Summary
Learning and memory are essential processes of both vertebrate and invertebrate nervous systems that allow animals to survive and reproduce. The neurotransmitter glutamate signals via ionotropic glutamate receptors (iGluRs) that have been linked to learning and memory formation [1, 2]; however, the signaling pathways that contribute to these behaviors are still not well understood. We therefore undertook a genetic and electrophysiological analysis of learning and memory in the nematode Caenorhabditis elegans. Here we show that two genes, nmr-1 and nmr-2, are predicted to encode the subunits of an NMDA-type (NMDAR) iGluR that is necessary for memory retention in C. elegans. We cloned nmr-2, generated a deletion mutation in the gene and show that like nmr-1 [3], nmr-2 is required for in vivo NMDA-gated currents. Using an associative learning paradigm that pairs starvation with the attractant NaCl [4], we also show that the memory of a learned avoidance response is dependent on NMR-1 and NMR-2, and that expression of NMDARs in a single pair of interneurons is sufficient for normal memory. Our results provide new insights into the molecular and cellular mechanisms underlying the memory of a learned event.
Results and Discussion
Associative learning in C. elegans
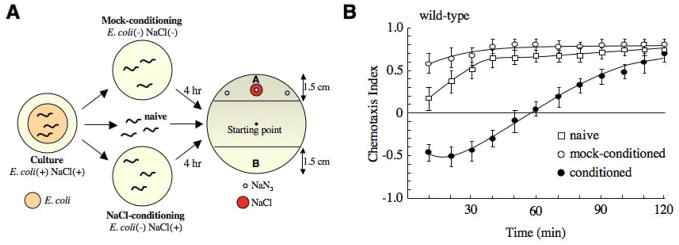
A number of learning paradigms have been developed in C. elegans [5–10], including salt chemotaxis learning where wild-type worms learn to avoid normally attractive NaCl if it is first paired with starvation [4] (Fig. 1). Thus, when tested in a chemotaxis assay, the chemotaxis index (CI) (see Experimental Procedures) of conditioned worms (starved in the presence of NaCl) 10 minutes after conditioning was approximately −0.5 compared to 0.6 observed for mock-conditioned worms (starved in the absence of NaCl). This learned avoidance behavior weakened with time, with most of the worms reaching the source of NaCl two hours post conditioning (CI ~ 0.75) (Fig. 1B). Interestingly, naive worms initially showed a greater avoidance of NaCl (CI ~ 0.2 at 10 minutes) compared to mock-conditioned worms (p<0.01), suggesting that starvation in the absence of salt enhances the attraction to NaCl.
Figure 1.
Salt chemotaxis learning in wild-type worms. (A) Schematic of the salt chemotaxis learning assay. The starting point of naive, mock-conditioned and conditioned worms at the beginning of the chemotaxis assay is indicated. Sodium azide (NaN3) was used to paralyze animals once they reached the source of NaCl. (B) Salt chemotaxis learning behavior in naive (n=5), mock-conditioned (n=5) and conditioned (n=6) wild-type worms.
Several gene products have been implicated in the learning of salt avoidance, including HEN-1, a protein with an LDL motif that is expressed in the bilateral pair of ASE salt-sensing neurons (ASER and ASEL) [11], CASY-1, the orthologue of calsyntenins/alcadeins that is specifically required in ASER [12], as well as proteins involved in the insulin-like signaling pathway [13] and the Go (GOA-1) and Gq (EGL-30) pathway [14]. However, no genes have been described that contribute to the memory of the learned event.
AMPA receptors (AMPARs) and NMDARs have been implicated in learning and memory in many organisms [1, 2]. In vertebrates, neural activity influences the cycling of AMPARs in and out of synapses. This dynamic behavior is thought to modify synaptic strength and may underlie cellular mechanisms of learning and memory, such as long-term potentiation (LTP) and long-term depression (LTD) [15, 16]. Modification of AMPAR trafficking is also thought to regulate synaptic plasticity and thus learning and memory in C. elegans [17] and Aplysia [18]. In addition, NMDARs have been implicated in associative learning and memory in Drosophila [19, 20] and Apis mellifera (honeybee) [21], and disrupting NMDAR function prevents LTP and leads to changes in learning and memory in mice [22–24] and Aplysia [25, 26]. However, linking iGluRs and memory formation to specific cells and neural circuits that control behavior is limited by the tremendous complexity of most nervous systems and the relative difficulty of achieving specific genetic perturbations. To overcome these difficulties, we undertook a genetic analysis of associative learning and memory in C. elegans.
