Abstract
Background
Autism spectrum disorders (ASD) are severe neurodevelopmental disorders with the male:female ratio of 4:1, implying the contribution of X chromosome genetic factors to the susceptibility of ASD. The ribosomal protein L10 (RPL10) gene, located on chromosome Xq28, codes for a key protein in assembling large ribosomal subunit and protein synthesis. Two non-synonymous mutations of RPL10, L206M and H213Q, were identified in four boys with ASD. Moreover, functional studies of mutant RPL10 in yeast exhibited aberrant ribosomal profiles. These results provided a novel aspect of disease mechanisms for autism – aberrant processes of ribosome biosynthesis and translation. To confirm these initial findings, we re-sequenced RPL10 exons and quantified mRNA transcript level of RPL10 in our samples.
Methods
141 individuals with ASD were recruited in this study. All RPL10 exons and flanking junctions were sequenced. Furthermore, mRNA transcript level of RPL10 was quantified in B lymphoblastoid cell lines (BLCL) of 48 patients and 27 controls using the method of SYBR Green quantitative PCR. Two sets of primer pairs were used to quantify the mRNA expression level of RPL10: RPL10-A and RPL10-B.
Results
No non-synonymous mutations were detected in our cohort. Male controls showed similar transcript level of RPL10 compared with female controls (RPL10-A, U = 81, P = 0.7; RPL10-B, U = 61.5, P = 0.2). We did not observe any significant difference in RPL10 transcript levels between cases and controls (RPL10-A, U = 531, P = 0.2; RPL10-B, U = 607.5, P = 0.7).
Conclusion
Our results suggest that RPL10 has no major effect on the susceptibility to ASD.
Background
Autism spectrum disorders (ASD) are complex neurobehavioral disorders characterized by impaired social interaction and language development and by repetitive and stereotyped behaviors and interests. Twin and family studies indicate that genetic factors contribute to the susceptibility to ASD[1,2]. However, only 10–25% of cases of autism harbour identified chromosome abnormalities and/or present with genetic syndromes; the remaining 90% are idiopathic[3]. The male:female ratio of autism is approximately 4:1, implying the contribution of X chromosome genetic factors to the susceptibility of ASD.
Ribosomal protein L10 (RPL10), also called QM, is a highly conserved component of the large ribosome subunit (60s) that plays a crucial role in protein synthesis[4]. In humans, the RPL10 gene is located on chromosome Xq28, within a candidate region for ASD[5]. Recently, Klauck et al. identified two non-synonymous mutations, L206M and H213Q, in the C-terminal domain of RPL10 in four boys with ASD from two independent families. Furthermore, functional studies using yeast strains expressing human mutant RPL10 cDNAs exhibited aberrant ribosomal profiles, suggesting that the mutations may actually have an effect on translation[6]. High expression of RPL10 was observed in mouse hippocampus, an important brain site for learning and memory, which are impaired in autism[7]. To investigate whether RPL10 is involved in the pathogenesis of autism, we sequenced all RPL10 exons and quantified RPL10 mRNA level in patients with ASD and controls.
Methods
Subjects
A total of 141 individuals with ASD were recruited by the Paris Autism Research International Sibpair (PARIS) study. All probands met DSM-IV diagnostic criteria for autism, Asperger syndrome or pervasive developmental disorder not otherwise specified (PDD-NOS)[8]. The patients were evaluated by experienced psychiatrists or child neurologists and assessed with the Autism Diagnostic Interview-Revised (ADI-R) [9] or the Asperger Syndrome Diagnostic Interview (ASDI) [10]. Totally, 129 subjects met full criteria for autistic disorder, 6 for Asperger Syndrome and 6 for PDD-NOS. 109 patients showed low IQ (<70). Laboratory tests to rule out medical causes of autism included standard karyotyping, fragile X testing, and metabolic screening. Brain imaging and EEG were performed when possible. Patients diagnosed with medical disorders such as fragile X syndrome or chromosomal abnormalities were excluded from the study. Of 141 patients, 88 were selected from the families with X chromosome inactivation skewing (XCI) from 289 ASD families based on the previous study[8]. The other 53 patients were from multiplex families with random XCI. There were 101 males and 40 females (78 subjects from multiplex families and 63 sporadic cases). All individuals were of European descent, except 5 sub-Saharan Africans, and 3 of mixed ethnicity. The study was approved by the research ethics boards of the collaborating institutions. Informed consent was obtained from all families participating in the study.
