Abstract
Although a hereditary contribution to emphysema has been long suspected, severe α1-antitrypsin deficiency remains the only conclusively proven genetic risk factor for chronic obstructive pulmonary disease (COPD). Recently, genome-wide linkage analysis has led to the identification of two promising candidate genes for COPD: TGFB1 and SERPINE2. Like multiple other COPD candidate gene associations, even these positionally identified genes have not been universally replicated across all studies. Differences in phenotype definition may contribute to nonreplication in genetic studies of heterogeneous disorders such as COPD. The use of precisely measured phenotypes, including emphysema quantification on high-resolution chest computed tomography scans, has aided in the discovery of additional genes for clinically relevant COPD-related traits. The use of computed tomography scans to assess emphysema and airway disease as well as newer genetic technologies, including gene expression microarrays and genome-wide association studies, has great potential to detect novel genes affecting COPD susceptibility, severity, and response to treatment.
Keywords: α1-antitrypsin deficiency, chronic obstructive pulmonary disease, genetic linkage, single-nucleotide polymorphism
For nearly two hundred years, a hereditary contribution to chronic obstructive pulmonary disease (COPD) has been suspected. In the early nineteenth century, the American physician James Jackson, Jr. observed that patients with emphysema were more likely than unaffected individuals to have parents with emphysema (1). A century and a half later, Laurell and Eriksson described serum α1-antitrypsin (AAT) deficiency as a risk factor for emphysema (2), the first report of a specific genetic contribution to COPD. Severe AAT deficiency remains the only conclusively proven genetic risk factor for COPD, although several other promising candidate genes for COPD unrelated to AAT deficiency have been recently identified (3, 4).
Like many other common chronic diseases, the etiology of COPD is complex, with contributions from multiple genetic and environmental factors. Unlike most other complex diseases, the major risk factor for COPD—cigarette smoking—is well known. The fact that smoking is a clear risk factor leads to dilemmas and opportunities in the study of COPD genetics. Because the major risk factor (environmental, genetic, or otherwise) has been identified, one may question the purpose of COPD genetics research, especially because the majority of COPD genes are likely to be of modest effect sizes, as opposed to the strong effect of AAT deficiency.
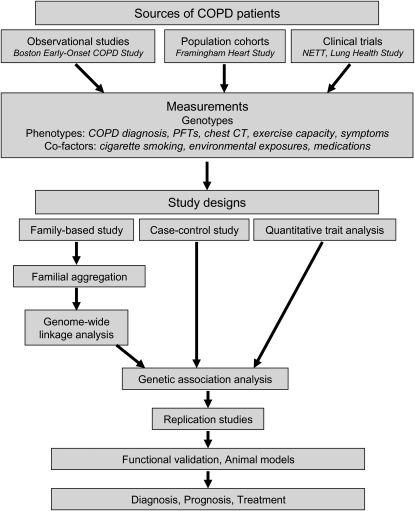
Genetics provides a unique tool to study the pathophysiology of COPD. Traditional laboratory or animal model studies may focus on a single gene or protein or on a few genes/proteins in combination, with these genes/proteins identified based on prior knowledge or suspected mechanisms of COPD pathogenesis. Human genetics studies may afford a complementary approach to corroborate the findings of these candidate gene studies (Figure 1). However, classical genetics tools (e.g., linkage analysis) and newer techniques (e.g., genome-wide association studies) allow for the comprehensive evaluation of the entire genome without prior assumptions regarding disease biology. These studies may lead to the identification of novel COPD candidate genes, which can be subjected to further in vitro or in vivo experimentation. These genes may identify novel targets for COPD therapies or biomarkers for the diagnosis and follow-up of patients with COPD.
Figure 1.
Integration of study designs in the genetic epidemiology of chronic obstructive pulmonary disease (COPD). NETT = National Emphysema Treatment Trial; PFT= pulmonary function test.
The study of COPD genetics can also provide a useful example to study other common chronic diseases. The main environmental risk factor of cigarette smoking can be easily assessed via patient interview or questionnaire (5) and is commonly categorized into current, former, or never smokers or quantified by pack-years of smoking (packs per day multiplied by number of years of smoking). Gene–environment interactions are likely to be important in many complex diseases (6); in COPD, these interactions can be explicitly modeled. The gene-interaction models can provide valuable insight into COPD pathogenesis and serve as an example for other complex disease genetic studies. It is possible that interactions with other environmental exposures, such as environmental tobacco smoke or nutritional factors, are relevant in COPD.
AAT DEFICIENCY
AAT is a serine protease inhibitor and the major inhibitor of neutrophil elastase in the lung. Additional functions of AAT that may be relevant to COPD pathogenesis have been described (7, 8). This 52-kD protein is encoded by the SERPINA1 gene on chromosome 14. The lung and liver diseases due to severe AAT deficiency are inherited in an autosomal recessive pattern. The majority of affected individuals inherit two copies of the mutant Z allele, which is termed protease inhibitor (PI) ZZ. The Z mutation is caused by a single nucleotide base pair change that leads to the substitution of lysine for glutamic acid at amino acid 342, affecting protein function. The Z protein polymerizes in the liver, causing reduced serum AAT levels and therefore less AAT to protect against proteolytic stress in the lung (9, 10). Individuals with a PI SZ genotype, a compound heterozygote of the Z allele with the mutant S allele (reduced AAT levels), are at risk for COPD (11). AAT deficiency may also be due to a combination of the Z allele with one of several rare null alleles (no functional protein) (12).
Individuals with severe AAT deficiency are at risk for emphysema at an early age, especially those who smoke cigarettes. Phenotypic expression of lung and liver disease in AAT deficiency is highly variable and is likely subject to other modifier genes and environmental exposures (13). Although AAT-deficient individuals have significantly increased risks for COPD, severe AAT deficiency accounts for only 1% to 2% of COPD cases in the United States and Europe (14). Heterozygous carriers of the Z allele (termed PI MZ) are more common, but it is not clear whether carriers are at an increased risk for COPD (15). As with the PI ZZ genotype, environmental and genetic factors may modify COPD risk in individuals with the PI MZ genotype.
Patients with COPD and severe AAT deficiency are identified based on serum AAT levels, followed by isoelectric focusing of serum to determine the PI phenotype and/or genotyping of the PI locus. Augmentation therapy with pooled human AAT may be beneficial in COPD due to severe AAT deficiency (16), particularly in patients with moderate airflow obstruction (17, 18). Recommendations for screening and testing and indications for augmentation therapy have been reviewed elsewhere (19). Case series have described lung volume reduction surgery (LVRS) as a treatment for severe emphysema due to AAT deficiency (20–22). However, data from 10 AAT-deficient patients randomized to LVRS in the National Emphysema Treatment Trial (NETT) suggest reduced benefit from LVRS in AAT-deficient patients compared with those without AAT deficiency (23).
GENOME-WIDE LINKAGE ANALYSES IN COPD
In a genome-wide linkage analysis, a set of short tandem repeat DNA markers is genotyped throughout the genome in a family-based study to identify chromosomal regions that segregate with the disease of interest. In COPD, the only reported linkage analyses have been performed in the Boston Early-Onset COPD Study (24–27). In this study, extended families were ascertained through a proband with severe airflow obstruction (FEV1 <40% predicted), age less than 53 years, and no severe AAT deficiency (28). These individuals may be expected to have a greater genetic contribution to COPD than subjects diagnosed at later ages. The most significant evidence for linkage was found on chromosome 2q for FEV1/FVC ratio, both pre- and postbronchodilator (25, 26). Suggestive evidence for linkage of prebronchodilator FEV1 was demonstrated on chromosome 12p. In the initial genome scans, significant linkage for postbronchodilator FEV1 was shown on chromosome 8p (26), although the evidence for linkage was attenuated when additional markers were studied (29). Chromosome 19q also had suggestive evidence for linkage for qualitative and quantitative traits (3, 24). No other linkage analyses have been published in COPD to confirm these results. However, an overlapping region on chromosome 2q has been linked to FEV1/FVC in families from the general population (30), pointing to the potential importance of a gene or genes in this region.
