Abstract
Programmed Death (PD)-1 is a transmembrane protein involved in the regulation of immunological tolerance. Multiple studies have reported an association between high levels of PD-1 expressed on T cell surfaces and exhaustion in lymphocyte populations when challenged by chronic viral infections, such as human immunodeficiency virus (HIV). By using model systems consisting of murine EL4 cells, which constitutively express PD-1, and primary murine CD8 T cells that express PD-1 upon T cell stimulation, we have identified two tissue-specific hypersensitive sites at the 5′ conserved region of the PD-1 locus. Gene reporter assays in CD8 T cells have shown that one of these sites has robust transcriptional activity in response to cell stimulation. Cell treatment with the calcineurin inhibitor cyclosporine A or a Nuclear Factor of Activated T Cells (NFAT)-specific inhibitor led to a sharp reduction in PD-1 expression in the constitutive and inducible systems. Furthermore, analysis of this region by chromatin immunoprecipitation assay revealed NFATc1 binding associated with gene activation in EL4 and primary CD8 T cells. Mutation of the NFATc1 binding site in PD-1 reporter constructs resulted in a complete loss of promoter activity. Together, these results demonstrate that PD-1 gene regulation occurs in part via the recruitment of NFATc1 to a novel regulatory element at the pdcd1 locus and provides the molecular mechanism responsible for the induction of PD-1 in response to T cell stimulation.
Keywords: T Cells, Cell Surface Molecules, Transcription Factors, Gene Regulation
Introduction
Programmed Death (PD)-1 (CD279) is a transmembrane protein that shares homology with the B7/CD28 family of T cell signaling molecules (1). PD-1 interactions with its ligands PD-L1 and/or PD-L2 provides a negative regulatory signal to CD4 and CD8 T cells that results ultimately in a phenotype termed T cell exhaustion (2, 3). Exhausted T cells have lost the ability to combat invading pathogens, synthesize cytokines, and proliferate (4-6). Specific examples include T cell responses towards chronic viral infections caused by human immunodeficiency virus (HIV) and lymphocytic choriomeningitis virus (LCMV) (7-11). Molecular blockade of the interaction between PD-1 and its ligands results in the reinvigoration of the exhausted T cell's effector functions, enabling cytotoxic action and cytokine release, suggesting that the PD-1 pathway is reversible (11, 12).
Depending on the model used, genetic studies showed that deletion of the PD-1 gene (pdcd1) in mice resulted in the manifestation of spontaneous autoimmune diseases, including arthritis, gastritis, and cardiomyopathy (13-15). Other studies have concluded that the PD-1/PD-L1 pathway also plays a role in the onset of diabetes and encephalomyelitis in mice (16, 17). In humans, polymorphisms within pdcd1 have been linked to multiple sclerosis, rheumatoid arthritis, Type 1 diabetes, and systemic lupus erythematosis (18-21). Additionally, aberrant PD-1 function and/or misregulation have been implicated in aiding tumor evasion from the immune system and playing a role in graft-versus-host disease (18-26). These studies suggest that PD-1 is likely to be critically involved in the development and maintenance of peripheral tolerance.
PD-1 expression is restricted to cells of the immune system. Early studies demonstrated that PD-1 was upregulated in thymocyte populations exposed to apoptotic stimuli (27). PD-1 is expressed on thymocytes at the early double negative and double positive stages; however, the full significance of PD-1 expression at these stages is not known. In addition to appearing on the surfaces of exhausted T cells, PD-1 is transiently expressed following T cell activation.
Upon T cell activation, family members of the nuclear factor of activated T cells (NFAT) are dephosphorylated and translocate to the nucleus where they can activate target genes (28). Nuclear translocation of NFAT is abrogated by inhibition of the phosphatase activity of calcineurin with immunophilin-interacting compounds such as cyclosporine A (29-31). Examination of cDNA microarray data comparing exhausted, effector, and memory CD8 T cells showed an increase in NFATc1 (also called NFAT2) expression, suggesting a possibility for its role in regulating PD-1 transcription (32).