nmr-2 encodes a protein with greatest sequence identity to vertebrate NR2 subunits
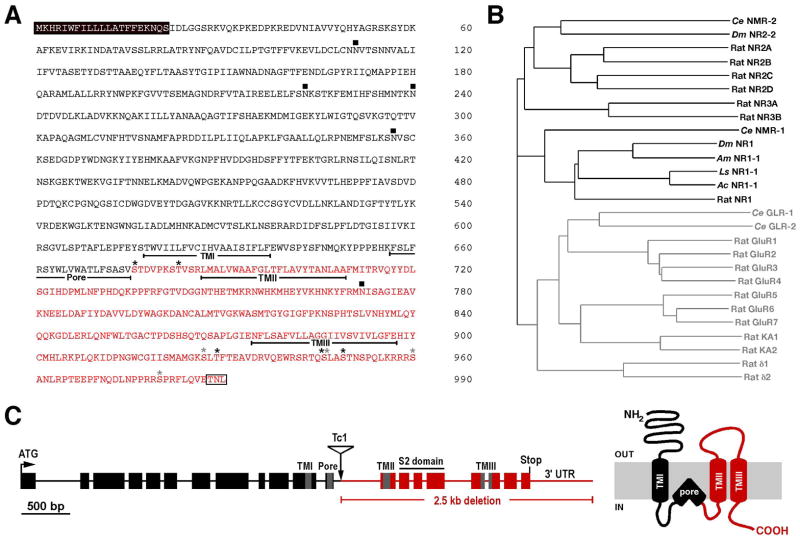
To determine whether glutamatergic signaling is required for salt chemotaxis learning in C. elegans, we first tested the role of the GLR-1 [27, 28] and GLR-2 [29] AMPAR subunits, and the NMR-1 NMDAR subunit [3]. In addition, we cloned and characterized a second gene, nmr-2, encoding a putative NMDAR subunit. The full-length nmr-2 cDNA is predicted to encode a 990 amino acid protein and includes an additional 513 bp compared to that predicted by GENEFINDER analysis [30]. NMR-2 is predicted to have a membrane topology similar to that of other iGluR subunits and greatest sequence identity to vertebrate NR2 subunits (Fig. 2A, B). To study the contribution of NMR-2 to neuronal function, we generated a deletion mutation in nmr-2 using standard techniques; first, screening for insertion of the Tc1 transposon in the nmr-2 locus, then identifying a rare imprecise excision event (Fig. 2C). The nmr-2(ak7) deletion removes approximately 2.5 kb of genomic sequence, including that predicted to encode transmembrane domains II and III and the S2 domain that forms part of the ligand binding pocket (Fig. 2C). Similar to nmr-1(ak4) mutants [3], nmr-2(ak7) mutants were viable and showed no gross defects in locomotion (data not shown).
Figure 2.
nmr-2 encodes a 990 a.a. protein with greatest sequence identity to vertebrate NR2 subunits. (A) Predicted protein sequence encoded by the nmr-2 gene. Indicated are the putative transmembrane domains (underlined); N-linked glycosylation sites (filled squares); PKA (gray asterisks) and PKC (black asterisks) phosphorylation sites; the putative signal sequence (black box); the region deleted by the ak7 mutation (red text); and the putative PDZ-domain binding motif (white box). (B) Phylogenetic tree of C. elegans (Ce), Rattus norvegicus (Rat), Drosophila melanogaster (Dm), Apis mellifera (Am), Aplysia californica (Ac) and Lymnaea stagnalis (Ls) iGluRs. NMDARs are highlighted in black and non-NMDARs in gray text. (C) Genomic organization of the nmr-2 locus with exons and introns represented as boxes and lines, respectively (left). The site of the Tc1 insert is indicated and the region deleted by its imprecise excision is shown in red. The approximate location of the sequence encoding the pore region and TMI-TMIII are highlighted in grey, and the S2 domain coding sequence is shown (black line). The predicted membrane topology of NMR-2 with the region deleted by the ak7 mutation shown in red (right).