GENETIC ASSOCIATION STUDIES IN COPD
In contrast to the genome-wide linkage studies mentioned previously, the published COPD genetic association studies have focused on candidate genes, identified based on presumed importance in COPD pathogenesis or location in a region of linkage. Most commonly, distributions of alleles or genotypes of one or more single nucleotide polymorphisms (SNPs) are compared in COPD cases versus control subjects without disease. Variations on this design have been used, including analysis of quantitative traits or family-based study designs. Polymorphisms in multiple genes have been associated with COPD, emphysema, or related traits. Genes that have been associated in two or more studies are listed in Table 1.
TABLE 1.
CANDIDATE GENES, BESIDES α1-ANTITRYPSIN (SERPINA1), WITH REPLICATED ASSOCIATIONS TO CHRONIC OBSTRUCTIVE PULMONARY DISEASE, EMPHYSEMA, OR RELATED TRAITS
| Gene | Initial Association | Replication | Do Not Support Association |
|---|---|---|---|
| α1-Antichymotrypsin (SERPINA3) | Poller, 1992 (80, 81) | Ishii, 2000 (82) | Sandford, 1998 (83); Benetazzo, 1999 (84) |
| β2-Adrenergic receptor (ADRB2) | Ho, 2001 (85) | Hegab, 2004 (72) | Joos, 2003 (86); Brogger, 2006 (87) |
| Defensin β1 (DEFB1) | Matsushita, 2002 (88) | Hu, 2004 (89) | Wallace, 2006 (90); Hersh, 2006 (29) |
| Microsomal epoxide hydrolase (EPHX1) | Smith, 1997 (91) | Sandford, 2001 (51); Xiao, 2004 (92); DeMeo, 2007 (62) | Yoshikawa, 2000 (93); Takeyabu, 2000 (94); Yim, 2000 (95) |
| Vitamin D binding protein (GC) | Horne, 1990 (96) | Schellenberg, 1998 (97); Ishii, 2001 (98); Ito, 2004 (99) | Kauffmann, 1983 (100); Kasuga, 2003 (101); Sandford, 2001 (51) |
| Glutathione S-transferase M1 (GSTM1) | Harrison, 1997 (102) | Baranova, 1997 (54); Cheng, 2004 (103) | Yim, 2000 (95); He, 2002 (104) |
| Glutathione S-transferase pi (GSTP1) | Ishii, 1999 (105) | He, 2002 (104) | Yim, 2002 (106); Cheng, 2004 (103); Xiao, 2004 (92) |
| Heme oxygenase (HMOX1) | Yamada, 2000 (53) | Guenegou, 2006 (107); Nakayama, 2006 (108) | He, 2002 (104) |
| Matrix metallopeptidase-9 (MMP9) | Minematsu, 2001 (52) | Zhou, 2004 (109) | Joos, 2002 (110) |
| Serpin peptidase inhibitor E2 (SERPINE2) | DeMeo, 2006 (4) | Zhu, 2007 (48) | Chappell, 2006 (49) |
| Extracellular superoxide dismutase (SOD3) | Juul, 2006 (111) | Young, 2006 (112) | |
| Surfactant protein B (SFTPB) | Guo, 2001 (75) | Seifart, 2002 (113) | |
| Transforming growth factor–β1 (TGF-β1) | Wu, 2004 (40) | Celedon, 2004 (3); Su, 2005 (114); Van Diemen, 2006 (41) | Yoon, 2006 (42); Ogawa 2007 (43) |
| Tumor necrosis factor (TNF) | Huang, 1997 (55) | Sakao, 2001 (115); Matheson, 2006 (116) | Higham, 2000 (117); Ishii, 2000 (118); Patuzzo, 2000 (119); Sandford, 2001 (51); Ferrarotti, 2003 (120); Chierakul, 2005 (121); Hegab, 2005 (122); Jiang, 2005 (123); Seifart, 2005 (124); Brogger, 2006 (87); Ruse, 2007 (125) |
To test systematically the replication validity of previously published COPD genetic associations, our group examined 29 variants in 12 COPD candidate genes (31). Genotyping was performed in two study populations: a family-based study of extended pedigrees from the Boston Early-Onset COPD Study and a case-control study comparing 304 subjects with emphysema and severe airflow obstruction from NETT with 441 smokers without airflow obstruction from the Normative Aging Study (32). In the Boston Early-Onset COPD Study families, a promoter SNP in tumor necrosis factor (TNF)-α, a coding variant in surfactant protein B (SFTPB Thr131Ile), and a repeat polymorphism near heme oxygenase-1 (HMOX1) were significantly associated with quantitative and qualitative spirometric traits. In the case-control analysis, the SFTPB Thr131Ile variant was significantly associated in a model that incorporated a gene-by-smoking interaction term. A different allele of the HMOX1 repeat was significant. The TNF promoter SNP was not replicated, but a coding SNP in microsomal epoxide hydrolase (EPHX1 His139Arg, termed the “fast” allele based on its presumed effect to increase enzyme activity [33]) was significant only in the case-control study.
The results of our study and the publications listed in Table 1 highlight that many COPD genetic associations have not been consistently replicated across all studies. Replication failure is a problem throughout complex trait genetics and is not unique to COPD (34, 35). Multiple factors are likely to explain the inconsistent replication in COPD genetic association studies (36). False-negative results may be the consequence of genotyping error or inadequately powered sample sizes. Spurious associations may result from genotyping error, multiple testing, or population stratification, which can arise from differences in allele frequency between cases and controls due to ethnic diversity and not true disease association. True genetic differences between study populations, termed genetic heterogeneity, may also lead to replication failure, particularly when comparing studies performed in different countries. Variation in case definition or in the phenotypes analyzed across studies is likely to be an important cause of nonreplication. This phenotypic variation may be particularly relevant for studies of COPD, a heterogeneous disease that includes components of emphysema and airway disease, often occurring in variable combinations in any given patient.
Two potential COPD susceptibility genes have been identified in the chromosomal regions found by the linkage analyses described previously. Transforming growth factor (TGF)-β1 is a widely expressed cytokine that has potential roles in airway disease and interstitial lung disease (37). The TGFβ1 gene is located on chromosome 19q, a region linked to COPD-related traits in the Boston Early-Onset COPD Study. Celedón and colleagues genotyped additional short tandem repeat markers on chromosome 19q, which led to increased evidence of linkage, especially in a stratified analysis limited to current and former smokers (3). Analysis of five SNP markers in TGFβ1 found that three SNPs, including one in the promoter (rs2241712), were significantly associated with FEV1 in the Boston Early-Onset COPD Study families. The association with the promoter SNP was replicated in the study comparing NETT cases with control smokers without airflow obstruction. A coding SNP (Leu10Pro, which may lead to higher circulating TGF-β1 levels [38]) and an additional promoter SNP were significant in the case-control study. Analysis of unlinked SNPs did not show evidence of population stratification between the cases and control subjects (39).
