Abstract
A key step in the activation of heterodimeric integrin adhesion receptors is the transmission of an agonist-induced cellular signal from the short α- and/or β-cytoplasmic tails to the extracellular domains of the receptor. The structural details of how the cytoplasmic tails mediate such an inside-out signaling process remain unclear. We report herein the NMR structures of a membrane-anchored cytoplasmic tail of the αIIb-subunit and of a mutant αIIb-cytoplasmic tail that renders platelet integrin αIIbβ3 constitutively active. The structure of the wild-type αIIb-cytoplasmic tail reveals a “closed” conformation where the highly conserved N-terminal membrane-proximal region forms an α-helix followed by a turn, and the acidic C-terminal loop interacts with the N-terminal helix. The structure of the active mutant is significantly different, having an “open” conformation where the interactions between the N-terminal helix and C-terminal region are abolished. Consistent with these structural differences, the two peptides differ in function: the wild-type peptide suppressed αIIbβ3 activation, whereas the mutant peptide did not. These results provide an atomic explanation for extensive biochemical/mutational data and support a conformation-based “on/off switch” model for integrin activation.
Keywords: NMR, cytoplasmic domain
By mediating numerous cell–cell and cell–matrix interactions, the integrin family of cell-surface receptors plays crucial roles in the development and health of all multicellular organisms (1–3). These noncovalent heterodimers (α and β) are expressed widely and regulate diverse biological processes such as matrix assembly, hemostasis, wound healing, inflammation, and tumor metastasis (1–3). Each subunit of an integrin consists of a single type I transmembrane domain, a large extracellular domain of several hundred amino acids, and typically, a short cytoplasmic tail of ≈20–70 residues (1, 2, 4). The extracellular domains interact with each other to form a complex binding site for a wide variety of ligands. Central to the function of the integrins is their capacity to undergo activation, a transition from a low to a high affinity/avidity state for their ligands. Such activation is tightly regulated through a process termed “inside-out signaling” (3, 4), i.e., agonist stimulation initiates intracellular changes that ultimately render the extracellular domain competent to bind ligands. The biological importance of such integrin activation is underscored by the major platelet integrin, αIIbβ3, which cannot engage its abundant plasma protein ligands unless the cell has been stimulated by an agonist. This transition depends on the transmission of a cellular signal, typically initiated by occupancy of a G protein-coupled receptor, to the short cytoplasmic tails of the αIIb- and/or β3-subunits, which is then propagated across the cell membrane to the extracellular ligand-binding domain. The structural basis of this inside-out signaling event remains poorly understood.
The cytoplasmic tails of the α-subunits of integrins are composed of a highly conserved N-terminal membrane-proximal region followed by a variable C terminus (refs. 1 and 2; Fig. 1A). This pattern is typified by the cytoplasmic tail of the integrin αIIb-subunit, which has often served as a prototype for structure-function analyses. Deletion of almost the entire αIIb-cytoplasmic tail (G991–E1008) constitutively activates αIIbβ3 (5), suggesting that this tail exerts a negative regulatory function and locks the αIIbβ3 in a low-affinity state. Mutation of its conserved N-terminal region enhances the affinity of αIIbβ3 for ligands, likely by altering the conformation of the αIIb-cytoplasmic tail (6, 7), which ultimately generates an inside-out signal to activate the receptor. However, the interpretation of such mutational data has remained conjectural, because the structure of the αIIb-cytoplasmic tail, as well as that of any of the α-subunits, is unknown. Recently, it has been shown that a lipid-modified peptide containing only the membrane-proximal region of αIIb (K989–R995) can penetrate the platelet membrane and induce αIIbβ3 activation (8). This observation attests to the significance of the N-terminal region of the αIIb-cytoplasmic tail in integrin activation and indicates that such peptides can adopt a functionally relevant conformation. To understand the mechanism by which the αIIb-cytoplasmic tail regulates integrin activation, we have determined the NMR structure of a lipid-modified (myristoylated) peptide corresponding to the αIIb-cytoplasmic tail as well as that of a double-substituted peptide P998A/P999A (see Fig. 1A), because these substitutions impart constitutive activity to αIIbβ3 (9). A critical issue in solving the structures of component peptides is their relationship to the native structure of the parent molecule. We address the issue by showing that these lipid-modified wild-type and mutant peptides retain the functional properties of intact receptors, with distinct effects on the activation states of αIIbβ3 in platelets. Our structures explain not only these functional differences but also other biochemical/mutational results, providing a basis for understanding integrin activation at the atomic level.
Figure 1.