nmr-1 and nmr-2 mutants are defective in salt chemotaxis learning
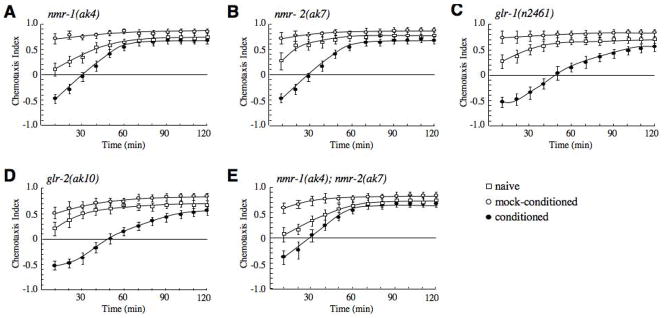
To test the role of both non-NMDA and NMDA iGluRs in learning and memory, we characterized salt chemotaxis learning in glr-1(n2461) [27], glr-2(ak10) [29], nmr-1(ak4) [3], and nmr-2(ak7) mutants. All single mutants showed normal chemotaxis to NaCl in mock-conditioned assays and avoided NaCl just after conditioning (Fig. 3A–D). Interestingly, the nmr-1 and nmr-2 mutants were unable to fully retain the memory of the learned behavior and recovered from the avoidance state (CI = 0 at 30 min) more rapidly than either wild type animals (CI = 0 at 60 min) (Fig. 1B) or AMPAR mutants (CI = 0 at 50 min). We also examined memory retention in double mutants. Worms with either the glr-1(n2461) or glr-2(ak10) mutation in combination with either nmr-1(ak4) or nmr-2(ak7) were not different from either the nmr-1 or nmr-2 single mutants (data not shown). Similarly, the nmr-1(ak4); nmr-2(ak7) double mutant was not significantly different from either single mutant (Fig. 3E). These data suggest that NMDARs, but not AMPA-type non-NMDARs, are required for the memory component of salt chemotaxis learning and that NMR-1 and NMR-2 may combine to form a functional heteromeric NMDAR.
Figure 3.
Retention of avoidance memory is impaired in nmr-1 and nmr-2 mutants. (A–E) Chemotaxis learning in nmr-1(ak4) (n=6) (A), nmr-2(ak7) (n=6) (B), glr-1(n2461) (n=4) (C), glr-2(ak10) (n=5) (D) and nmr-1(ak4); nmr-2(ak7) (n=4) (E). nmr-1 and nmr-2 single mutants, and the nmr-1; nmr-2 double mutant were statistically different from wild-type at 40 min (p<0.001).
nmr-1 and nmr-2 mutants can sense food and starvation
To ensure that the memory defects observed in nmr-1 and nmr-2 mutants were not due to an inability to sense starvation, we tested the behavior of both well-fed and starved mutants in the basal and enhanced slowing response. Sawin et al. [31] showed that well-fed animals move more slowly in the presence of food than in the absence of food (basal slowing response). Furthermore, when starved animals encounter food the slowing response is even greater (enhanced slowing response). Both the basal and enhanced slowing response was normal in the nmr-1(ak4) and nmr-2(ak7) mutants (data not shown) indicating that these worms can normally sense starvation.
NMDA-gated currents are dependent on both nmr-1 and nmr-2
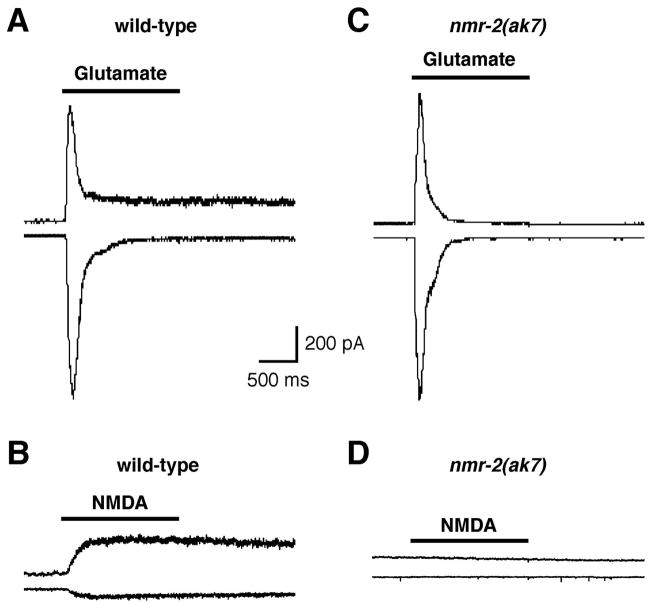
To test the hypothesis that functional NMDARs in C. elegans require both NMR-1 and NMR-2, we measured glutamate- and NMDA-gated currents in AVA interneurons of wild-type worms and nmr-2 mutants. In wild-type worms, glutamate elicited a rapidly activating inward or outward current that quickly desensitized when the membrane potential was held at either −60 mV or +40 mV, respectively (Fig. 4A). This rapid current component is mediated by GLR-1/GLR-2 AMPARs [29]. A smaller, more slowly desensitizing current component that is known to be dependent on NMR-1 [3] was also observed. The slower, outwardly rectifying current could be isolated using the specific agonist NMDA (Fig. 4B). Similar to that found for nmr-1 mutants [3], glutamate elicited a rapidly activating and inactivating current in nmr-2(ak7) worms (Fig. 4C); however, NMDA-gated currents were not observed (Fig. 4D). These data further support the notion that NMR-1 and NMR-2 form a heteromeric NMDAR.