Despite PD-1's role in immune responses and its association with disease, little is known about the regulation of this critical gene. Here, an unbiased approach was taken to determine the mechanism of PD-1 regulation during T cell activation as a first step in understanding the regulatory pathway that controls this gene during complex immune responses. The results have identified two 5′ elements that are hypersensitive to DNase I, function together to regulate expression following T cell activation, and when activated, bear marks of transcriptionally active chromatin. We also show that one of these elements binds and uses NFATc1 as its critical factor in regulating PD-1 expression following activation. These studies therefore describe key elements contributing to the transcriptional regulatory mechanism that initiates and defines the PD-1 pathway.
Material and Methods
Cells and Culture
The murine B and T cell lymphoma cell lines, A20 and EL4, were obtained from the American Type Culture Collection (Manassas, VA). Cells were maintained in RPMI 1640 supplemented with 5% fetal bovine serum (Hyclone, Inc., Logan, UT), 5% bovine calf serum (Hyclone), 100 U/ml penicillin/streptomycin, 4.5 g/L glucose, 1.0 mM sodium pyruvate, and 10 mM HEPES. Primary splenocytes were obtained from C57BL/6 mice and CD8 T cells were isolated by negative selection using a magnetic bead purification kit from Miltenyi Biotec, Inc. (Auburn, CA). This process results in CD8 T lymphocyte populations of ∼95 percent purity as determined by flow cytometry. Where indicated cells were treated with the following: 50 ng/ml phorbol 12-myristate 13-acetate (PMA) (Sigma, Inc., St. Louis, MO), 2 mM ionomycin (Sigma), 1 mg/ml cyclosporin A (CsA) (Sigma, Inc.), and 2 mM cell-permeable VIVIT (EMD Biosciences, Inc., San Diego, CA).
Mice
C57BL/6 mice were used for these studies and treated and cared for according to approved protocols.
Real-time RT-PCR analysis
RNA was prepared using the RNeasy kit (Qiagen, Inc., Valencia, CA). Two micrograms of total RNA were used for each reverse transcriptase reaction carried out using the GeneAmp RNA PCR kit (Applied Biosystems, Inc., Foster City, CA). Real-time PCR was used to quantify the amount of PD-1 mRNA and results were normalized to that of 18S ribosomal RNA. Each experiment was performed at least three times and standard error bars were calculated. A list of the primers used in this manuscript can be found in Supplemental Table 1.
DNase I Hypersensitivity Assays
DNase I hypersensitivity assays were carried out as described by Lu et al (33). In brief, nuclei were isolated from 2 × 107 cells and treated with increasing concentrations of DNase I enzyme (Worthington Biochemical, Inc., Lakewood, NJ). Following digestion, genomic DNA was isolated and digested with the appropriate restriction enzymes (NEB, Inc., Ipswich, MA). DNA was subjected to gel electrophoresis and subsequent blotting to a nitrocellulose membrane overnight. Following blotting, membranes were hybridized with a gene-specific radiolabeled probe and the results were visualized by autoradiography.
The protocol for the PCR-based hypersensitivity assay was adapted from Schoenborn et al. (34). Briefly, nuclei were harvested as above, treated with increasing concentrations of DNase I, and genomic DNA was isolated following phenol/chloroform extraction and ethanol precipitation. Real-time PCR was carried out using PCR amplicons spanning portions of the PD-1 locus and relative hypersensitivity was determined by quantifying the level of retained DNA following DNase I treatment. Relative fold change was normalized to samples that received no DNase I treatment and a control fragment that remained refractory to DNase I cleavage. These assays were performed on three individually prepared sets of DNA. The data were averaged and plotted with standard error bars.
Gene Reporter Assays
The pGL3basic plasmid (Promega, Inc., Madison, WI) containing the firefly reporter gene was used in transient transfection transcriptional reporter assays. PCR fragments containing the PD-1 promoter region with conserved region B or both B and C were cloned into the vector using an XhoI restriction site. In all assays, the results were normalized to the signal obtained for an expression vector constitutively expressing the Renilla luciferase gene (pRLTK) that was cotransfected. Transfections were carried out by electroporation in the case of EL4 cells and by nucleofection (Amaxa, Inc., Gaithersburg, MD) for primary CD8 T cells. At least three independent experiments were performed and standard error bars were calculated based on the combined errors from the data obtained in each experiment.