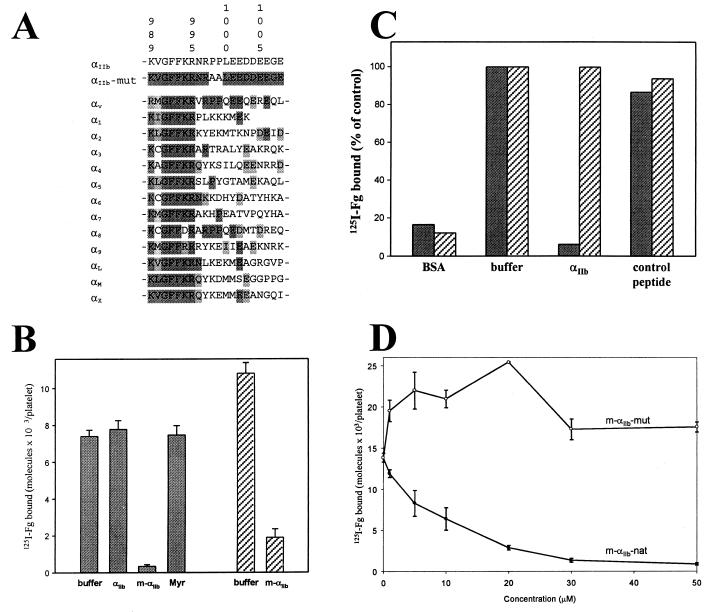
Sequence and function of the integrin αIIb-cytoplasmic domain. (A) Sequence alignment of various human integrin α-cytoplasmic domains with both wild-type and mutant αIIb. The sequence identity and similarity are highlighted in decreased darkness. Residue numbers correspond to αIIb. (B) Effect of αIIb-cytoplasmic peptides on the binding of fibrinogen (Fg) to stimulated platelets. Human platelets in Tyrode's buffer alone (buffer), with or without 50 μM (final concentration) of nonmyristoylated αIIb K989–E1008 (αIIb), m-αIIb-wt (m-αIIb), and myristic acid (Myr) were preincubated for 30 min with 300 nM 125I-fibrinogen. The platelets were then stimulated with either 10 μM ADP/20 μM epinephrine (shaded bars) or 50 μM thrombin receptor agonist peptide (hatched bars). After 15 min, 125I-fibrinogen binding was measured. Incubation of nonstimulated platelets with cytoplasmic peptides or with myristic acid had no effect on fibrinogen binding (data not shown). (C) Effects of αIIb-cytoplasmic peptides on the binding of fibrinogen to immunocaptured forms of αIIbβ3. Both forms of the receptor, platelet αIIbβ3 (shaded bars) or sr-αIIbβ3 (hatched bars), were immunocaptured onto immobilized monoclonal antibody AP3. The captured receptors were preincubated for 30 min with either buffer alone (buffer) or 50 μM nonmyristoylated αIIb K989–E1008 (αIIb). Peptide specificity was confirmed with a nonrelated acidic peptide (control peptide; GDKNADGWIEFEEL). Results are expressed as a percentage of maximal fibrinogen binding in the absence of peptides. (D) Divergent effects of myristoylated wild-type and mutant αIIb-cytoplasmic peptides on the binding of fibrinogen to stimulated platelets. Platelets were preincubated with varying concentrations of either m-αIIb-wt (closed circles) or m-αIIb-mut peptides (open circles), and then 125I-fibrinogen binding to ADP/epinephrine-stimulated platelets was measured.
Methods
Peptide Synthesis and Purification.
Peptides were synthesized on 4-methylbenzhydrylamine resins (α-carboxamides) or on appropriate Boc-aminoacyl-OCH2-Pam resins (carboxylates). The synthesis followed a manual stepwise in situ neutralization/activation protocol by using tert-butyloxycarbonyl (Boc) protection, with the efficiency of coupling being monitored by the quantitative ninhydrin method (10). Myristoylation of the N terminus of peptides was performed with myristic acid and 1,3-diisopropylcarbodiimide (1:0.95 mol:mol) in a mixture of dichloromethane:dimethylformamide (4:1, vol/vol), following standard solution methods (11). Synthetic peptides were cleaved from the resin and deprotected by hydrofluoric acid, extracted into aqueous acetic acid, and then purified by C18 reverse-phase HPLC (12). The purity of each peptide was confirmed to be >98%, as assessed by analytical HPLC and mass spectroscopy (12).
Fibrinogen Binding to Platelets.
Gel-filtered human platelets were suspended in Tyrode's buffer containing 1 mM Ca2+ to a final concentration of 3 × 108 per ml (13). Peptides were then added to the platelets up to 50 μM for a 30-min incubation at 22°C, together with 300 nM 125I-labeled fibrinogen. The platelets were then stimulated with either 10 μM ADP/20 μM epinephrine or 50 μM thrombin receptor agonist peptide. After 15 min, 125I-fibrinogen binding was measured as described (13, 14).
Fibrinogen Binding to Immunocaptured Receptor.