In a case-control study from New Zealand, the Leu10Pro-coding SNP in TGFβ1 was associated with COPD (40). A general population study from the Netherlands found significant COPD associations with three TGFβ1 SNPs, including the promoter SNP in the study by Celedón and colleagues and the Leu10Pro-coding SNP (41). The association between TGFβ1 SNPs and COPD has not been consistently replicated in other studies (42, 43).
The SERPINE2 gene is located on chromosome 2q in a region linked to FEV1/FVC ratio in the Boston Early-Onset COPD Study. Based on a microarray experiment (44), DeMeo and colleagues reported that this gene is highly expressed during mouse lung development (4). Genes important in lung development may predispose to emphysema through effects on airspace size or injury repair. SERPINE2 expression was associated with measures of pulmonary function in a microarray study of emphysematous lung tissue from patients with LVRS (45). The mouse and human gene expression results were integrated with human genetic association data, initially by genotyping SERPINE2 SNPs in members of the Boston Early-Onset COPD Study families. Significant associations with multiple SNPs were confirmed in an analysis of the NETT COPD cases and community control subjects, identifying a risk haplotype. The potential role of this novel COPD gene in disease pathogenesis has yet to be determined; however, this study demonstrates the power of integrating gene expression and genotype data as well as human and murine studies. Animal models are an important tool in COPD genetics research and have been reviewed elsewhere (46, 47).
In a multicenter, family-based study of COPD in North America and Europe, Zhu and colleagues genotyped 25 SNPs in SERPINE2, finding six SNPs to be significantly associated with COPD and spirometric measures of lung function (48). Five of the six SNPs were associated with airflow obstruction among COPD cases in a large case-control population from Norway. Three of the five replicated SNPs overlapped with the SNPs found to be significantly associated by DeMeo and colleagues (4)
The association of SERPINE2 SNPs with COPD was not replicated in a case-control study from the United Kingdom (49). These cases had a broader range of airflow limitation than the severely affected cases from the Boston Early-Onset COPD Study, NETT, and the family-based sample in the study by Zhu and colleagues (48). This difference in phenotype is one potential explanation for the discordant results of the SERPINE2 association studies.
GENETIC STUDIES OF QUANTITATIVE COPD PHENOTYPES
Phenotypic differences between subjects in COPD genetic association studies extend beyond the SERPINE2 example and are likely an important cause of nonreplication in the field. In published genetics studies, researchers have defined COPD based on clinical diagnosis (50), baseline pulmonary function (25), lung function decline (51), emphysema (52, 53), or chronic bronchitis (54, 55). Although some genetic mechanisms may be common to this entire range of COPD phenotypes, other genes may be more relevant to a specific COPD subtype. The analysis of quantitative COPD-related traits as intermediate phenotypes may aid in genetic studies by reducing phenotypic heterogeneity. Several researchers have analyzed spirometric measures as quantitative traits (3, 4, 56); however, reduced lung function may be the final consequence of multiple pathophysiologic processes.
Exercise capacity and symptoms of dyspnea are important outcomes for patients with emphysema, and exercise capacity has been shown to be a predictor of response to LVRS (57). These traits often show wide variability between patients with similar levels of lung function. To search for a genetic contribution to this phenotypic variation, we genotyped DNA polymorphisms in 22 candidate genes in 304 NETT subjects (56). By randomly dividing the population into a test set and a replication set to guard against false-positive results, we identified variants in four genes—EPHX1, SFTPB, TGFβ1, and latent TGFβ binding protein-4—that were associated with exercise capacity (measured using cycle ergometry and 6-minute walk test distance), dyspnea symptoms, BODE (body mass index, airflow obstruction, dyspnea, exercise capacity) score (58), and carbon monoxide diffusing capacity. The association between a promoter SNP in TGFβ1 and dyspnea symptoms was replicated in the Boston Early-Onset COPD Study families.
In severe α1-antritrypsin deficiency, emphysema distribution has been classically described as basilar predominant, although apical-predominant emphysema is sometimes observed (59). It is possible that genetic factors influence emphysema distribution in individuals without severe AAT deficiency. In addition to the subjective assessment of emphysema severity and distribution by radiologists, high-resolution chest CT (HRCT) scans can allow for quantitative densitometric measurements of emphysema severity and distribution (60). HRCT-derived intermediate phenotypes are starting to be used in COPD genetics studies. Ito and colleagues genotyped a SNP in the promoter region of matrix metallopeptidase-9 in 84 Japanese patients with COPD and 85 control smokers (61). Although these researchers found no association with COPD susceptibility, patients carrying the T allele had a predilection for upper-lobe–predominant emphysema as determined by quantitative HRCT analysis.
In NETT, upper-lobe–predominant emphysema distribution emerged as an important predictor of response to LVRS (57). DeMeo and colleagues examined variants in 22 candidate genes in 282 subjects in the NETT Genetics Ancillary Study with available CT scan data (62). Variants in two genes—glutathione S-transferase pi (GSTP1) and EPHX1—showed significant association with emphysema distribution based on computerized densitometry analysis (−950 Hounsfield units) and radiologist scoring of emphysema. Coding variants in both genes (GSTP1 Ile105Val and EPHX1 His139Arg) were associated with upper-lobe–predominant emphysema. In an analysis comparing the subset of 171 NETT subjects with upper-lobe–predominant emphysema to Normative Aging Study control subjects, the EPHX1 His139Arg SNP was significantly associated with COPD susceptibility, despite the reduced sample size.
RARE VARIANTS IN COPD GENETICS
Another avenue into understanding COPD genetics is to leverage the lessons learned from other monogenic disorders, besides AAT deficiency, that include emphysema as part of their clinical phenotypes. One such disorder is autosomal dominant cutis laxa (63), which can result from mutations in the gene encoding elastin (64, 65), an important component of the lung extracellular matrix. Sequencing of the six terminal exons of elastin in 116 Boston Early-Onset COPD Study probands led to the discovery of a novel coding variant in one subject (66). In this proband's extended pedigree, all adult carriers of the mutation had airflow obstruction. This variant was found in 1.25% of 318 NETT participants and in 0.55% of control smokers, although this difference was not statistically significant. Cellular studies demonstrated a functional effect of this variant on the elastin protein, related to elastic fiber assembly, susceptibility to proteolysis, and decreased interaction with cellular receptors. Besides cutis laxa, emphysema has been infrequently associated with other connective tissues disorders, including Ehlers-Danlos syndrome and Marfan syndrome (46).
SUMMARY AND FUTURE DIRECTIONS
The use of precise intermediate phenotypes has tremendous potential to aid in the discovery and confirmation of genetic susceptibility factors for COPD and its subtypes. Quantitative HRCT measurements of emphysema severity and distribution are starting to be applied to human genetics studies, and the use of these measurements can be expected to increase in the future. High-throughput analysis of airway disease on HRCT has lagged behind quantification of emphysema severity and distribution but may become a reality for large-scale genetic epidemiology studies. Besides serving as quantitative traits for COPD genetics studies, these HRCT measurements may have direct clinical relevance because emphysema distribution is a major determinant of a patient's eligibility for LVRS. Predisposition to exacerbations and response to therapies for COPD—including supplemental oxygen, LVRS (67), and inhaled and systemic medications—are important outcomes for patients, outcomes likely to have genetic contributions. Future studies of well characterized populations of patients with COPD, incorporating prospective collection of DNA samples in COPD clinical trials, are necessary to find these pharmacogenetic mechanisms.