Chromatin Immunoprecipitation (ChIP) Assays
ChIP assays were performed as previously described (35). In brief, chromatin was isolated from 4 × 107 cells. Chromatin was pre-cleared using protein A beads and immunoprecipitation (IP) with appropriate antibodies was carried out overnight. Anti-H3K9Ac, anti-H3K18Ac, and anti-H3K4me3 were purchased from Millipore, Inc. (Billerica, MA). Anti-NFATc1 and anti-NFATc2 were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Following the IP and subsequent washing steps, DNA was isolated and subjected to real-time PCR analysis and quantitated based on a standard curve to that primer set. The values for ChIP immunoprecipitated DNA were normalized to that of input DNA and then plotted as fold over the normalized values of irrelevant antibody control (anti-TCR). Each ChIP analysis was performed a minimum of three times from independent chromatin preparations and standard error bars were calculated.
Electrophoretic Mobility Shift Assays (EMSAs)
Nuclear extracts were prepared from EL4 cells and probes and binding reactions were prepared and carried out as described (36-38). Antibody supershifts were conducted with an anti-NFATc1 antibody (Santa Cruz). DNA protein complexes were resolved on 5% polyacrylamide gels (69:1 :: acrylamide:bisacrylamide) in a taurine-based gel buffer system. Dried gels were exposed and the results were visualized using autoradiography.
Statistics
Student T tests were used to assess the significance of differences in all quantitative results.
Results
PD-1 is transiently induced on activated CD8 T cells and is constitutively expressed in cells exhibiting the exhausted phenotype (8-11). To develop systems in which PD-1 expression could be studied, a number of lymphocytic cell lines were analyzed for expression of PD-1. The EL4 T lymphoma cell line was identified as constitutively expressing high levels of PD-1 mRNA, which could be further induced following stimulation with PMA and ionomycin (Fig. 1a). In contrast, the A20 B lymphoma line was found to be negative for PD-1 expression (Fig. 1a). Additionally, ex vivo isolated CD8 T cells from C57BL/6 mice could achieve significant levels of PD-1 expression following stimulation with PMA and ionomycin (Fig. 1b). In the experiments that follow, the cell lines EL4 and A20 will be used to initially identify and assess PD-1 regulatory elements, while ex vivo stimulated CD8 T cells will be used to confirm the findings in primary cells.
Figure 1. Analysis of PD-1 transcription in murine cell lines and primary T cells.
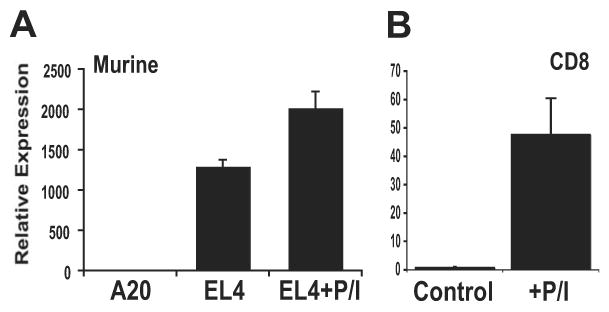
(A) Expression levels of pdcd1 transcripts were determined in cell lines by quantitative reverse transcriptase (qRT)-PCR. Where indicated, EL4 cells were treated with PMA/ionomycin overnight (+P/I). Expression levels were normalized to qRT-PCR values obtained for 18s ribosomal RNA. (B) CD8 T cells were isolated as described under Materials and Methods and left untreated or treated with PMA/ionomycin overnight. Following incubation, the cells were harvested and the levels of PD-1 mRNA were determined as above. The data are expressed as fold PD-1 expression over untreated/control CD8 T cells. Each experiment was performed a minimum of three times and standard error bars were calculated.
Two hypersensitive sites are located in the 5′ conserved regions of PD-1
Complex gene regulation involves the combinatorial action of cis-acting elements, including promoters, enhancers, locus control regions, and boundary elements. To identify potential conserved regulatory elements encoded in the PD-1 locus, the University of California Santa Cruz genome browser (www.genome.ucsc.edu) was employed to screen for regions that encode high mammalian sequence homology. Two highly conserved regions located 5′ to the transcriptional start site were identified through BLAST homology sequences and were termed conserved regions B (CR-B) and C (CR-C) with CR-A representing the first exon of PD-1 (Fig. 2a).