Fibrinogen binding to αIIbβ3 or soluble recombinant αIIbβ3 (sr-αIIbβ3; ref. 15), lacking the transmembrane and cytoplasmic tails of both subunits, was performed by using an immunocapture assay similar to that described (16). A monoclonal antibody directed against β3 (AP3) was coated overnight onto microtiter wells (Immunlon 2, Dynatech). The wells were washed and blocked with 1% BSA, and then αIIbβ3 was captured from a detergent extract of stimulated platelets (16) or sr-αIIbβ3 was captured from cell culture supernatants (15). Unbound material was removed by washes, and the wells were preincubated with modified Tyrode's buffer (16) containing 50 μM test peptide. After 30 min, 300 nM 125I- fibrinogen was added, and the wells were incubated for 4 h. The wells were then washed, and bound radioactivity was counted. Nonspecific binding was estimated by using BSA-coated wells and AP3-coated wells, both of which were not exposed to receptor.
Sample Preparation and NMR Spectroscopy.
The myristoylated peptides—the myristoylated cytoplasmic tail of the integrin αIIb-subunit (m-αIIb-wt) and the myristoylated P998A/P999A mutant of the αIIb-cytoplasmic tail (m-αIIb-mut)—were dissolved in an acetate buffer (50 mM sodium acetate/50 mM NaCl, pH 5.0) containing 150 mM perdeuterated dodecylphosphocholine (DPC) micelles and 10% (vol/vol) 2H2O. All the peptide concentrations for NMR studies were ≈3 mM. NMR experiments were performed on Varian Inova 500- and 600-MHz instruments. For resonance assignment and nuclear Overhauser effect (NOE) analyses, the following experiments were performed at 25°C: double-quantum filtered correlation spectroscopy, total correlation spectroscopy, and NOE spectroscopy (NOESY; ref. 17). The mixing times were 100 ms for NOESY and 50–70 ms for total correlation spectroscopy. All the data were processed on an SGI Octane workstation (Mountain View, CA) by using nmrpipe (18) and analyzed by using pipp (19). Total 1H-resonance assignments were made at both pH 5.0 and pH 6.3 in DPC micelles with a combination of NOESY, double-quantum filtered correlation spectroscopy, and total correlation spectroscopy by using conventional sequential assignment procedures (17). The significant line-broadening prevented the accurate measurement of the small helical JHN-Hα in the N-terminal region. The resolved C-terminal residues all exhibited ≈7–8 Hz for JHN-Hα, consistent with their loop feature. The structures of m-αIIb-wt and m-αIIb-mut were calculated on an SGI Octane workstation by using xplor (version 3.2; ref. 20). The target function that is minimized during simulated annealing is comprised of quadratic harmonic potential terms for covalent geometry, square-well quadratic potentials for the experimental distance and for torsion angle restraints, and a quadratic van der Waals repulsion term for the nonbonded contacts. No hydrogen-bonding, electrostatic, or 6–12 Lennard–Jones empirical potential energy terms were included in the target function. The distance restraints were grouped into four distance ranges—1.8–2.5 Å, 1.8–3.5 Å, 1.8–5.0 Å, and 1.8–6.0 Å—corresponding to strong, medium, weak, and very weak NOEs, respectively. Table 1 shows the detailed statistics for calculated structures.
Table 1.
Structural statistics for wild-type and mutant αIIb
| Parameter | Wild type | Mutant |
|---|---|---|
| NOE distance constraints | ||
| All | 388 | 254 |
| Sequential (|i − j| = 1) | 129 | 59 |
| Medium (1 < |i − j| ≤ 5) | 98 | 48 |
| Long range (|i − j| > 5) | 29 | 0 |
| Intraresidue | 132 | 147 |
| Largest RMSD from exponential distance restraint, Å | 0.099 | 0.215 |
| RMSD from idealized covalent geometry | ||
| Bonds, Å | 0.005 | 0.003 |
| Angles, degrees | 0.857 | 0.329 |
| Impropers, degrees | 0.474 | 0.240 |
| EL–J, kcal⋅mol−1* | −34.0 | −34.1 |
| procheck (Ramachandran plot), residues in allowed regions, % | 100 | 98.1 |
| Most favored regions, % | 32.9 | 53.3 |
| Additionally allowed regions, % | 66.2 | 39.4 |
| Generously allowed regions, % | 0.9 | 5.4 |
| Disallowed regions, % | 0.0 | 1.9 |
| Average RMSD to the mean structure | Residues 989–1,005 | Residues 989–1,002 |
| Backbone, Å | 0.46 | 0.47 |
| Heavy atoms, Å | 1.33 | 1.15 |
Statistics were derived from the ensemble of 15 final simulated annealing structures. RMSD, root-mean-square deviation.
EL–J is the Lennard–Jones van der Waals energy value calculated with the charmm empirical energy function and is not included in the target function for simulated annealing or restrained minimization.
CD Spectroscopy.