Most genetic association studies in COPD have focused on a single gene or a limited set of genes. The International HapMap project has characterized variation at millions of SNPs throughout the genome in individuals of European-American, African, and Asian descent (68). These publicly available data (www.hapmap.org), coupled with advances in high-throughput genotyping technologies, have made feasible the simultaneous analysis of hundreds of thousands of SNPs to capture variation throughout the genome. These genome-wide association studies have the potential to uncover novel genetic associations for COPD and related phenotypes and to confirm and expand previous candidate gene results. Although genome-wide association studies have not been performed in COPD, this technique has been successfully applied to identify genes for other complex diseases with broad public health impact, including age-related macular degeneration (69), obesity (70), and type 2 diabetes (71).
Most of the COPD genetics studies discussed in this article have been performed in individuals of European or Asian descent, with occasional studies performed in other countries, such as Egypt (72). Two studies have demonstrated significant familial aggregation of pulmonary function measurements in African Americans, in sibships from Maryland (73), and in twins from North Carolina (74). However, genetic linkage and association studies for COPD or lung function have not been reported in African Americans. One study performed in Mexicans has demonstrated association with COPD susceptibility for variants in the genes encoding several surfactant proteins (75); no other studies in Hispanics have been reported. Given the increasing recognition of COPD morbidity and mortality in African American (76) and Hispanic populations (77), additional COPD genetics studies in ethnic minorities are clearly warranted.
In this article, we have focused on variation in an individual's DNA as it relates to COPD susceptibility and related traits. However, studies of gene expression profiling, using RNA or cDNA microarrays, are another avenue of active investigation in COPD and emphysema (45, 78, 79). Gene expression profiling was used to aid in the identification of SERPINE2 as a COPD susceptibility gene (4). Further studies that integrate gene expression and genetic variation, in addition to the various study designs described in this article, are likely to uncover additional genes important in COPD pathogenesis, with the hope of leading to improved diagnosis, monitoring, and treatment of this common chronic illness.
The National Emphysema Treatment Trial (NETT) is supported by contracts with the National Heart, Lung, and Blood Institute (N01HR76101, N01HR76102, N01HR76103, N01HR76104, N01HR76105, N01HR76106, N01HR76107, N01HR76108, N01HR76109, N01HR76110, N01HR76111, N01HR76112, N01HR76113, N01HR76114, N01HR76115, N01HR76116, N01HR76118, and N01HR76119), the Centers for Medicare and Medicaid Services (CMS), and the Agency for Healthcare Research and Quality (AHRQ).
Supported by NIH grants HL080242, HL072918, HL071393, HL075478, HL068926, U01HL065899, and P01HL083069 and by a grant from the Alpha-1 Foundation.
Conflict of Interest Statement: C.P.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. D.L.D. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. E.K.S. has received grant support, consulting fees, and honoraria from GlaxoSmithKline for studies of COPD genetics and honoraria from Wyeth, Bayer, and AstraZeneca for lectures on COPD genetics.
References
- 1.Knudson RJ. James Jackson, Jr., the young pulmonologist who described familial emphysema: an historical footnote. Chest 1985;87:673–676. [DOI] [PubMed] [Google Scholar]
- 2.Laurell CB, Eriksson S. The electrophoretic alpha1-globulin pattern of serum in alpha1-antitrypsin deficiency. Scand J Clin Lab Invest 1963;15:132–140. [Google Scholar]
- 3.Celedón JC, Lange C, Raby BA, Litonjua AA, Palmer LJ, DeMeo DL, Reilly JJ, Kwiatkowski DJ, Chapman HA, Laird N, et al. The transforming growth factor-β1 (TGFB1) gene is associated with chronic obstructive pulmonary disease (COPD). Hum Mol Genet 2004;13:1649–1656. [DOI] [PubMed] [Google Scholar]
- 4.DeMeo DL, Mariani TJ, Lange C, Srisuma S, Litonjua AA, Celedón JC, Lake SL, Reilly JJ, Chapman HA, Mecham BH, et al. The SERPINE2 gene is associated with chronic obstructive pulmonary disease. Am J Hum Genet 2006;78:253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferris BG. Epidemiology Standardization Project (American Thoracic Society). Am Rev Respir Dis 1978;118:1–120. [PubMed] [Google Scholar]
- 6.Hunter DJ. Gene-environment interactions in human diseases. Nat Rev Genet 2005;6:287–298. [DOI] [PubMed] [Google Scholar]
- 7.Petrache I, Fijalkowska I, Zhen L, Medler TR, Brown E, Cruz P, Choe KH, Taraseviciene-Stewart L, Scerbavicius R, Shapiro L, et al. A novel antiapoptotic role for alpha1-antitrypsin in the prevention of pulmonary emphysema. Am J Respir Crit Care Med 2006;173:1222–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lomas DA. The selective advantage of alpha1-antitrypsin deficiency. Am J Respir Crit Care Med 2006;173:1072–1077. [DOI] [PubMed] [Google Scholar]
- 9.Lomas DA, Evans DL, Finch JT, Carrell RW. The mechanism of Z alpha 1-antitrypsin accumulation in the liver. Nature 1992;357:605–607. [DOI] [PubMed] [Google Scholar]
- 10.Lomas DA, Parfrey H. Alpha1-antitrypsin deficiency. 4: molecular pathophysiology. Thorax 2004;59:529–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahl M, Hersh CP, Ly NP, Berkey CS, Silverman EK, Nordestgaard BG. The protease inhibitor PI*S allele and COPD: a meta-analysis. Eur Respir J 2005;26:67–76. [DOI] [PubMed] [Google Scholar]
- 12.Lee JH, Brantly M. Molecular mechanisms of alpha1-antitrypsin null alleles. Respir Med 2000;94:S7–S11. [DOI] [PubMed] [Google Scholar]
- 13.DeMeo DL, Silverman EK. Alpha1-antitrypsin deficiency. 2: genetic aspects of alpha(1)-antitrypsin deficiency: phenotypes and genetic modifiers of emphysema risk. Thorax 2004;59:259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lieberman J, Winter B, Sastre A. Alpha 1-antitrypsin Pi-types in 965 COPD patients. Chest 1986;89:370–373. [DOI] [PubMed] [Google Scholar]
- 15.Hersh CP, Dahl M, Ly NP, Berkey CS, Nordestgaard BG, Silverman EK. Chronic obstructive pulmonary disease in {alpha}1-antitrypsin PI MZ heterozygotes: a meta-analysis. Thorax 2004;59:843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dirksen A, Dijkman JH, Madsen F, Stoel B, Hutchison DC, Ulrik CS, Skovgaard LT, Kok-Jensen A, Rudolphus A, Seersholm N, et al. A randomized clinical trial of alpha(1)-antitrypsin augmentation therapy. Am J Respir Crit Care Med 1999;160:1468–1472. [DOI] [PubMed] [Google Scholar]
- 17.Seersholm N, Wencker M, Banik N, Viskum K, Dirksen A, Kok-Jensen A, Konietzko N; Wissenschaftliche Arbeitsgemeinschaft zur Therapie von Lungenerkrankungen (WATL) α1-AT study group. Does α1-antitrypsin augmentation therapy slow the annual decline in FEV1 in patients with severe hereditary α1-antitrypsin deficiency? Eur Respir J 1997;10:2260–2263. [DOI] [PubMed] [Google Scholar]