Figure 2. Two DNase I hypersensitive sites are located in 5′ conserved regions of the pdcd1 locus.
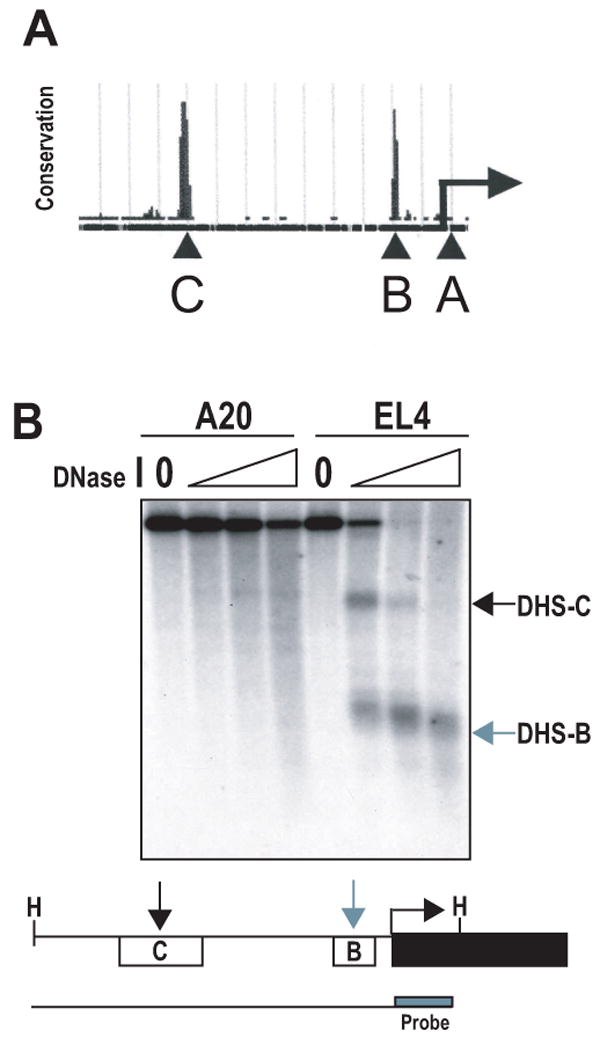
(A) The UCSC genome browser was used to identify regions of high DNA sequence conservation in the region 5′ of the PD-1 transcriptional start site (arrow). Two regions were identified and termed B and C (as seen labeled above with capital letters). (B) Nuclei from EL4 and A20 cells were isolated and treated with increasing concentrations of DNase I. Genomic DNA was obtained and digested with HindIII (H) to generate the full-length genomic DNA fragment (line below schematic). Digested DNA was then subjected to Southern blot analysis with a probe specific for the 3′ end of the HindIII fragment (box labeled in schematic). The resulting DNase I hypersensitive sites (DHS) map to conserved regions B and C located 5′ of the pdcd1 gene and are indicated by arrows.
Sensitivity of intact chromatin to DNase I digestion is a hallmark of accessible chromatin and potential regulatory function. To ascertain the significance of the conserved sequences, DNase I hypersensitivity assays were performed on A20 (PD-1 negative) and EL4 (PD-1 positive) lymphocytes. The results highlighted two DNase I hypersensitive regions (termed DHS-B and DHS-C), which were located within four kilobases of the transcriptional start site. These sites mapped to CR-B and CR-C (Fig. 2b). Importantly, the hypersensitive sites were present in the PD-1 expressing cell line, EL4, but not in the null cell line, A20, suggesting that these conserved regions may regulate PD-1 expression.
To evaluate these sites in primary CD8 T cells, a quantitative-PCR based assay was employed (34) to analyze DNase I hypersensitivity in ex vivo isolated cells where total cell numbers can be limiting. In this assay, DNase I hypersensitivity is measured by the inability to amplify a region of DNA following purification from nuclei treated with nuclease. A reduction or loss of PCR product/signal signifies that a region was sensitive to DNase I. As compared to unstimulated ex vivo isolated CD8 T cells, amplicons spanning CR-B and CR-C were diminished significantly following ex vivo stimulation (Fig. 3b, c). The intensity of the control amplicon remained relatively unchanged following stimulation.