CD spectra were acquired by using a Jasco-600 spectrometer (Easton, MD) as described (12). Samples for far-UV studies were prepared by dissolving about 0.5 mg/ml peptides in the same buffer used for NMR studies. All samples were scanned four times and averaged to obtain a high signal-to-noise representation of the spectrum. Each spectrum was corrected for solvent contribution and then analyzed by applying a previously described algorithm (12).
Results and Discussion
NMR structural analyses were performed with two synthetic peptides: m-αIIb-wt and m-αIIb-mut. The sequences of these peptides are shown in Fig. 1A. We developed the myristoylation approach for studying these short peptides, because the myristoyl tail is a surrogate transmembrane domain that anchors proteins/domains onto the membrane surface, inducing compact conformations (21–23) and thus mimicking the cytoplasmic domains of intact receptors. Furthermore, the hydrophobic acyl chain overcomes the inability of peptides to penetrate the plasma membrane and allows the delivery of the peptides into cells for functional studies (Fig. 1 B–D). Platelets exhibit negligible binding of 125I-fibrinogen unless the cells are stimulated with agonists that activate αIIbβ3. In the analyses shown in Fig. 1B, the platelets were stimulated with ADP + epinephrine or with thrombin receptor agonist peptide. Fibrinogen binding induced by either of these agonists was suppressed to nonspecific background levels by the m-αIIb-wt peptide. In contrast, nonmyristoylated αIIb-cytoplasmic peptide and myristic acid itself had no effect. However, in an immunocapture assay, nonmyristoylated αIIb-peptide could inhibit the binding of fibrinogen to αIIbβ3 (Fig. 1C). In these experiments, the capacity of nonmyristoylated αIIb-peptide to inhibit the binding of fibrinogen to activated platelet αIIbβ3 and to sr-αIIbβ3 was compared. sr-αIIbβ3 is a soluble recombinant form of αIIbβ3 that lacks the transmembrane and cytoplasmic domains of both subunits and spontaneously assumes an activated conformation (15). When preincubated with αIIbβ3, nonmyristoylated αIIb-peptide dramatically reduced the binding of fibrinogen to nonspecific levels (Fig. 1C, BSA lanes), while having no effect on the capacity of sr-αIIbβ3 to bind fibrinogen. These results indicate that the biological effects exerted by the αIIb-cytoplasmic peptides are mediated through the cytoplasmic tails of the activated receptor. With intact platelets, myristoylation was required for interaction with the cell, whereas the nonmyristoylated peptide was functional with purified receptor. Finally, Fig. 1D shows that, although m-αIIb-wt inhibited the fibrinogen binding to platelets in a concentration-dependent manner, the mutant peptide (P998A/P999A) m-αIIb-mut had no inhibitory activity. Rather, m-αIIb-mut slightly augmented fibrinogen binding to stimulated platelets. These differences in functional activities were not caused by differences in the amount of each peptide associated with the platelets; when fluorescently labeled peptides were used, equal amounts of both myristoylated peptides were delivered into platelets (data not shown). Thus, the myristoylated peptides are functionally active counterparts of the cytoplasmic tail of αIIb within the intact receptor; although wild-type αIIbβ3 resides in a resting state and does not bind fibrinogen, the P998A/P999A mutant receptor is constitutively active, binding fibrinogen without requiring an activating stimulus (9).
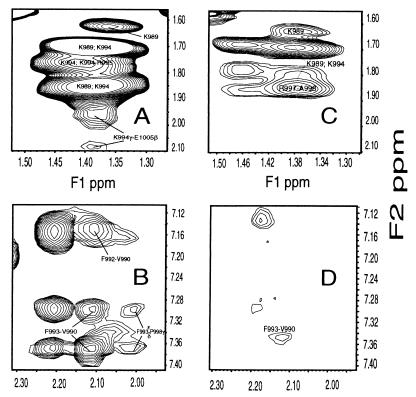
The myristoylated peptides were dissolved in membrane-mimic DPC micelles for NMR studies. DPC has the same zwitterionic phosphocholine head group as the dominant phospholipids of most eukaryotic membranes and provides a favorable balance between desirable NMR properties and biochemical compatibility with membrane-associated proteins (24–26). The myristoyl group has been shown to insert into the lipid bilayer or DPC micelles stabilizing peptide structures (21–23). Indeed, both myristoylated m-αIIb-wt and m-αIIb-mut had folded structures, as judged by CD and NOESY data. Importantly, a number of long range NOEs were observed for m-αIIb-wt but not for m-αIIb-mut (Table 1 and Fig. 2 A–D). Critical long-range NOEs for m-αIIb-wt involve a cluster of residues: F993/P1000, F993/D1004, K994/D1004, K994/E1005, R997/D1003, R997/D1004, and P1000/D1004. As a comparison, although the nonmyristoylated αIIb-wt dissolved in DPC also exhibits the helical dNN(i,i+1) NOEs in the N-terminal K989–R997 region, few such long-range contacts and fewer dαN(i,i+3) and dαβ(i,i+3) NOEs in the N-terminal helical region were observed, indicating a partially folded structure and weaker association with the DPC. The myristoyl group anchors the peptide into the micelles for tight binding and promotes a stable tertiary conformation (O.V., T.H., E.F.P., and J.Q., unpublished work), thereby mimicking the cytoplasmic domain of the intact receptor. This mimicry is strongly supported by the aforementioned functional data that indicate that the myristoylated peptides are functionally consistent with their intact receptors.
