interactions between magnesium hydrate and the phosphate backbone of the HDV ribozyme using Raman crystallography. J Am Chem Soc. 2008;130:9670–9672. doi: 10.1021/ja801861s. [DOI] [PubMed] [Google Scholar]
- 31.Majoube M. Vibrational spectra of guanine. A normal coordinate analysis. J Chim Phys. 1984;81:303–315. [Google Scholar]
- 32.Duguid J, Bloomfield VA, Benevides J, Thomas GJ., Jr Raman spectroscopy of DNA-metal complexes. I Interactions and conformational effects of the divalent cations: Mg, Ca, Sr, Ba, Mn, Co, Ni, Cu, Pd, and Cd. Biophys J. 1993;65:1916–1928. doi: 10.1016/S0006-3495(93)81263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juneau K, Podell E, Harrington DJ, Cech TR. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA--solvent interactions. Structure. 2001;9:221–231. doi: 10.1016/s0969-2126(01)00579-2. [DOI] [PubMed] [Google Scholar]
- 34.Lemieux S, Chartrand P, Cedergren R, Major F. Modeling active RNA structures using the intersection of conformational space: application to the lead-activated ribozyme. RNA. 1998;4:739–749. doi: 10.1017/s1355838298971266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yajima R, Proctor DJ, Kierzek R, Kierzek E, Bevilacqua PC. A conformationally restricted guanosine analog reveals the catalytic relevance of three structures of an RNA enzyme. Chem Biol. 2007;14:23–30. doi: 10.1016/j.chembiol.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Hoogstraten CG, Legault P, Pardi A. NMR solution structure of the lead-dependent ribozyme: evidence for dynamics in RNA catalysis. J Mol Biol. 1998;284:337–350. doi: 10.1006/jmbi.1998.2182. [DOI] [PubMed] [Google Scholar]
- 37.Wedekind JE, McKay DB. Crystal structure of a lead-dependent ribozyme revealing metal binding sites relevant to catalysis. Nat Struct Biol. 1999;6:261–268. doi: 10.1038/6700. [DOI] [PubMed] [Google Scholar]
- 38.Wedekind JE, McKay DB. Crystal structure of the leadzyme at 1.8 A resolution: metal ion binding and the implications for catalytic mechanism and allo site ion regulation. Biochemistry. 2003;42:9554–9563. doi: 10.1021/bi0300783. [DOI] [PubMed] [Google Scholar]
- 39.Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang S, Karbstein K, Peracchi A, Beigelman L, Herschlag D. Identification of the hammerhead ribozyme metal ion binding site responsible for rescue of the deleterious effect of a cleavage site phosphorothioate. Biochemistry. 1999;38:14363–14378. doi: 10.1021/bi9913202. [DOI] [PubMed] [Google Scholar]
- 41.Heckman JE, Lambert D, Burke JM. Photocrosslinking detects a compact, active structure of the hammerhead ribozyme. Biochemistry. 2005;44:4148–4156. doi: 10.1021/bi047858b. [DOI] [PubMed] [Google Scholar]
- 42.Blount KF, Uhlenbeck OC. The structure-function dilemma of the hammerhead ribozyme. Annu Rev Biophys Biomol Struct. 2005;34:415–440. doi: 10.1146/annurev.biophys.34.122004.184428. [DOI] [PubMed] [Google Scholar]
- 43.Lambert D, Heckman JE, Burke JM. Three conserved guanosines approach the reaction site in native and minimal hammerhead ribozymes. Biochemistry. 2006;45:7140–7147. doi: 10.1021/bi052457x. [DOI] [PubMed] [Google Scholar]
- 44.Scott WG, Finch JT, Klug A. The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell. 1995;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- 45.McKay DB. Structure and function of the hammerhead ribozyme: an unfinished story. RNA. 1996;2:395–403. [PMC free article] [PubMed] [Google Scholar]
- 46.Shih IH, Been MD. Ribozyme cleavage of a 2,5-phosphodiester linkage: mechanism and a restricted divalent metal-ion requirement. RNA. 1999;5:1140–1148. doi: 10.1017/s1355838299990763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perrotta AT, Been MD. A single nucleotide linked to a switch in metal ion reactivity preference in the HDV ribozymes. Biochemistry. 2007;46:5124–5130. doi: 10.1021/bi602569x. [DOI] [PubMed] [Google Scholar]