- 18.Survival and FEV1 decline in individuals with severe deficiency of α1-antitrypsin. The Alpha-1-Antitrypsin Deficiency Registry Study Group. Am J Respir Crit Care Med 1998;158:49–59. [DOI] [PubMed] [Google Scholar]
- 19.American Thoracic Society/European Respiratory Society Statement. Standards for the diagnosis and management of individuals with alpha-1 antitrypsin deficiency. Am J Respir Crit Care Med 2003;168:818–900. [DOI] [PubMed] [Google Scholar]
- 20.Cassina PC, Teschler H, Konietzko N, Theegarten D, Stamatis G. Two-year results after lung volume reduction surgery in alpha1-antitrypsin deficiency versus smoker's emphysema. Eur Respir J 1998;12:1028–1032. [DOI] [PubMed] [Google Scholar]
- 21.Gelb AF, McKenna RJ, Brenner M, Fischel R, Zamel N. Lung function after bilateral lower lobe lung volume reduction surgery for alpha1-antitrypsin emphysema. Eur Respir J 1999;14:928–933. [DOI] [PubMed] [Google Scholar]
- 22.Tutic M, Bloch KE, Lardinois D, Brack T, Russi EW, Weder W. Long-term results after lung volume reduction surgery in patients with alpha1-antitrypsin deficiency. J Thorac Cardiovasc Surg 2004;128:408–413. [DOI] [PubMed] [Google Scholar]
- 23.Stoller JK, Gildea TR, Ries AL, Meli YM, Karafa MT. Lung volume reduction surgery in patients with emphysema and alpha-1 antitrypsin deficiency. Ann Thorac Surg 2007;83:241–251. [DOI] [PubMed] [Google Scholar]
- 24.Silverman EK, Mosley JD, Palmer LJ, Barth M, Senter JM, Brown A, Drazen JM, Kwiatkowski DJ, Chapman HA, Campbell EJ, et al. Genome-wide linkage analysis of severe, early-onset chronic obstructive pulmonary disease: airflow obstruction and chronic bronchitis phenotypes. Hum Mol Genet 2002;11:623–632. [DOI] [PubMed] [Google Scholar]
- 25.Silverman EK, Palmer LJ, Mosley JD, Barth M, Senter JM, Brown A, Drazen JM, Kwiatkowski DJ, Chapman HA, Campbell EJ, et al. Genomewide linkage analysis of quantitative spirometric phenotypes in severe early-onset chronic obstructive pulmonary disease. Am J Hum Genet 2002;70:1229–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmer LJ, Celedon JC, Chapman HA, Speizer FE, Weiss ST, Silverman EK. Genome-wide linkage analysis of bronchodilator responsiveness and post-bronchodilator spirometric phenotypes in chronic obstructive pulmonary disease. Hum Mol Genet 2003;12:1199–1210. [DOI] [PubMed] [Google Scholar]
- 27.DeMeo DL, Celedon JC, Lange C, Reilly JJ, Chapman HA, Sylvia JS, Speizer FE, Weiss ST, Silverman EK. Genome-wide linkage of forced mid-expiratory flow in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2004;170:1294–1301. [DOI] [PubMed] [Google Scholar]
- 28.Silverman EK, Chapman HA, Drazen JM, Weiss ST, Rosner B, Campbell EJ, O'Donnell WJ, Reilly JJ, Ginns L, Mentzer S, et al. Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease: risk to relatives for airflow obstruction and chronic bronchitis. Am J Respir Crit Care Med 1998;157:1770–1778. [DOI] [PubMed] [Google Scholar]
- 29.Hersh CP, DeMeo DL, Raby BA, Litonjua AA, Sylvia JS, Sparrow D, Reilly JJ, Silverman EK. Genetic linkage and association analysis of COPD-related traits on chromosome 8p. COPD 2006;3:189–194. [DOI] [PubMed] [Google Scholar]
- 30.Malhotra A, Peiffer AP, Ryujin DT, Elsner T, Kanner RE, Leppert MF, Hasstedt SJ. Further evidence for the role of genes on chromosome 2 and chromosome 5 in the inheritance of pulmonary function. Am J Respir Crit Care Med 2003;168:556–561. [DOI] [PubMed] [Google Scholar]
- 31.Hersh CP, DeMeo DL, Lange C, Litonjua AA, Reilly JJ, Kwiatkowski D, Laird N, Sylvia JS, Sparrow D, Speizer FE, et al. Attempted replication of reported chronic obstructive pulmonary disease candidate gene associations. Am J Respir Cell Mol Biol 2005;33:71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bell B, Rose CL, Damon H. The Normative Aging Study: an interdisciplinary and longitudinal study of health and aging. Aging Hum Dev 1972;3:5–17. [Google Scholar]
- 33.Hassett C, Aicher L, Sidhu JS, Omiecinski CJ. Human microsomal epoxide hydrolase: genetic polymorphism and functional expression in vitro of amino acid variants. Hum Mol Genet 1994;3:421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirschhorn JN, Altshuler D. Once and again-issues surrounding replication in genetic association studies. J Clin Endocrinol Metab 2002;87:4438–4441. [DOI] [PubMed] [Google Scholar]
- 35.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet 2001;29:306–309. [DOI] [PubMed] [Google Scholar]
- 36.Colhoun HM, McKeigue PM, Davey Smith G. Problems of reporting genetic associations with complex outcomes. Lancet 2003;361:865–872. [DOI] [PubMed] [Google Scholar]
- 37.Bartram U, Speer CP. The role of transforming growth factor beta in lung development and disease. Chest 2004;125:754–765. [DOI] [PubMed] [Google Scholar]
- 38.Yokota M, Ichihara S, Lin TL, Nakashima N, Yamada Y. Association of a T29→C polymorphism of the transforming growth factor-beta1 gene with genetic susceptibility to myocardial infarction in Japanese. Circulation 2000;101:2783–2787. [DOI] [PubMed] [Google Scholar]
- 39.Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet 1999;65:220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu L, Chau J, Young RP, Pokorny V, Mills GD, Hopkins R, McLean L, Black PN. Transforming growth factor-beta1 genotype and susceptibility to chronic obstructive pulmonary disease. Thorax 2004;59:126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Diemen CC, Postma DS, Vonk JM, Bruinenberg M, Nolte IM, Boezen HM. Decorin and TGF-beta1 polymorphisms and development of COPD in a general population. Respir Res 2006;7:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoon HI, Silverman EK, Lee HW, Yoo CG, Lee CT, Chung HS, Kim YW, Han SK, Shim YS, Yim JJ. Lack of association between COPD and transforming growth factor-beta1 (TGFB1) genetic polymorphisms in Koreans. Int J Tuberc Lung Dis 2006;10:504–509. [PubMed] [Google Scholar]
- 43.Ogawa E, Ruan J, Connett JE, Anthonisen NR, Pare PD, Sandford AJ. Transforming growth factor-beta1 polymorphisms, airway responsiveness and lung function decline in smokers. Respir Med 2007;101:938–943. [DOI] [PubMed] [Google Scholar]
- 44.Mariani TJ, Reed JJ, Shapiro SD. Expression profiling of the developing mouse lung: insights into the establishment of the extracellular matrix. Am J Respir Cell Mol Biol 2002;26:541–548. [DOI] [PubMed] [Google Scholar]
- 45.Spira A, Beane J, Pinto-Plata V, Kadar A, Liu G, Shah V, Celli B, Brody JS. Gene expression profiling of human lung tissue from smokers with severe emphysema. Am J Respir Cell Mol Biol 2004;31:601–610. [DOI] [PubMed] [Google Scholar]
- 46.Hersh CP, DeMeo DL, Silverman EK. Chronic obstructive pulmonary disease. In: Silverman EK, Shapiro SD, Lomas DA, Weiss ST, editors. Respiratory genetics. New York: Hodder Arnold; 2005. pp. 253–296.