Figure 3. DNase I hypersensitive sites are located in the 5′ conserved region of the pdcd1 locus in primary CD8 T cells expressing PD-1.
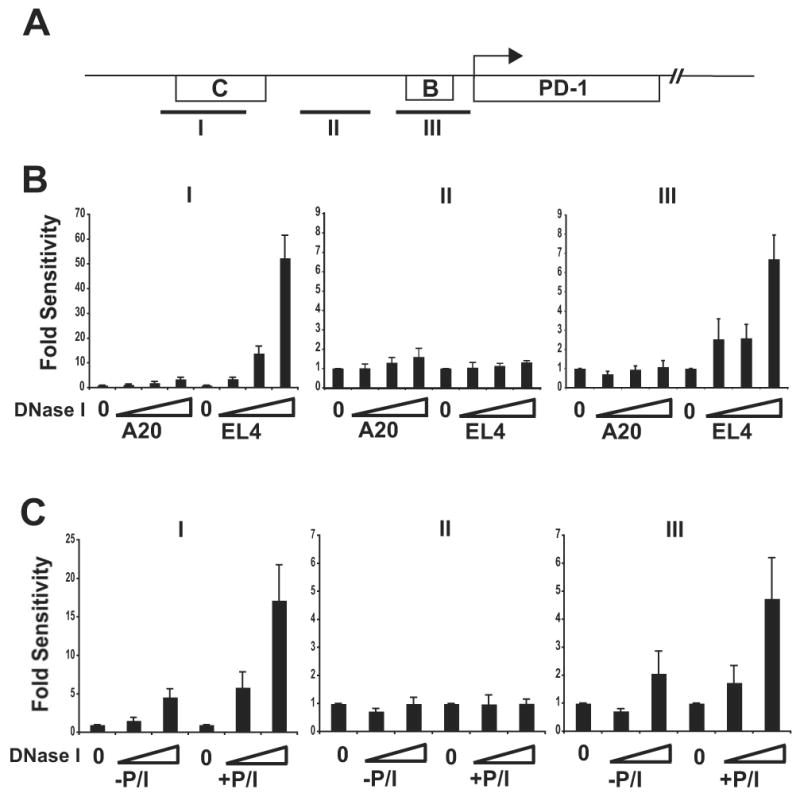
(A) A schematic of conserved regions of PD-1 and primer set amplicons are shown. (B) Nuclei from A20, EL4, and (C) primary CD8 T (+/- PMA/ionomycin) cells were isolated and treated with increasing concentrations of DNase I. Genomic DNA was isolated and subjected to real-time PCR analysis with primer sets (I, II, and III) spanning portions of the pdcd1 gene. Relative hypersensitivity was assayed by normalization of DNase I treated DNA to that of the genomic (untreated) DNA control and the data are plotted as fold sensitivity. Increasing DNase I concentration is indicated along the X-axis. The data shown are the average (-/+ standard error) of three independent experiments.
Conserved regions B and C encode transcriptionally active sequences
To determine the functional relevance of the regions encoding the hypersensitive sites identified by DNase I hypersensitivity analysis, luciferase promoter reporter vectors containing the PD-1 promoter region with either CR-B or CR-B and C were constructed and transiently transfected into A20 and EL4 cells (Fig. 4). Cells transfected with CR-B induced a moderate level of luciferase activity in both cell types, while a more robust level of promoter activity was seen only in EL4 cells transfected with CR-B and C. In contrast, the addition of CR-C to CR-B led to a notable decrease in transcriptional activity of the reporter vector in A20 cells. In conjunction with the data in Figures 2 and 3, these results strongly suggest that conserved regions B and C contain regulatory elements necessary for PD-1 promoter activity in EL4 cells.
Figure 4. Conserved regions B and C encode transcriptionally active sequences.