- 48.Das SR, Piccirilli JA. General acid catalysis by the hepatitis delta virus ribozyme. Nat Chem Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- 49.Perrotta AT, Shih I, Been MD. Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science. 1999;286:123–126. doi: 10.1126/science.286.5437.123. [DOI] [PubMed] [Google Scholar]
- 50.Nakano S, Cerrone AL, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: classifying the catalytic and structural metal ion sites within a multichannel reaction mechanism. Biochemistry. 2003;42:2982–2994. doi: 10.1021/bi026815x. [DOI] [PubMed] [Google Scholar]
- 51.Burkard ME, Turner DH. NMR structures of r(GCAGGCGUGC)2 and determinants of stability for single guanosine-guanosine base pairs. Biochemistry. 2000;39:11748–11762. doi: 10.1021/bi000720i. [DOI] [PubMed] [Google Scholar]
- 52.Moody EM, Bevilacqua PC. Folding of a stable DNA motif involves a highly cooperative network of interactions. J Am Chem Soc. 2003;125:16285–16293. doi: 10.1021/ja038897y. [DOI] [PubMed] [Google Scholar]
- 53.Krasovska MV, Sefcikova J, Reblova K, Schneider B, Walter NG, Sponer J. Cations and hydration in catalytic RNA: molecular dynamics of the hepatitis delta virus ribozyme. Biophys J. 2006;91:626–638. doi: 10.1529/biophysj.105.079368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka Y, Hori T, Tagaya M, Sakamoto T, Kurihara Y, Katahira M, Uesugi S. Imino proton NMR analysis of HDV ribozymes: nested double pseudoknot structure and Mg2+ ion-binding site close to the catalytic core in solution. Nucleic Acids Res. 2002;30:766–774. doi: 10.1093/nar/30.3.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jeoung YH, Kumar PK, Suh YA, Taira K, Nishikawa S. Identification of phosphate oxygens that are important for self-cleavage activity of the HDV ribozyme by phosphorothioate substitution interference analysis. Nucleic Acids Res. 1994;22:3722–3727. doi: 10.1093/nar/22.18.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prabhu NS, Dinter-Gottlieb G, Gottlieb PA. Single substitutions of phosphorothioates in the HDV ribozyme G73 define regions necessary for optimal self-cleaving activity. Nucleic Acids Res. 1997;25:5119–5124. doi: 10.1093/nar/25.24.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fauzi H, Kawakami J, Nishikawa F, Nishikawa S. Analysis of the cleavage reaction of a trans-acting human hepatitis delta virus ribozyme. Nucleic Acids Res. 1997;25:3124–3130. doi: 10.1093/nar/25.15.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strobel SA, Jones FD, Oyelere AK, Ryder SP. Biochemical detection of adenosine and cytidine ionization within RNA by interference analysis. Nucleic Acids Res Suppl. 2003:229–230. doi: 10.1093/nass/3.1.229. [DOI] [PubMed] [Google Scholar]
- 59.Nishikawa F, Shirai M, Nishikawa S. Site-specific modification of functional groups in genomic hepatitis delta virus (HDV) ribozyme. Eur J Biochem. 2002;269:5792–5803. doi: 10.1046/j.1432-1033.2002.03280.x. [DOI] [PubMed] [Google Scholar]
- 60.Shih IH, Been MD. Involvement of a cytosine side chain in proton transfer in the rate-determining step of ribozyme self-cleavage. Proc Natl Acad Sci U S A. 2001;98:1489–1494. doi: 10.1073/pnas.98.4.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jencks WP. Catalysis in chemistry and enzymology. GcGraw-Hill; New York: 1969. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The basis for assignments in the HDV Raman difference spectrum
Quantum mechanical calculations of pentahydrate Mg2+ bound inner sphere to N7 of a guanine
Table S1: Positions (cm−1) and assignment of major Raman difference bands observed in Raman difference spectra of [HDV pH 7.5] minus [HDV pH 5.0] and [CMP pH 6.0] minus [CMP pH 3.0]
Figure S1: Sequence of the HDV ribozyme used in this study
Figure S2: Raman difference spectrum of [CMP pH 6.0] minus [CMP pH 3.0]
Figure S3: Model used in quantum mechanical calculations of a pentahydrate Mg2+ bound inner sphere to N7 of a guanine.
This material is available free of charge via the Internet at hppt://pubs.acs.org.