- 47.Shapiro SD. Transgenic and gene-targeted mice as models for chronic obstructive pulmonary disease. Eur Respir J 2007;29:375–378. [DOI] [PubMed] [Google Scholar]
- 48.Zhu G, Warren L, Aponte J, Gulsvik A, Bakke P, The International COPD Genetics Network (ICGN) Investigators, Anderson WH, Lomas DA, Silverman EK, Pillai SG. The SERPINE2 gene is associated with chronic obstructive pulmonary disease in two large populations. Am J Respir Crit Care Med 2007;176:167–173. [DOI] [PubMed] [Google Scholar]
- 49.Chappell S, Daly L, Morgan K, Baranes TG, Roca J, Rabinovich R, Millar A, Donnelly SC, Keatings V, MacNee W, et al. The SERPINE2 gene and chronic obstructive pulmonary disease. Am J Hum Genet 2006;79:184–186 (author reply: 186–187). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seersholm N, Wilcke JTR, Kok-Jensen A, Dirksen A. Risk of hospital admission for obstructive pulmonary disease in α1-antitrypsin heterozygotes of phenotype PiMZ. Am J Respir Crit Care Med 2000;161:81–84. [DOI] [PubMed] [Google Scholar]
- 51.Sandford AJ, Chagani T, Weir TD, Connett JE, Anthonisen NR, Paré PD. Susceptibility genes for rapid decline of lung function in the lung health study. Am J Respir Crit Care Med 2001;163:469–473. [DOI] [PubMed] [Google Scholar]
- 52.Minematsu N, Nakamura H, Tateno H, Nakajima T, Yamaguchi K. Genetic polymorphism in matrix metalloproteinase-9 and pulmonary emphysema. Biochem Biophys Res Commun 2001;289:116–119. [DOI] [PubMed] [Google Scholar]
- 53.Yamada N, Yamaya M, Okinaga S, Nakayama K, Sekizawa K, Shibahara S, Sasaki H. Microsatellite polymorphism in the heme oxygenase-1 gene promoter is associated with susceptibility to emphysema. Am J Hum Genet 2000;66:187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baranova H, Perriot J, Albuisson E, Ivaschenko T, Baranov VS, Hemery B, Mouraire P, Riol N, Malet P. Peculiarities of the GSTM1 0/0 genotype in French heavy smokers with various types of chronic bronchitis. Hum Genet 1997;99:822–826. [DOI] [PubMed] [Google Scholar]
- 55.Huang S-L, Su C-H, Chang S-C. Tumor necrosis factor-α gene polymorphism in chronic bronchitis. Am J Respir Crit Care Med 1997;156:1436–1439. [DOI] [PubMed] [Google Scholar]
- 56.Hersh CP, DeMeo DL, Lazarus R, Celedón JC, Raby BA, Benditt JO, Criner G, Make B, Martinez FJ, Scanlon PD, et al. Genetic association analysis of functional impairment in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2006;173:977–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fishman A, Martinez F, Naunheim K, Piantadosi S, Wise R, Ries A, Weinmann G, Wood DE. A randomized trial comparing lung-volume-reduction surgery with medical therapy for severe emphysema. N Engl J Med 2003;348:2059–2073. [DOI] [PubMed] [Google Scholar]
- 58.Celli BR, Cote CG, Marin JM, Casanova C, Montes de Oca M, Mendez RA, Pinto Plata V, Cabral HJ. The body-mass index, airflow obstruction, dyspnea, and exercise capacity index in chronic obstructive pulmonary disease. N Engl J Med 2004;350:1005–1012. [DOI] [PubMed] [Google Scholar]
- 59.Parr DG, Stoel BC, Stolk J, Stockley RA. Pattern of emphysema distribution in α1-antitrypsin deficiency influences lung function impairment. Am J Respir Crit Care Med 2004;170:1172–1178. [DOI] [PubMed] [Google Scholar]
- 60.Muller NL, Coxson H. Chronic obstructive pulmonary disease. 4: imaging the lungs in patients with chronic obstructive pulmonary disease. Thorax 2002;57:982–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ito I, Nagai S, Handa T, Muro S, Hirai T, Tsukino M, Mishima M. Matrix metalloproteinase-9 promoter polymorphism associated with upper lung dominant emphysema. Am J Respir Crit Care Med 2005;172:1378–1382. [DOI] [PubMed] [Google Scholar]
- 62.Demeo DL, Hersh CP, Hoffman EA, Litonjua AA, Lazarus R, Sparrow D, Benditt JO, Criner G, Make B, Martinez FJ, et al. Genetic determinants of emphysema distribution in the National Emphysema Treatment Trial. Am J Respir Crit Care Med 2007;176:42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Corbett E, Glaisyer H, Chan C, Madden B, Khaghani A, Yacoub M. Congenital cutis laxa with a dominant inheritance and early onset emphysema. Thorax 1994;49:836–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tassabehji M, Metcalfe K, Hurst J, Ashcroft GS, Kielty C, Wilmot C, Donnai D, Read AP, Jones CJ. An elastin gene mutation producing abnormal tropoelastin and abnormal elastic fibres in a patient with autosomal dominant cutis laxa. Hum Mol Genet 1998;7:1021–1028. [DOI] [PubMed] [Google Scholar]
- 65.Zhang MC, He L, Giro M, Yong SL, Tiller GE, Davidson JM. Cutis laxa arising from frameshift mutations in exon 30 of the elastin gene (ELN). J Biol Chem 1999;274:981–986. [DOI] [PubMed] [Google Scholar]
- 66.Kelleher CM, Silverman EK, Broekelmann T, Litonjua AA, Hernandez M, Sylvia JS, Stoler J, Reilly JJ, Chapman HA, Speizer FE, et al. A functional mutation in the terminal exon of elastin in severe, early-onset chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol 2005;33:355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hersh CP, DeMeo DL, Reilly JJ, Silverman EK. Xenobiotic metabolizing enzyme gene polymorphisms predict response to lung volume reduction surgery. Respir Res 2007;8:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.The International HapMap Consortium. The International HapMap Project. Nature 2003;426:789–796. [DOI] [PubMed] [Google Scholar]
- 69.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, et al. Complement factor H polymorphism in age-related macular degeneration. Science 2005;308:385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Herbert A, Gerry NP, McQueen MB, Heid IM, Pfeufer A, Illig T, Wichmann HE, Meitinger T, Hunter D, Hu FB, et al. A common genetic variant is associated with adult and childhood obesity. Science 2006;312:279–283. [DOI] [PubMed] [Google Scholar]
- 71.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007;445:881–885. [DOI] [PubMed] [Google Scholar]
- 72.Hegab AE, Sakamoto T, Saitoh W, Massoud HH, Massoud HM, Hassanein KM, Sekizawa K. Polymorphisms of IL4, IL13, and ADRB2 genes in COPD. Chest 2004;126:1832–1839. [DOI] [PubMed] [Google Scholar]
- 73.Cotch MF, Beaty TH, Munoz A, Cohen BH. Estimating familial aggregation while adjusting for covariates: application to pulmonary function data from black and white sibships. Ann Epidemiol 1992;2:317–324. [DOI] [PubMed] [Google Scholar]
- 74.Whitfield KE, Wiggins SA, Belue R, Brandon DT. Genetic and environmental influences on forced expiratory volume in African Americans: the Carolina African-American Twin Study of Aging. Ethn Dis 2004;14:206–211. [PubMed] [Google Scholar]
- 75.Guo X, Lin HM, Lin Z, Montano M, Sansores R, Wang G, DiAngelo S, Pardo A, Selman M, Floros J. Surfactant protein gene A, B, and D marker alleles in chronic obstructive pulmonary disease of a Mexican population. Eur Respir J 2001;18:482–490. [DOI] [PubMed] [Google Scholar]
- 76.National Heart, Lung, and Blood Institute. Morbidity & mortality: 2002. chart book on cardiovascular, lung, and blood diseases. Bethesda (MD): National Institutes of Health; 2002.