Figure 4.
NMR-2 is required for NMDA-gated currents in the AVA interneuron. Currents in response to 1 mM glutamate (A, C) or 1 mM NMDA (B, D) recorded from the AVA interneuron held at either −60 or +40 mV in either wild-type (A, B) or nmr-2(ak7) (C, D) worms.
NMDARs function in the RIM interneurons to facilitate memory retention
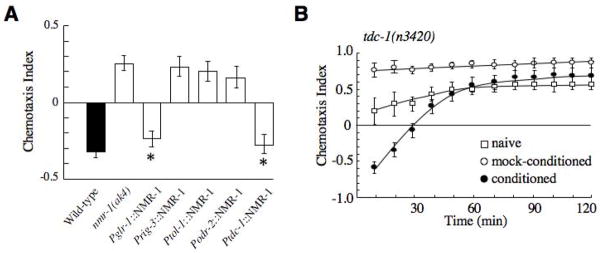
We next determined in which neurons NMDARs function to facilitate the retention of avoidance memory. nmr-1 and nmr-2 are co-expressed in a limited number of neurons [32], including the command interneurons AVA, AVD, AVE and PVC that form part of the neural circuit that regulates both forward and backward movement [33], the RIM interneurons, and the AVG pioneer neuron. We expressed wild-type nmr-1 in a subset of these neurons in transgenic nmr-1(ak4) mutants using cell-specific promoters and tested these worms in the salt chemotaxis learning assay. The behavior was restored in transgenic nmr-1 mutants that expressed nmr-1 under the regulation of the glr-1 promoter that drives expression in all cells that normally express NMDARs (Fig. 5A). However, expressing nmr-1 in either AVA (rig-3 promoter), AVD (tol-1 promoter), or AVG (odr-2 promoter) did not rescue the memory defect of nmr-1(ak4) worms (Fig. 5A). Interestingly, avoidance behavior in transgenic mutants that expressed nmr-1 in the RIM interneurons using the tdc-1 promoter was not significantly different than wild-type worms (Fig. 5A). Together, these data suggest that NMDARs expressed in the RIM interneurons play a crucial role in memory retention in the salt chemotaxis-learning paradigm.
Figure 5.
NMDARs are required in the RIM interneurons to facilitate memory retention. (A) Chemotaxis index 40 minutes post conditioning in wild-type worms (black), nmr-1(ak4) mutants and transgenic nmr-1 mutants (white) that expressed wild-type nmr-1 under the regulation of various cell specific promoters (n=4–5). * Significantly different from nmr-1(ak4), p<0.001. (B) Chemotaxis learning in tdc-1(n3420) mutants (n=6). tdc-1 mutants were statistically different from wild-type at 40 min (p<0.005).
To determine whether RIM synaptic activity is important for memory retention, we assessed salt chemotaxis learning in tdc-1(n3420) mutants. tdc-1 encodes a tyrosine decarboxylase that is expressed in RIM and necessary for both tyramine and octopamine biosynthesis and neurotransmission [34]. Interestingly, tdc-1 mutants showed the same memory retention defects as NMDAR mutants (Fig. 5B), suggesting that signaling downstream of RIM occurs via neurotransmission rather than electrical coupling through gap junctions.
NMR-1 and NMR-2 are essential for memory retention of a learned avoidance behavior
Using C. elegans, we have taken a genetic approach to identify the cellular and molecular requirements for an associative learning behavior. Interestingly, we showed that the NMDAR subunits NMR-1 and NMR-2, but not the GLR-1 and GLR-2 AMPAR subunits, are required for the memory of a learned avoidance response. Thus, in salt chemotaxis learning [4], nmr-1 and nmr-2 single mutants learned to avoid NaCl after starvation conditioning; however, their memory of this association was impaired and chemotaxis toward NaCl recovered more rapidly than in wild-type animals. This is the first evidence that NMDARs are required for memory retention in C. elegans and provides insight into the genes and neural circuits that regulate a fundamental process that is conserved across species.