Figure 2.
Selected regions of two dimensional 1H NOESY for m-αIIb-wt and m-αIIb-mut at 25°C and pH 5.0. (A) Portion of aliphatic region for m-αIIb-wt, showing K994–E1005 contact. (B) Portion of aromatic region for m-αIIb-wt, showing F993–P998 contact. (C) Region corresponding to that shown in A for m-αIIb-mut. (D) Region corresponding to that shown in B for m-αIIb-mut. Note that other than loss of long-range NOEs for the mutant, the mutant had fewer sequential NOEs than the wild type (see text).
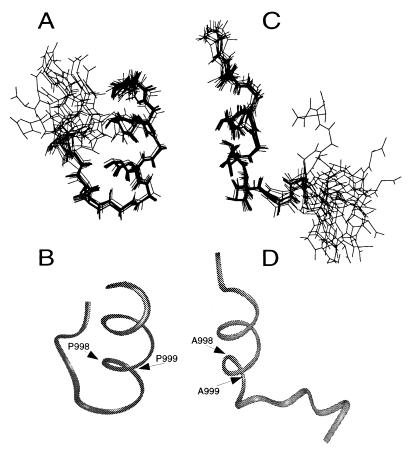
The superpositions of the 15 best structures for m-αIIb-wt and m-αIIb-mut are shown in Fig. 3 A and C, respectively. The quality of the calculated structures is demonstrated by the overall structural statistics (Table 1). Both wild-type and mutant structures have α-helical features in their N-terminal part (Fig. 3 B and D). Because the N-terminal part is highly conserved among the integrin α-subunits (Fig. 1A), we anticipate that this region adopts a similar structure in other integrins. For the wild-type peptide, the helix terminates at P999, followed by a turn, which allows the acidic C-terminal loop to fold back and interact with the positively charged N-terminal region, resulting in a “closed” conformation (Fig. 4B). The interactions between the N and C termini are primarily electrostatic, i.e., through salt bridges between R997/D1003, K994/D1004, and K994/E1005 (Fig. 4A). In addition, the aliphatic side chain of D1004 makes hydrophobic contact with the aromatic ring of F993, contributing to the stabilization of tertiary structure between the N-terminal helix and the C-terminal loop. For the structure of m-αIIb-mut, although the N-terminal α-helix is retained as compared with wild-type m-αIIb-wt (Figs. 3D and 4B), the double mutation disrupts the turn, thereby preventing interactions between the N-terminal helix and C-terminal loop, resulting in an “open” conformation (Fig. 4B). The structure of the mutant is apparently less rigid, with a significantly reduced number of NOE constraints (Table 1) as a result of disruption of N- and C-terminal contacts.
Figure 3.
Illustration of m-αIIb-wt and m-αIIb-mut structures. (A) Backbone superpositions of the 15 best m-αIIb-wt structures. (B) Backbone ribbon diagram of minimized average m-αIIb-wt structure in the same view as in A. (C) Backbone superpositions of m-αIIb-mut structures. (D) Backbone ribbon diagram of minimized average m-αIIb-mut structure in the same view as in C. The structural statistics are listed in Table 1.
Figure 4.
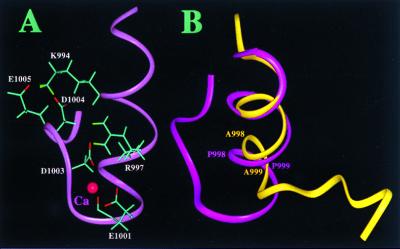
Structural highlights of the cytoplasmic domain of αIIb. (A) Structure of the m-αIIb-wt showing the side chains of K994, R997, E1001, D1003, D1004, and E1005 for the salt-bridge network. A proposed Ca2+-binding site is also shown in the figure involving R997, E1001, D1003, and D1004, which were identified by comparing the two-dimensional 1H total correlation spectroscopy spectra of Ca2+-free and Ca2+-bound forms (Ca2+ was added up to 5-fold). The binding of Ca2+ to the αIIb-tail has been shown by biochemical studies (12, 27). (B) Backbone overlay of m-αIIb-wt (pink) and m-αIIb-mut (yellow), showing the structural difference. The C-terminal loop of m-αIIb-wt folds back to interact with the N-terminal helix, whereas in the mutant, the C-terminal part is highly flexible and makes no interactions with the helix. The C-terminal part (D1003–E1008) of the m-αIIb-mut is disordered.