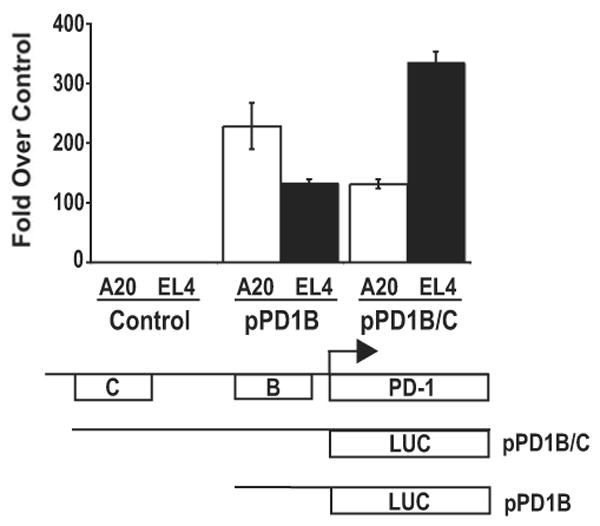
Luciferase reporter gene (pGL3) constructs containing conserved region B, B and C, or vector alone were transiently cotransfected into EL4 or A20 cells with a constitutive renilla luciferase expression vector. Following 24 hours, cells were lysed and assayed for luciferase activity as described in Materials and Methods. Samples were normalized to the renilla luciferase activity and the average values from three independent experiments were plotted as fold over vector alone.
Active chromatin modifications are present at the PD-1 regulatory regions and promoter in PD-1 expressing cells
Induction of reporter activity in the PD-1 negative A20 cells suggested that an additional mechanism(s) might play a role in the regulation of PD-1. Transcriptional regulatory regions are associated with a combination of specific posttranslational modifications on their nucleosomes (39). These modifications are indicative of the potential activation state of the local chromatin structure. To identify such marks for PD-1, ChIP assays for histone modifications associated with gene expression were carried out in PD-1 expressing and non-expressing cell lines (Fig. 5a). Compared to A20 cells, the upstream sequences of the PD-1 gene in EL4 cells displayed high levels of histone acetylation at lysines 9 and 18 of histone 3 (H3K9Ac and H3K18Ac). Histone acetylation levels were highest at CR-B, but still robust at CR-C and the sequences separating the two regions. Histone H3 lysine 4 trimethylation (H3K4me3) marks were highest at CR-B, the site closest to the transcriptional start site. This latter mark is often associated with actively transcribing genes, and typically peaks at transcription start sites (40).
Figure 5. High levels of H3 acetylation and H3K4 trimethylation are found at the PD-1 regulatory regions in PD-1 expressing cells.
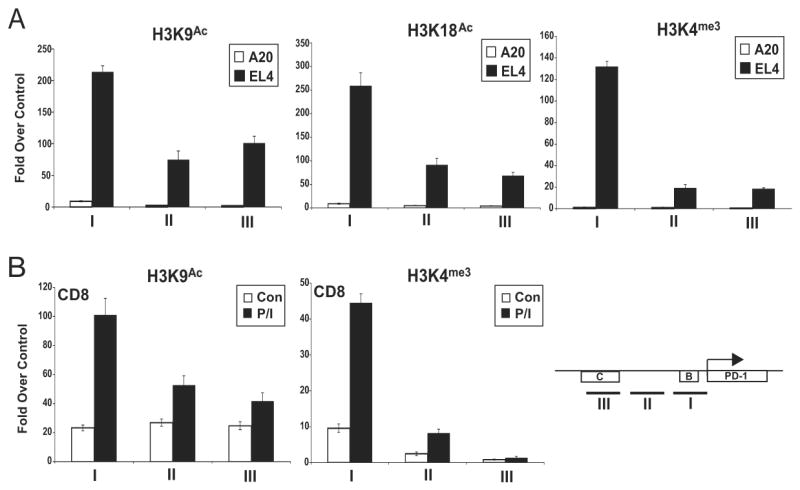
Chromatin was isolated from (A) A20 and EL4; or (B) primary CD8 T cells (Con) and primary CD8 T cells treated with PMA/ionomycin (P/I). ChIP assays were conducted with the indicated antibodies, and quantitative real-time PCR was used to determine the levels of chromatin modifications at CR-B, the intervening region between CR-B and CR-C and CR-C. The locations of the PCR amplicons are shown in the schematic. Quantitative PCR results were normalized to input DNA amounts and plotted as fold over an irrelevant antibody control (anti-TCR). The results presented were the average (-/+ standard error) of three independent experiments.