The NMR-1 and NMR-2 subunits are co-expressed in the same subset of neurons and are predicted to form a functional heteromeric NMDA-type iGluR [32]. In support of this hypothesis, we showed that memory defects of the nmr-1; nmr-2 double mutant were identical to both single mutants, and that like nmr-1 [3], nmr-2 is required for NMDA-gated currents in the AVA interneurons. NMR-1 and NMR-2 are expressed in 5 pairs of interneurons [3, 32] and are required in only one of these, the RIMs, for memory retention of salt avoidance. The RIM interneurons receive input from the ASE salt-sensing neurons via the AIY interneurons. Ablating either AIY or RIM changes the behavior of worms in starved conditions. Wild-type worms transferred to a food free environment initially execute a high frequency of direction changes (reversals), which gradually diminishes over time [35]. In contrast, worms lacking either AIY or RIM maintain a high reversal frequency under starved conditions [36]. Furthermore, modifying reversal behavior has been implicated in navigation processes during taxis behaviors [37–39]. Thus, NMDARs in the RIM interneurons may maintain the association between NaCl and starvation by experience dependent modification of the reversal frequency. We also showed that tdc-1 mutants have the same memory defects as nmr-1 and nmr-2 mutants, suggesting that the signaling pathway downstream of RIM involves either tyramine or octopamine neurotransmission. Interestingly, octopamine has been shown to modulate associative learning in insects [40–43] and our results suggest that similar mechanisms may exist in C. elegans.
NMDARs are thought to facilitate associated learning and memory by acting as coincidence detectors [44]. Thus, activation of vertebrate NMDARs requires two events to happen simultaneously: depolarization of the postsynaptic cell that relieves a voltage dependent Mg2+ block of the channel pore; and ligand-binding to the receptor causing channel opening. NMDA-gated currents in C. elegans are outwardly rectifying consistent with a voltage dependent Mg2+ block on the receptor [3]. Interestingly, although GLR-1 and GLR-2 are expressed in the same neurons as NMR-1 and NMR-2, the GLR-1/GLR-2 AMPARs do not appear to have a central role in salt chemotaxis learning and memory. This suggests that other non-NMDA-type iGluR subunits, e.g., GLR-4 or GLR-5 that are co-expressed with NMR-1 and NMR-2 [32], may have critical roles in these processes. Contrary to salt chemotaxis learning, GLR-1 is necessary for long-term habituation to vibration stimuli, but a role for NMDARs in this form of learning has not been described [17]. Our findings suggest that two independent signaling pathways regulate the memory of these different learning processes – habituation and associative learning. Further genetic analyses of salt chemotaxis learning, including the identification of interacting molecules acting upstream or downstream in the pathway, will help elucidate the neuronal mechanisms of learning and memory acquisition in C. elegans and may lead to a better understanding of these important behaviors in more complex organisms.
Experimental Procedures
General methods and strains
Animals were grown at 20 °C unless otherwise noted. All strains were derivatives of the Bristol strain N2 (wild-type). The mutants used in this study were glr-1(n2461), glr-2(ak10), nmr-1(ak4), nmr-2(ak7), and tdc-1(n3420). Transgenic strains were generated by microinjection to achieve germ-line transformation as previously described [45]. The nmr-2(ak7) deletion mutation was generated by imprecise excision of the Tc1 transposon from the nmr-2 locus. PCR was used to identify Tc1 insertion and subsequent excision. Electrophysiological recordings in vivo from the interneuron AVA were made as previously described [3, 46]. The paired glutamate- and NMDA-gated currents for wild-type and nmr-2(ak7) were recorded from the same AVA neuron.
Salt chemotaxis learning assay
Details of the learning assay have been previously described [4]. The animals were washed with 10 mM MOPS buffer, placed on a conditioning plate (10 mM MOPS-NH40 [pH 7.2], 50 mM NaCl, 3% agar) or a mock-conditioned plate (10 mM MOPS-NH4 [pH 7.2], 3% agar) and incubated at 20 °C for 4 hr. The animals were again collected and chemotaxis was assayed by placing them at the center of a 6 cm plate on chemotaxis agar (10 mM MOPS-NH4 [pH 7.2], 3% agar) on which a salt gradient had been formed for 19–23 hr by placing an agar plug containing 50 mM NaCl at one end of the plate. Thereafter, the number of animals was counted every 10 min for a total of 4 hrs. The chemotaxis index was calculated as previously described [13], (A−B)/(A+B) where A was the number of animals on the NaCl side of the plate and B was the number of animals on the opposite side (Fig. 1A). To account for worms that died or were not able to move, animals that remained at the starting point were not counted. Student’s t test or ANOVA was used to determine statistical significance. Error bars throughout represent the SEM.