Considering the importance of F992, F993, K994, and R997 in stabilizing the N-terminal helix and its interaction with the C-terminal loop for the wild-type αIIb-cytoplasmic tail, it is now clear why mutations of these residues in the helix activate αIIbβ3 (6); such mutations are likely to disrupt the closed conformation. Even for the G991A mutation, the additional methyl group would introduce a steric clash with the nearby V990 and F992, which results in a structural perturbation that moderately activates αIIbβ3 (6). In contrast to the aforementioned amino acids, the side chain of R995 is highly exposed, making no intrasubunit contacts. Thus, the R995A mutation, which results in a constitutively active receptor (6), cannot be explained by alterations in αIIb structure. Rather, the exposure of the guanidinium group of R995 supports the speculation that R995 may form a salt bridge with D723 of the β3-cytoplasmic tail (6).
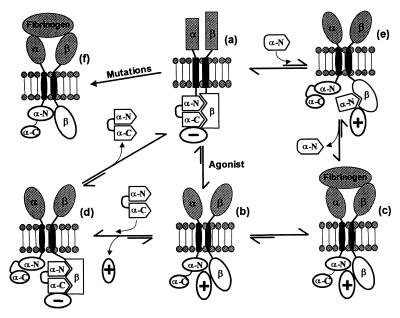
The structural basis for the constitutive ligand-binding activity of the P998A/P999A mutant of αIIbβ3 is evident in the NMR structure of m-αIIb-mut and supports the “conformational switch model” depicted in Fig. 5. According to this model, the acidic C-terminal loop acts as a switch to close or open the conformation of the αIIb-cytoplasmic tail, thereby regulating integrin activation. The closed switch is the conformation of the α-subunit in wild-type αIIbβ3 and in the m-αIIb-wt peptide. Intramolecular interactions between the C-terminal and N-terminal regions create a stable closed structure/switch. This closed switch maintains αIIbβ3 in the latent state, possibly by interacting with the β3-subunit, with a negative regulator (Fig. 5a), or with both. It is noteworthy that studies of complex formation have implicated both the C- and N-terminal regions of the αIIb-cytoplasmic tail in interactions with β3-cytoplasmic tail (6, 12, 27, 28). Opening of the switch by either protein–protein interaction (Fig. 5b) or selective mutations in the αIIb-tail (Fig. 5f) occurs through a conformational change that exposes the N-terminal helix of the αIIb-cytoplasmic tail and activates the receptor. How exactly the conformational change associated with opening of the switch induces a transmembrane signaling must remain conjectural. One possibility is that this transmission is intrinsic to the conformational state of the αIIb-cytoplasmic tail alone and is a dynamic and energetically driven process. As elegantly shown for another membrane receptor structure, FhuA (29), subtle changes in the ligand-binding site of FhuA induce substantial structural perturbation in the periplasmic face, but the transmembrane regions in the free and ligand-bound forms remain very similar, suggesting a dynamic and energy-transducing process. Also relevant is the recent report by Ottemann et al. (30), which shows that subtle changes in the juxtapositioning of two receptor subunits can propagate conformational changes across cell membranes. Because the cytoplasmic tails of αIIb and β3 may interact with each other, changes in their spatial relationship induced by the conformational switch of αIIb-tail is likely involved in mediating the transmembrane signaling. It is apparent from our structures that the closed and open conformations for the wild-type and mutant αIIb could interact with the β3 differently (Fig. 5 a and b), which could lead to inactive or active states of the receptor. This model also explains the inhibitory function of m-αIIb-wt: for example, the delivery of the “inhibitory” m-αIIb-wt into the cell competes with the active αIIb-tail for interacting with the β3-tail (Fig. 5d), which results in the binding of a negative regulator and release of the active signaling molecule. The αIIb-tail in the intact receptor then undergoes an exchange process with αIIb-peptide for binding to β3, which releases the peptide and shifts the equilibrium to the inactive αIIbβ3 (Fig. 5a). The structural change between the wild type and the P998A/P999A mutant suggests that the C-terminal acidic loop of the αIIb-tail exerts an inhibitory constraint on αIIbβ3 activation. Indeed, the truncated αIIb-peptide with the removal of the C-terminal inhibitory constraint activates αIIbβ3 (8), which can be appreciated in our model: the C-terminally truncated αIIb-peptide competes with the inactive αIIb-tail for binding to β3-tail, leading to the recognition of a positive regulator (Fig. 5e) and shifting the equilibrium to the active αIIbβ3 (Fig. 5c). It should be noted that αIIb-binding proteins have been identified (31, 32); these proteins may be responsible for responding to or inducing this conformational switch (3). However, the conformational switch may be entirely intrinsic to the cytoplasmic tails of αIIb and β3, and positive or negative regulators may not be required to maintain the active and inactive states of the receptor. Recent studies also showed that synthetic cell-permeable β-peptides can also affect integrin activation (33, 34), suggesting that there may be more than one switch per integrin.