Sequencing
All RPL10 exons and flanking junctions were amplified using the primers described previously[6]. Amplicons were directly sequenced using the Big Dye version 3.1 in ABI 3100 sequencers (Applied Biosystems, Foster City, CA)
Real-Time Quantitative PCR
A sample of 48 patients (34 males and 14 females, of which 35 patients IQ<70) and 27 controls (15 males and 12 females, all IQ>90) were available for B lymphoblastoid cell lines (BLCL). Total RNA was isolated from BLCL using the NucleoSpin® RNA II kit. Oligo(dT)-primed cDNA prepared from 5 μg of BLCL RNA using Superscript II (Invitrogen, Carlsbad, CA) was used as template for quantitative PCR with SYBR Green on an ABI PRISM 7500 instrument (Applied Biosystems, Foster City, CA). Two sets of primer pairs were used to quantify the mRNA expression level of RPL10: RPL10-A, spanning exons 4 and 5, and RPL10-B, spanning exons 6 and 7. The forward primer of RPL10-A was 5'-ATA TGA GCA GCT GTC CTC TGA AG-3' and the reverse was 5'-CCA TCT TTG CCA CAA CTT TTT ACC-3'. The forward primer of RPL10-B was 5'-AGA ACA AGG AGC ATG TGA TTG AG-3' and the reverse was 5'-CTT CTT TGA GAT GTG GAT CTT CTG-3'. GAPDH was used as an endogenous control. The forward primer of GAPDH was 5'-GAT GAC ATC AAG AAG GTG GTG-3' and the reverse was 5'-GTC ATA CCA GGA AAT GAG CTT G-3'. The efficiencies of three primer sets were measured using a dilution series of cDNA. The raw threshold cycle (Ct) values were converted to linear form REL (relative expression level) by the 2-ΔΔCt method to quantify the relative gene expression[11]. The REL of each transcript was normalized to GAPDH and relative to the mean Ct value of control males. All the reactions were performed in triplicate. Melting curves were analyzed for each reaction to ensure the specificity of the amplicons.
Statistics
Statistic significance was calculated for RPL10 mRNA transcript levels of different groups using non parametric Mann-Whitney U-test in SPSS10.0. The significance lever for all statistical tests was P < 0.05.
Results
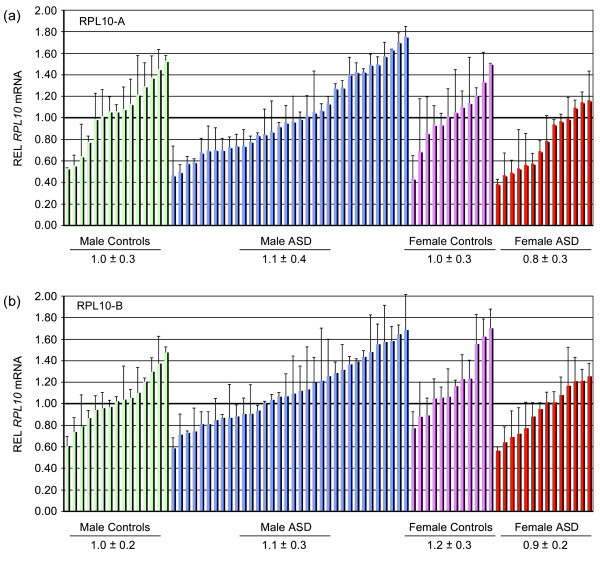
No non-synonymous mutations were detected in our cohort. The mRNA level of RPL10 was compared in four subgroups: male controls, male ASD, female controls and female ASD using two sets of primer pairs RPL10-A and RPL10-B (Figure 1). In controls, the transcript level of RPL10 was not significantly different between males and females (RPL10-A, U = 81, P = 0.7; RPL10-B, U = 61.5, P = 0.2). When cases and controls were compared, we did not observe any significant difference in RPL10 transcript levels (RPL10-A, U = 531, P = 0.2; RPL10-B, U = 607.5, P = 0.7). Female cases had a somewhat lower expression level of RPL10 compared with female controls (RPL10-A, U = 49, P = 0.06; RPL10-B, U = 47.5, P = 0.07). However, this trend disappeared after correction for multiple tests.