Plasmids
The various promoter fusions to nmr-1 coding sequences were constructed using the GATEWAY system (Invitrogen). To construct entry vectors carrying a promoter sequence, the promoter regions were amplified by PCR from C. elegans genomic DNA and then inserted into the pDONR201 vector by site specific recombination. Promoter fragments were: 5.3 kb glr-1; 4 kb rig-3; 5.5 kb tol-1; 5 kb odr-2; and 4.5 kb tdc-1. To generate destination vectors, nmr-1 coding sequences were amplified from first strand cDNA and subcloned into the KpnI sites of the pPDDEST vector. The oligonucleotides used to amplify nmr-1 were 5′-CAGATATGTTCCGAATATCAGTTA-3′ (sense) and 5′-CACATAAAATCTAGTTGATCTTGCT-3′ (antisense). The cosmid T01C3 contains an open reading frame predicted to encode an NMDAR subunit (NMR-2). We identified the authentic 5′ end of nmr-2 by PCR amplification from first strand C. elegans cDNA using spice leader SL1-specific oligonucleotides. Analysis of the predicted NMR-2 protein sequence was done using the ExPASy Proteomics suite of programs [47]. Sequence data for the nmr-2 cDNA is available in GenBank accession number EU588979.
Acknowledgments
We thank the Caenorhabditis Genetics Center for strains, which is funded by the National Institutes of Health: National Center for Resources. We thank A. Fraser and A. Coulson for providing cosmid clones. We are appreciative of the Worm Genome Consortium for providing the C. elegans genome sequence and proteome for its rapid annotation. This research was made possible by support from NIH Grant NS35812 (A.V.M.), by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture and Science of Japan (R.H.), and by KAKENHI (Grant-in-Aid for Scientific Research) on Priority Areas “Systems Genomics” from the Ministry of Education, Culture, Sports, Science and Technology of Japan (Y.I.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lynch MA. Long-term potentiation and memory. Physiol Rev. 2004;84:87–136. doi: 10.1152/physrev.00014.2003. [DOI] [PubMed] [Google Scholar]
- 2.Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 3.Brockie PJ, Mellem JE, Hills T, Madsen DM, Maricq AV. The C. elegans glutamate receptor subunit NMR-1 is required for slow NMDA-activated currents that regulate reversal frequency during locomotion. Neuron. 2001;31:617–630. doi: 10.1016/s0896-6273(01)00394-4. [DOI] [PubMed] [Google Scholar]
- 4.Saeki S, Yamamoto M, Iino Y. Plasticity of chemotaxis revealed by paired presentation of a chemoattractant and starvation in the nematode Caenorhabditis elegans. J Exp Biol. 2001;204:1757–1764. doi: 10.1242/jeb.204.10.1757. [DOI] [PubMed] [Google Scholar]
- 5.de Bono M, Maricq AV. Neuronal substrates of complex behaviors in C. elegans. Annu Rev Neurosci. 2005;28:451–501. doi: 10.1146/annurev.neuro.27.070203.144259. [DOI] [PubMed] [Google Scholar]
- 6.Hedgecock EM, Russell RL. Normal and mutant thermotaxis in the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1975;72:4061–4065. doi: 10.1073/pnas.72.10.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohri A, Kodama E, Kimura KD, Koike M, Mizuno T, Mori I. Genetic control of temperature preference in the nematode Caenorhabditis elegans. Genetics. 2005;169:1437–1450. doi: 10.1534/genetics.104.036111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rankin CH, Beck CD, Chiba CM. Caenorhabditis elegans: a new model system for the study of learning and memory. Behav Brain Res. 1990;37:89–92. doi: 10.1016/0166-4328(90)90074-o. [DOI] [PubMed] [Google Scholar]
- 9.Wen JY, Kumar N, Morrison G, Rambaldini G, Runciman S, Rousseau J, van der Kooy D. Mutations that prevent associative learning in C. elegans. Behav Neurosci. 1997;111:354–368. doi: 10.1037//0735-7044.111.2.354. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Lu H, Bargmann CI. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 2005;438:179–184. doi: 10.1038/nature04216. [DOI] [PubMed] [Google Scholar]
- 11.Ishihara T, Iino Y, Mohri A, Mori I, Gengyo-Ando K, Mitani S, Katsura I. HEN-1, a secretory protein with an LDL receptor motif, regulates sensory integration and learning in Caenorhabditis elegans. Cell. 2002;109:639–649. doi: 10.1016/s0092-8674(02)00748-1. [DOI] [PubMed] [Google Scholar]