Figure 5.
Model for the regulation of ligand binding to αIIbβ3. In resting platelets, αIIbβ3 is maintained in an inactive/resting state (a) by a negative regulator (− circle). The negative regulator may be the interaction between the α/β-cytoplasmic tails and/or an intracellular protein. The α-cytoplasmic tail adopts a closed conformation with its N terminus (α-N) interacting with its acidic C terminus (α-C). Platelet stimulation (Agonist) leads to the association of intracellular constituents (positive regulator, + circle) with the cytoplasmic domain of αIIbβ3, generating an “activating conformational signal” (b) such that fibrinogen binds to the receptor (c). m-αIIb-wt converts the b state back into the inactive a state by forming an intermediate d, an a → b → d → a pathway. The N-terminal αIIb-cytoplasmic peptide perturbs the α/β-complex shown in a, generating an intermediate e, which shifts the equilibrium to c state for binding of fibrinogen, the a → e → c pathway. Selective mutations (see text) in either the N or C terminus of αIIb or β3 can also lead directly to the expression of a constitutively active receptor (f), bypassing the a → b → c pathway.
In summary, we have determined the solution NMR structures of the myristoylated peptides corresponding to the αIIb-cytoplasmic tail and to its P998A/P999A mutant, which renders αIIbβ3 constitutively active. The myristoylation represents a unique approach to anchor a cytoplasmic domain to the micelle surface, mimicking a membrane-anchored cytoplasmic domain in an intact receptor. Importantly, we also have shown that these same lipid-modified peptides are functional mimetics of the intact receptor and can alter the activation state of αIIbβ3. The three-dimensional folding of the two peptides is significantly different with the C-terminal region folding back and interacting with the highly conserved N-terminal activating region in wild-type αIIb but not in the mutant. These data provide a basis for understanding the integrin activation at the atomic level, suggest a conformation-switch mechanism for regulating αIIbβ3 activation, and may reflect the basis of a general mechanism for integrin activation.
Acknowledgments
We thank Julie Peterson and Peter Newman for providing sr-αIIbβ3, Frank Delaglio and Dan Garrett for NMR software, and Yanwu Yang, Chuck Sanders, and Sambasivarao Nanduri for useful discussions. The work was supported in part by National Institutes of Health grants (to J.Q. and E.F.P.) and an American Heart Association Scientist Development grant (to T.H.). O.V. is supported by a National Institutes of Health postdoctoral fellowship. The NMR studies were performed in the Cleveland Center for Structural Biology, which is supported by the State of Ohio and the Cleveland Foundation.
Abbreviations
- NOE
nuclear Overhauser effect
- NOESY
NOE spectroscopy
- m-αIIb-wt
myristoylated cytoplasmic tail of the integrin αIIb-subunit
- m-αIIb-mut
myristoylated P998A/P999A mutant of the αIIb cytoplasmic tail
- sr-αIIbβ3
soluble recombinant αIIbβ3
- DPC
dodecylphosphocholine
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1DPK and 1DPQ).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040548197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040548197
References
- 1.Hynes R O. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz M A, Schaller M D, Ginsberg M H. Annu Rev Cell Biol. 1995;11:549–599. doi: 10.1146/annurev.cb.11.110195.003001. [DOI] [PubMed] [Google Scholar]
- 3.Ruoslahti E, Engvall E. J Clin Invest. 1997;99:1149–1152. doi: 10.1172/JCI119269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett J S. Trends Cardiovasc Res. 1996;6:31–36. doi: 10.1016/1050-1738(95)00126-3. [DOI] [PubMed] [Google Scholar]
- 5.O'Toole T E, Mandelman D, Forsyth J, Shattil S J, Plow E F, Ginsberg M H. Science. 1991;254:845–847. doi: 10.1126/science.1948065. [DOI] [PubMed] [Google Scholar]
- 6.Hughes P E, Diaz-Gonzalez F, Leong L, Wu C, McDonald J A, Shattil S J, Ginsberg M H. J Biol Chem. 1996;271:6571–6574. doi: 10.1074/jbc.271.12.6571. [DOI] [PubMed] [Google Scholar]