- 77.Adams SG, Anzueto A, Pugh JA, Lee S, Hazuda HP. Mexican American elders have similar severities of COPD despite less tobacco exposure than European American elders. Respir Med 2006;100:1966–1972. [DOI] [PubMed] [Google Scholar]
- 78.Ning W, Li CJ, Kaminski N, Feghali-Bostwick CA, Alber SM, Di YP, Otterbein SL, Song R, Hayashi S, Zhou Z, et al. Comprehensive gene expression profiles reveal pathways related to the pathogenesis of chronic obstructive pulmonary disease. Proc Natl Acad Sci USA 2004;101:14895–14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Golpon HA, Coldren CD, Zamora MR, Cosgrove GP, Moore MD, Tuder RM, Geraci MW, Voelkel NF. Emphysema lung tissue gene expression profiling. Am J Respir Cell Mol Biol 2004;31:595–600. [DOI] [PubMed] [Google Scholar]
- 80.Poller W, Faber JP, Scholz S, Weidinger S, Bartholome K, Olek K, Eriksson S. Mis-sense mutation of alpha 1-antichymotrypsin gene associated with chronic lung disease. Lancet 1992;339:1538. [DOI] [PubMed] [Google Scholar]
- 81.Poller W, Faber JP, Weidinger S, Tief K, Scholz S, Fischer M, Olek K, Kirchgesser M, Heidtmann HH. A leucine-to-proline substitution causes a defective alpha 1-antichymotrypsin allele associated with familial obstructive lung disease. Genomics 1993;17:740–743. [DOI] [PubMed] [Google Scholar]
- 82.Ishii T, Matsuse T, Teramoto S, Matsui H, Hosoi T, Fukuchi Y, Ouchi Y. Association between alpha-1-antichymotrypsin polymorphism and susceptibility to chronic obstructive pulmonary disease. Eur J Clin Invest 2000;30:543–548. [DOI] [PubMed] [Google Scholar]
- 83.Sandford AJ, Chagani T, Weir TD, Pare PD. Alpha 1-antichymotrypsin mutations in patients with chronic obstructive pulmonary disease. Dis Markers 1998;13:257–260. [DOI] [PubMed] [Google Scholar]
- 84.Benetazzo MG, Gile LS, Bombieri C, Malerba G, Massobrio M, Pignatti PF, Luisetti M. alpha 1-antitrypsin TAQ I polymorphism and alpha 1-antichymotrypsin mutations in patients with obstructive pulmonary disease. Respir Med 1999;93:648–654. [DOI] [PubMed] [Google Scholar]
- 85.Ho L-I, Harn H-J, Chen C-J, Tsai N-M. Polymorphism of the β2-adrenoceptor in COPD in Chinese subjects. Chest 2001;120:1493–1499. [DOI] [PubMed] [Google Scholar]
- 86.Joos L, Weir TD, Connett JE, Anthonisen NR, Woods R, Pare PD, Sandford AJ. Polymorphisms in the beta2 adrenergic receptor and bronchodilator response, bronchial hyperresponsiveness, and rate of decline in lung function in smokers. Thorax 2003;58:703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brogger J, Steen VM, Eiken HG, Gulsvik A, Bakke P. Genetic association between COPD and polymorphisms in TNF, ADRB2 and EPHX1. Eur Respir J 2006;27:682–688. [DOI] [PubMed] [Google Scholar]
- 88.Matsushita I, Hasegawa K, Nakata K, Yasuda K, Tokunaga K, Keicho N. Genetic variants of human beta-defensin-1 and chronic obstructive pulmonary disease. Biochem Biophys Res Commun 2002;291:17–22. [DOI] [PubMed] [Google Scholar]
- 89.Hu RC, Xu YJ, Zhang ZX, Ni W, Chen SX. Correlation of HDEFB1 polymorphism and susceptibility to chronic obstructive pulmonary disease in Chinese Han population. Chin Med J (Engl) 2004;117:1637–1641. [PubMed] [Google Scholar]
- 90.Wallace AM, He JQ, Burkett KM, Ruan J, Connett JE, Anthonisen NR, Pare PD, Sandford AJ. Contribution of alpha- and beta-defensins to lung function decline and infection in smokers: an association study. Respir Res 2006;7:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Smith CA, Harrison DJ. Association between polymorphism in gene for microsomal epoxide hydrolase and susceptibility to emphysema. Lancet 1997;350:630–633. [DOI] [PubMed] [Google Scholar]
- 92.Xiao D, Wang C, Du MJ, Pang BS, Zhang HY, Xiao B, Liu JZ, Weng XZ, Su L, Christiani DC. Relationship between polymorphisms of genes encoding microsomal epoxide hydrolase and glutathione S-transferase P1 and chronic obstructive pulmonary disease. Chin Med J (Engl) 2004;117:661–667. [PubMed] [Google Scholar]
- 93.Yoshikawa M, Hiyama K, Ishioka S, Maeda H, Maeda A, Yamakido M. Microsomal epoxide hydrolase genotypes and chronic obstructive pulmonary disease in Japanese. Int J Mol Med 2000;5:49–53. [DOI] [PubMed] [Google Scholar]
- 94.Takeyabu K, Yamaguchi E, Suzuki I, Nishimura M, Hizawa N, Kamakami Y. Gene polymorphism for microsomal epoxide hydrolase and susceptibility to emphysema in a Japanese population. Eur Respir J 2000;15:891–894. [DOI] [PubMed] [Google Scholar]
- 95.Yim JJ, Park GY, Lee CT, Kim YW, Han SK, Shim YS, Yoo CG. Genetic susceptibility to chronic obstructive pulmonary disease in Koreans: combined analysis of polymorphic genotypes for microsomal epoxide hydrolase and glutathione S-transferase M1 and T1. Thorax 2000;55:121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Horne SL, Cockcroft DW, Dosman JA. Possible protective effect against chronic obstructive airways disease by the GC2 allele. Hum Hered 1990;40:173–176. [DOI] [PubMed] [Google Scholar]
- 97.Schellenberg D, Paré PD, Weir TD, Spinelli JJ, Walker BAM, Sandford AJ. Vitamin D binding protein variants and the risk of COPD. Am J Respir Crit Care Med 1998;157:957–961. [DOI] [PubMed] [Google Scholar]
- 98.Ishii T, Keicho N, Teramoto S, Azuma A, Kudoh S, Fukuchi Y, Ouchi Y, Matsuse T. Association of Gc-globulin variation with susceptibility to COPD and diffuse panbronchiolitis. Eur Respir J 2001;18:753–757. [DOI] [PubMed] [Google Scholar]
- 99.Ito I, Nagai S, Hoshino Y, Muro S, Hirai T, Tsukino M, Mishima M. Risk and severity of COPD is associated with the group-specific component of serum globulin 1F allele. Chest 2004;125:63–70. [DOI] [PubMed] [Google Scholar]
- 100.Kauffmann F, Kleisbauer JP, Cambon-De-Mouzon A, Mercier P, Constans J, Blanc M, Rouch Y, Feingold N. Genetic markers in chronic air-flow limitation: a genetic epidemiologic study. Am Rev Respir Dis 1983;127:263–269. [DOI] [PubMed] [Google Scholar]