Figure 1.
Quantitative RT-PCR of RPL10 in patients with ASD and controls. The level of RPL10 mRNA was quantified with two sets of primer pairs RPL10-A (a) and RPL10-B (b). Each column represents the value of relative expression level (REL) of an individual. The horizontal bar at 1 indicates the mean REL of male controls used as a calibrator. Four subgroups are shown in different colours. The REL value of each subgroup is expressed as mean ± SD.
Discussion
RPL10 belongs to the L10e family of ribosomal proteins and is hypothesized to be necessary for ribosome assembling and function. The identification of two missense mutations in highly conserved positions of RPL10 across species and functional studies in yeast strains expressing human wild-type/mutant RPL10 cDNAs suggested a novel aspect of disease mechanisms for autism – aberrant processes of ribosome biosynthesis and translation. To confirm these initial findings, we sequenced all RPL10 exons and flanking junctions in 141 ASD patients. No missense mutation was identified. We performed this study in European population as well as Klauck et al[6] in order to exclude the possible sample stratification. Other factors, such as the sample size and highly genetic heterogeneity of ASD, should be considered when explaining the absence of RPL10 mutations in our sample. To further address the expression issue, mRNA transcript level of RPL10 was quantified in 48 patients and 27 controls using the method of SYBR Green quantitative PCR. Male controls showed the same transcript level of RPL10 compared with female controls, supporting the evidence that RPL10 is subject to X chromosome inactivation [12]. No statistical significance in expression level was found between cases and controls, suggesting there was no difference in RPL10 expression between two groups. Taken together, our study could not confirm the association between RPL10 and ASD.
Three limitations should be considered in this study. First, our sample of patients screened for RPL10 (n = 141, 101 males and 40 females) was smaller compared to the previous study (n = 345, 268 males and 77 females)[6]. We had 56% of chance to detect at least one mutation in our sample. Our previous study indicated XCI profile could be a useful criteria to prioritize families for mutation screening of X-linked candidate genes in ASD, so we selected 88 individuals from the families with XCI skewing (≥ 70:30) and 53 patients from multiplex families for mutation screening of RPL10[8]. However, we did not detect any functional mutations. Second, our mutation screening was restricted to exons and therefore was not appropriate to detect the presence of variants altering the expression of RPL10 in promoter regions or other regulatory regions. However, we recently performed a high-throughput genotyping of 91 patients from this sample using the Illumina human1M-duo beadchip, and we could not detect any genomic imbalance within or close to RPL10 (unpublished data). Thirdly, we performed the quantification of RPL10 mRNA level in B lymphoblastoid cell lines and therefore we could have missed alterations specific to brain. Thus, further studies on other bigger samples are warranted and the promoter regions and other regulatory regions should be investigated.
Conclusion
The present study did not find any non-synonymous mutations in our cohort, neither abnormal RPL10 transcript levels in ASD patients compared to controls, suggesting that RPL10 has no major effect on the susceptibility to ASD.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
XG carried out sequencing and quantitative PCR testing, and contributed to manuscript writing. RD performed the statistical analysis. FF aided in quantitative PCR testing. CMD, PC, and HGB aided in sample storage and preparations. CB, GN, HA, MR, ICG, SK, MCMS, CG and ML contributed to the sample collection. TB participated in the design of the study and manuscript writing.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
We thank the patients and their families for participating in this study. This work was supported by the Pasteur Institute, INSERM, Assistance Publique-Hôpitaux de Paris, CNRS, FP6 EUSynapse, FP6 AUTISM MOLGEN, FP6 ENI-NET, Fondation France Télécom, Fondation de France, Fondation biomédicale de la Mairie de Paris, and Fondation pour la Recherche Médicale, Fondation FondaMentale.