- 12.Ikeda DD, Duan Y, Matsuki M, Kunitomo H, Hutter H, Hedgecock EM, Iino Y. CASY-1, an ortholog of calsyntenins/alcadeins, is essential for learning in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2008;105:5260–5265. doi: 10.1073/pnas.0711894105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomioka M, Adachi T, Suzuki H, Kunitomo H, Schafer WR, Iino Y. The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron. 2006;51:613–625. doi: 10.1016/j.neuron.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 14.Matsuki M, Kunitomo H, Iino Y. Goalpha regulates olfactory adaptation by antagonizing Gqalpha-DAG signaling in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2006;103:1112–1117. doi: 10.1073/pnas.0506954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groc L, Choquet D. AMPA and NMDA glutamate receptor trafficking: multiple roads for reaching and leaving the synapse. Cell Tissue Res. 2006;326:423–438. doi: 10.1007/s00441-006-0254-9. [DOI] [PubMed] [Google Scholar]
- 16.Malinow R, Malenka RC Cold Spring Harbor Laboratory, N.Y.U.S.A.m.c.o. AMPA receptor trafficking and synaptic plasticity. Annual review of neuroscience. 2002;25 doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- 17.Rose JK, Kaun KR, Chen SH, Rankin CH. GLR-1, a non-NMDA glutamate receptor homolog, is critical for long-term memory in Caenorhabditis elegans. J Neurosci. 2003;23:9595–9599. doi: 10.1523/JNEUROSCI.23-29-09595.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Q, Roberts AC, Glanzman DL. Synaptic facilitation and behavioral dishabituation in Aplysia: dependence on release of Ca2+ from postsynaptic intracellular stores, postsynaptic exocytosis, and modulation of postsynaptic AMPA receptor efficacy. J Neurosci. 2005;25:5623–5637. doi: 10.1523/JNEUROSCI.5305-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu CL, Xia S, Fu TF, Wang H, Chen YH, Leong D, Chiang AS, Tully T. Specific requirement of NMDA receptors for long-term memory consolidation in Drosophila ellipsoid body. Nat Neurosci. 2007;10:1578–1586. doi: 10.1038/nn2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia S, Miyashita T, Fu TF, Lin WY, Wu CL, Pyzocha L, Lin IR, Saitoe M, Tully T, Chiang AS. NMDA receptors mediate olfactory learning and memory in Drosophila. Curr Biol. 2005;15:603–615. doi: 10.1016/j.cub.2005.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Si A, Helliwell P, Maleszka R. Effects of NMDA receptor antagonists on olfactory learning and memory in the honeybee (Apis mellifera) Pharmacol Biochem Behav. 2004;77:191–197. doi: 10.1016/j.pbb.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 22.Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 23.Morris RG, Anderson E, Lynch GS, Baudry M. Selective impairment of learning and blockade of long-term potentiation by an N-methyl-D-aspartate receptor antagonist, AP5. Nature. 1986;319:774–776. doi: 10.1038/319774a0. [DOI] [PubMed] [Google Scholar]
- 24.Nakazawa K, McHugh TJ, Wilson MA, Tonegawa S. NMDA receptors, place cells and hippocampal spatial memory. Nat Rev Neurosci. 2004;5:361–372. doi: 10.1038/nrn1385. [DOI] [PubMed] [Google Scholar]
- 25.Ezzeddine Y, Glanzman DL. Prolonged habituation of the gill-withdrawal reflex in Aplysia depends on protein synthesis, protein phosphatase activity, and postsynaptic glutamate receptors. J Neurosci. 2003;23:9585–9594. doi: 10.1523/JNEUROSCI.23-29-09585.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy GG, Glanzman DL. Mediation of classical conditioning in Aplysia californica by long-term potentiation of sensorimotor synapses. Science. 1997;278:467–471. doi: 10.1126/science.278.5337.467. [DOI] [PubMed] [Google Scholar]
- 27.Hart AC, Sims S, Kaplan JM. Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature. 1995;378:82–85. doi: 10.1038/378082a0. [DOI] [PubMed] [Google Scholar]
- 28.Maricq AV, Peckol E, Driscoll M, Bargmann CI. Mechanosensory signalling in C. elegans mediated by the GLR-1 glutamate receptor [published erratum appears in Nature 1996 Feb 22;379(6567):749] Nature. 1995;378:78–81. doi: 10.1038/378078a0. [DOI] [PubMed] [Google Scholar]