- 101.Kasuga I, Pare PD, Ruan J, Connett JE, Anthonisen NR, Sandford AJ. Lack of association of group specific component haplotypes with lung function in smokers. Thorax 2003;58:790–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Harrison DJ, Cantlay AM, Rae F, Lamb D, Smith CA. Frequency of glutathione S-transferase M1 deletion in smokers with emphysema and lung cancer. Hum Exp Toxicol 1997;16:356–360. [DOI] [PubMed] [Google Scholar]
- 103.Cheng SL, Yu CJ, Chen CJ, Yang PC. Genetic polymorphism of epoxide hydrolase and glutathione S-transferase in COPD. Eur Respir J 2004;23:818–824. [DOI] [PubMed] [Google Scholar]
- 104.He J-Q, Ruan J, Connett JE, Anthonisen NR, Paré PD, Sandford AJ. Antioxidant gene polymorphisms and susceptibility to a rapid decline in lung function in smokers. Am J Respir Crit Care Med 2002;166:323–328. [DOI] [PubMed] [Google Scholar]
- 105.Ishii T, Matsuse T, Teramoto S, Matsui H, Miyao M, Hosoi T, Takahashi H, Fukuchi Y, Ouchi Y. Glutathione S-transferase P1 (GSTP1) polymorphism in patients with chronic obstructive pulmonary disease. Thorax 1999;54:693–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yim JJ, Yoo CG, Lee CT, Kim YW, Han SK, Shim YS. Lack of association between glutathione S-transferase P1 polymorphism and COPD in Koreans. Lung 2002;180:119–125. [DOI] [PubMed] [Google Scholar]
- 107.Guenegou A, Leynaert B, Benessiano J, Pin I, Demoly P, Neukirch F, Boczkowski J, Aubier M. Association of lung function decline with the heme oxygenase-1 gene promoter microsatellite polymorphism in a general population sample: results from the European Community Respiratory Health Survey (ECRHS), France. J Med Genet 2006;43:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nakayama K, Kikuchi A, Yasuda H, Ebihara S, Sasaki T, Ebihara T, Yamaya M. Heme oxygenase-1 gene promoter polymorphism and decline in lung function in Japanese men. Thorax 2006;61:921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhou M, Huang SG, Wan HY, Li B, Deng WW, Li M. Genetic polymorphism in matrix metalloproteinase-9 and the susceptibility to chronic obstructive pulmonary disease in Han population of south China. Chin Med J (Engl) 2004;117:1481–1484. [PubMed] [Google Scholar]
- 110.Joos L, He JQ, Shepherdson MB, Connett JE, Anthonisen NR, Pare PD, Sandford AJ. The role of matrix metalloproteinase polymorphisms in the rate of decline in lung function. Hum Mol Genet 2002;11:569–576. [DOI] [PubMed] [Google Scholar]
- 111.Juul K, Tybjaerg-Hansen A, Marklund S, Lange P, Nordestgaard BG. Genetically increased antioxidative protection and decreased chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2006;173:858–864. [DOI] [PubMed] [Google Scholar]
- 112.Young RP, Hopkins R, Black PN, Eddy C, Wu L, Gamble GD, Mills GD, Garrett JE, Eaton TE, Rees MI. Functional variants of antioxidant genes in smokers with COPD and in those with normal lung function. Thorax 2006;61:394–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Seifart C, Plagens A, Brodje D, Muller B, von Wichert P, Floros J. Surfactant protein B intron 4 variation in German patients with COPD and acute respiratory failure. Dis Markers 2002;18:129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Su ZG, Wen FQ, Feng YL, Xiao M, Wu XL. Transforming growth factor-beta1 gene polymorphisms associated with chronic obstructive pulmonary disease in Chinese population. Acta Pharmacol Sin 2005;26:714–720. [PubMed] [Google Scholar]
- 115.Sakao S, Tatsumi K, Igari H, Shino Y, Shirasawa H, Kuriyama T. Association of tumor necrosis factor alpha gene promoter polymorphism with the presence of chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2001;163:420–422. [DOI] [PubMed] [Google Scholar]
- 116.Matheson MC, Ellis JA, Raven J, Walters EH, Abramson MJ. Association of IL8, CXCR2 and TNF-alpha polymorphisms and airway disease. J Hum Genet 2006;51:196–203. [DOI] [PubMed] [Google Scholar]
- 117.Higham MA, Pride NB, Alikhan A, Morrell NW. Tumour necrosis factor-alpha gene promoter polymorphism in chronic obstructive pulmonary disease. Eur Respir J 2000;15:281–284. [DOI] [PubMed] [Google Scholar]
- 118.Ishii T, Matsuse T, Teramoto S, Matsui H, Miyao M, Hosoi T, Takahashi H, Fukuchi Y, Ouchi Y. Neither IL-1beta, IL-1 receptor antagonist, nor TNF-alpha polymorphisms are associated with susceptibility to COPD. Respir Med 2000;94:847–851. [DOI] [PubMed] [Google Scholar]
- 119.Patuzzo C, Gile LS, Zorzetto M, Trabetti E, Malerba G, Pignatti PF, Luisetti M. Tumor necrosis factor gene complex in COPD and disseminated bronchiectasis. Chest 2000;117:1353–1358. [DOI] [PubMed] [Google Scholar]
- 120.Ferrarotti I, Zorzetto M, Beccaria M, Gile LS, Porta R, Ambrosino N, Pignatti PF, Cerveri I, Pozzi E, Luisetti M. Tumour necrosis factor family genes in a phenotype of COPD associated with emphysema. Eur Respir J 2003;21:444–449. [DOI] [PubMed] [Google Scholar]
- 121.Chierakul N, Wongwisutikul P, Vejbaesya S, Chotvilaiwan K. Tumor necrosis factor-alpha gene promoter polymorphism is not associated with smoking-related COPD in Thailand. Respirology 2005;10:36–39. [DOI] [PubMed] [Google Scholar]
- 122.Hegab AE, Sakamoto T, Saitoh W, Nomura A, Ishii Y, Morishima Y, Iizuka T, Kiwamoto T, Matsuno Y, Massoud HH, et al. Polymorphisms of TNFalpha, IL1beta, and IL1RN genes in chronic obstructive pulmonary disease. Biochem Biophys Res Commun 2005;329:1246–1252. [DOI] [PubMed] [Google Scholar]
- 123.Jiang L, He B, Zhao MW, Ning LD, Li XY, Yao WZ. Association of gene polymorphisms of tumour necrosis factor-alpha and interleukin-13 with chronic obstructive pulmonary disease in Han nationality in Beijing. Chin Med J (Engl) 2005;118:541–547. [PubMed] [Google Scholar]
- 124.Seifart C, Plagens A, Dempfle A, Clostermann U, Vogelmeier C, von Wichert P, Seifart U. TNF-alpha, TNF-beta, IL-6, and IL-10 polymorphisms in patients with lung cancer. Dis Markers 2005;21:157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ruse CE, Hill MC, Tobin M, Neale N, Connolly MJ, Parker SG, Wardlaw AJ. Tumour necrosis factor gene complex polymorphisms in chronic obstructive pulmonary disease. Respir Med 2007;101:340–344. [DOI] [PubMed] [Google Scholar]