Contributor Information
Xiaohong Gong, Email: gongxh@fudan.edu.cn.
Richard Delorme, Email: delorme@creteil.inserm.fr.
Fabien Fauchereau, Email: fabienf@pasteur.fr.
Christelle M Durand, Email: Christelle.Durand@bordeaux.inserm.fr.
Pauline Chaste, Email: pchaste@pasteur.fr.
Catalina Betancur, Email: Catalina.Betancur@im3.inserm.fr.
Hany Goubran-Botros, Email: hgbotros@pasteur.fr.
Gudrun Nygren, Email: gudrun.m.nygren@vgregion.se.
Henrik Anckarsäter, Email: Henrik.Anckarsater@med.lu.se.
Maria Rastam, Email: maria.rastam@pediat.gu.se.
I Carina Gillberg, Email: carina.gillberg@pediat.gu.se.
Svenny Kopp, Email: svenny.kopp@vgregion.se.
Marie-Christine Mouren-Simeoni, Email: marie-christine.mouren-simeoni@rdb.ap-hop-paris.fr.
Christopher Gillberg, Email: Christopher.gillberg@pediat.gu.se.
Marion Leboyer, Email: leboyer@im3.inserm.fr.
Thomas Bourgeron, Email: thomasb@pasteur.fr.
References
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- Szatmari P, Jones MB, Zwaigenbaum L, MacLean JE. Genetics of autism: overview and new directions. J Autism Dev Disord. 1998;28:351–368. doi: 10.1023/A:1026096203946. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- Nguyen YH, Mills AA, Stanbridge EJ. Assembly of the QM protein onto the 60S ribosomal subunit occurs in the cytoplasm. J Cell Biochem. 1998;68:281–285. doi: 10.1002/(SICI)1097-4644(19980201)68:2<281::AID-JCB14>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Vincent JB, Melmer G, Bolton PF, Hodgkinson S, Holmes D, Curtis D, Gurling HM. Genetic linkage analysis of the X chromosome in autism, with emphasis on the fragile × region. Psychiatr Genet. 2005;15:83–90. doi: 10.1097/00041444-200506000-00004. [DOI] [PubMed] [Google Scholar]
- Klauck SM, Felder B, Kolb-Kokocinski A, Schuster C, Chiocchetti A, Schupp I, Wellenreuther R, Schmotzer G, Poustka F, Breitenbach-Koller L, Poustka A. Mutations in the ribosomal protein gene RPL10 suggest a novel modulating disease mechanism for autism. Mol Psychiatry. 2006;11:1073–1084. doi: 10.1038/sj.mp.4001883. [DOI] [PubMed] [Google Scholar]
- Kolb-Kokocinski A, Mehrle A, Bechtel S, Simpson JC, Kioschis P, Wiemann S, Wellenreuther R, Poustka A. The systematic functional characterisation of Xq28 genes prioritises candidate disease genes. BMC Genomics. 2006;7:29. doi: 10.1186/1471-2164-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Bacchelli E, Blasi F, Toma C, Betancur C, Chaste P, Delorme R, Durand CM, Fauchereau F, Botros HG, et al. Analysis of X chromosome inactivation in autism spectrum disorders. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:830–835. doi: 10.1002/ajmg.b.30688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CRM, Le Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- Gillberg CRM, Wentz E. The Asperger Syndrome (and high-functioning autism) Diagnostic Interview (ASDI): a preliminary study of a new structured clinical interview. Autism. 2001;5:57–66. doi: 10.1177/1362361301005001006. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]