- 29.Mellem JE, Brockie PJ, Zheng Y, Madsen DM, Maricq AV. Decoding of Polymodal Sensory Stimuli by Postsynaptic Glutamate Receptors in C. elegans. Neuron. 2002;36:933–944. doi: 10.1016/s0896-6273(02)01088-7. [DOI] [PubMed] [Google Scholar]
- 30.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, Burton J, Connell M, Copsey T, Cooper J, et al. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 31.Sawin ER, Ranganathan R, Horvitz HR. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26:619–631. doi: 10.1016/s0896-6273(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 32.Brockie PJ, Madsen DM, Zheng Y, Mellem J, Maricq AV. Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC-42. J Neurosci. 2001;21:1510–1522. doi: 10.1523/JNEUROSCI.21-05-01510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chalfie M, Sulston JE, White JG, Southgate E, Thomson JN, Brenner S. The neural circuit for touch sensitivity in Caenorhabditis elegans. J Neurosci. 1985;5:956–964. doi: 10.1523/JNEUROSCI.05-04-00956.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alkema MJ, Hunter-Ensor M, Ringstad N, Horvitz HR. Tyramine Functions independently of octopamine in the Caenorhabditis elegans nervous system. Neuron. 2005;46:247–260. doi: 10.1016/j.neuron.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 35.Hills T, Brockie PJ, Maricq AV. Dopamine and glutamate control area-restricted search behavior in Caenorhabditis elegans. J Neurosci. 2004;24:1217–1225. doi: 10.1523/JNEUROSCI.1569-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gray JM, Hill JJ, Bargmann CI. A circuit for navigation in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2005;102:3184–3191. doi: 10.1073/pnas.0409009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pierce-Shimomura JT, Morse TM, Lockery SR. The fundamental role of pirouettes in Caenorhabditis elegans chemotaxis. J Neurosci. 1999;19:9557–9569. doi: 10.1523/JNEUROSCI.19-21-09557.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryu WS, Samuel AD. Thermotaxis in Caenorhabditis elegans analyzed by measuring responses to defined Thermal stimuli. J Neurosci. 2002;22:5727–5733. doi: 10.1523/JNEUROSCI.22-13-05727.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zariwala HA, Miller AC, Faumont S, Lockery SR. Step response analysis of thermotaxis in Caenorhabditis elegans. J Neurosci. 2003;23:4369–4377. doi: 10.1523/JNEUROSCI.23-10-04369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farooqui T, Robinson K, Vaessin H, Smith BH. Modulation of early olfactory processing by an octopaminergic reinforcement pathway in the honeybee. J Neurosci. 2003;23:5370–5380. doi: 10.1523/JNEUROSCI.23-12-05370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammer M, Menzel R. Multiple sites of associative odor learning as revealed by local brain microinjections of octopamine in honeybees. Learn Mem. 1998;5:146–156. [PMC free article] [PubMed] [Google Scholar]
- 42.Schwaerzel M, Monastirioti M, Scholz H, Friggi-Grelin F, Birman S, Heisenberg M. Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J Neurosci. 2003;23:10495–10502. doi: 10.1523/JNEUROSCI.23-33-10495.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Unoki S, Matsumoto Y, Mizunami M. Participation of octopaminergic reward system and dopaminergic punishment system in insect olfactory learning revealed by pharmacological study. Eur J Neurosci. 2005;22:1409–1416. doi: 10.1111/j.1460-9568.2005.04318.x. [DOI] [PubMed] [Google Scholar]
- 44.Bourne HR, Nicoll R. Molecular machines integrate coincident synaptic signals. Cell. 1993;72:65–75. doi: 10.1016/s0092-8674(05)80029-7. [DOI] [PubMed] [Google Scholar]
- 45.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. Embo J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Francis MM, Maricq AV. Electrophysiological analysis of neuronal and muscle function in C. elegans. Methods Mol Biol. 2006;351:175–192. doi: 10.1385/1-59745-151-7:175. [DOI] [PubMed] [Google Scholar]
- 47.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]