- 7.O'Toole T E, Katagiri Y, Faull R J, Peter K, Tamura R, Quaranta V, Loftus J C, Shattil S J, Ginsberg M H. J Cell Biol. 1994;124:1047–1059. doi: 10.1083/jcb.124.6.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stephens G, O'Luanaigh N, Reilly D, Harriott P, Walker B, Fitzgerald D, Moran N. J Biol Chem. 1998;273:20317–20322. doi: 10.1074/jbc.273.32.20317. [DOI] [PubMed] [Google Scholar]
- 9.Leisner T M, Wencel-Drake J D, Wang W, Lam S C-T. J Biol Chem. 1999;274:12945–12949. doi: 10.1074/jbc.274.18.12945. [DOI] [PubMed] [Google Scholar]
- 10.Schmolzer M, Alewood P, Jones A, Alewood D, Kent S B. Int J Pept Protein Res. 1993;40:180–193. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 11.Bodanszky M. The Practice of Peptide Synthesis. New York: Springer; 1984. [Google Scholar]
- 12.Haas T A, Plow E F. J Biol Chem. 1996;271:6017–6026. doi: 10.1074/jbc.271.11.6017. [DOI] [PubMed] [Google Scholar]
- 13.Tranqui L, Andrieux A, Hudry-Clergeon G, Ryckewaert J J, Soyez S, Chapel A, Ginsberg M H, Plow E F, Marguerie G. J Cell Biol. 1989;108:2519–2527. doi: 10.1083/jcb.108.6.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Souza S E, Ginsberg M H, Matsueda G R, Plow E F. Nature (London) 1991;350:66–68. doi: 10.1038/350066a0. [DOI] [PubMed] [Google Scholar]
- 15.Peterson J A, Visentin G P, Newman P J, Aster R H. Blood. 1998;92:2053–2063. [PubMed] [Google Scholar]
- 16.Du X, Plow E F, Frelinger A L, III, O'Toole T E, Loftus J C, Ginsberg M H. Cell. 1991;65:409–416. doi: 10.1016/0092-8674(91)90458-b. [DOI] [PubMed] [Google Scholar]
- 17.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 18.Delaglio F, Grzesiek S, Vuister G W, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 19.Garrett D S, Powers R, Gronenborn A M, Clore G M. J Magn Reson. 1991;95:214–220. doi: 10.1016/j.jmr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Brünger A T. xplor. New Haven: Yale Univ.; 1993. , Version 3.1. [Google Scholar]
- 21.Losonczi J A, Prestegard J H. Biochemistry. 1998;37:706–716. doi: 10.1021/bi9717791. [DOI] [PubMed] [Google Scholar]
- 22.Michielin O, Vergeres G, Ramsden J J. J Am Chem Soc. 1999;121:6523–6526. [Google Scholar]
- 23.Sankaram M B. Biophys J. 1994;67:105–112. doi: 10.1016/S0006-3495(94)80459-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henry G D, Sykes B D. Methods Enzymol. 1994;239:515–535. doi: 10.1016/s0076-6879(94)39020-7. [DOI] [PubMed] [Google Scholar]
- 25.Vinogradova O, Soennichsen F D, Sanders C R. J Biomol NMR. 1998;4:381–386. doi: 10.1023/a:1008289624496. [DOI] [PubMed] [Google Scholar]
- 26.MacKenzie K R, Prestegard J H, Engleman D M. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 27.Vallar L, Melchior C, Plancon S, Drobecq H, Lippens G, Regnault V, Kieffer N. J Biol Chem. 1999;274:17257–17266. doi: 10.1074/jbc.274.24.17257. [DOI] [PubMed] [Google Scholar]
- 28.Muir T W, Williams M J, Ginsberg M H, Kent S B. Biochemistry. 1994;33:7701–7708. doi: 10.1021/bi00190a025. [DOI] [PubMed] [Google Scholar]
- 29.Locher K P, Rees B, Koebnik R, Mitschler A, Moulinier L, Rosenbusch J P, Moras D. Cell. 1998;95:771–778. doi: 10.1016/s0092-8674(00)81700-6. [DOI] [PubMed] [Google Scholar]
- 30.Ottemann K M, Xiao W, Shin Y-K, Koshland D E., Jr Science. 1999;285:1751–1754. doi: 10.1126/science.285.5434.1751. [DOI] [PubMed] [Google Scholar]
- 31.Naik U P, Patel P M, Parise L V. J Biol Chem. 1997;272:4651–4654. doi: 10.1074/jbc.272.8.4651. [DOI] [PubMed] [Google Scholar]
- 32.Horwitz A, Duggan K, Buck C, Beckerle M C, Burridge K. Nature (London) 1986;320:531–536. doi: 10.1038/320531a0. [DOI] [PubMed] [Google Scholar]
- 33.Buttery P C, Mallawaarachchi C M, Milner R, Doherty P, ffrench-Constant C. Biochem Biophys Res Commun. 1999;259:121–127. doi: 10.1006/bbrc.1999.0726. [DOI] [PubMed] [Google Scholar]
- 34.Liu K Y, Timmons S, Lin Y Z, Hawiger J. Proc Natl Acad Sci USA. 1996;93:11819–11824. doi: 10.1073/pnas.93.21.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]