To examine the chromatin structure of the PD-1 locus in primary cells, CD8 T cells were isolated from the spleens of C57BL/6 mice and ChIP assays were performed with chromatin obtained from PMA/ionomycin treated and untreated cells. High levels of H3K9Ac and H3K4me3 could be detected in the CD8 T cells stimulated to produce PD-1 when compared to untreated controls (Fig. 5b). These data indicate that the 5′ upstream regions of the pdcd1 locus exhibit hallmarks of active genes in both cell lines and primary CD8 T cells.
NFAT inhibition results in a reduction of transcriptional activation and PD-1 expression
Several pieces of information suggested that NFAT might be involved in PD-1 expression. First, agents that block T cell activation such as the NFAT inhibitor cyclosporine A (CsA) have previously been found to cause a decrease in PD-1 expression in mice undergoing transplantation responses (41). Second, transcripts of NFATc1 were observed at higher levels in exhausted CD8 T cells when compared to naïve, effector, and memory phenotypes (32). Lastly, sequence analysis of the 5′ upstream region of PD-1, including CR-B and CR-C, using Genomatix (www.genomatix.de) revealed the occurrence of numerous potential NFAT binding sites. To begin to examine a role for NFAT in PD-1 regulation, CsA was employed. CsA inhibits the transcription of NFAT regulated genes by preventing the dephosphorylation of NFAT by calcineurin and its subsequent translocation to the nucleus (28-31). EL4 cells, which constitutively produce PD-1, displayed a robust decrease in transcription after CsA treatment (Fig. 6a). Similarly, primary CD8 T cells, which produce PD-1 upon stimulation with PMA/Ionomycin, showed a marked reduction of expression when treated with CsA (Fig. 6a). Because CsA inhibits calcineurin function, there was the possibility that the inhibitory effect was not directly attributable to NFAT. To circumvent this issue, a cell permeable form of the NFAT specific inhibitor peptide VIVIT was used (42). The results showed that VIVIT specifically inhibited the induction of PD-1 in both EL4 and primary CD8 T cells (Fig. 6a).
Figure 6. NFAT inhibition results in a reduction of PD-1 transcription.
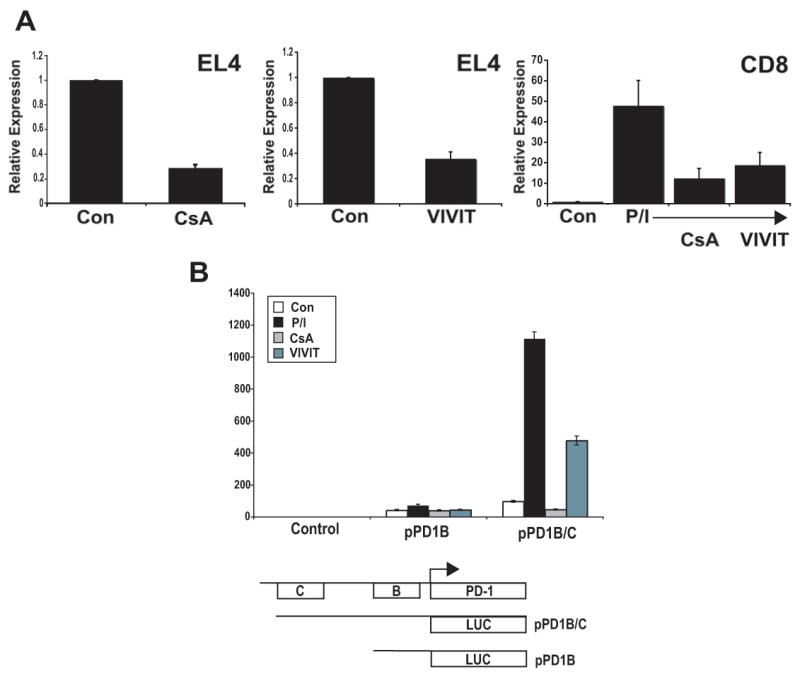
(A) NFAT inhibitors reduce PD-1 expression. EL4 cells and primary CD8 T cells (Control, Con; and PMA/Ionomycin treated, P/I) were treated with CsA and VIVIT for 2 days and 5 days, respectively. Following cell treatment, total RNA was harvested and the level of pdcd1 transcripts determined as in Figure 1. (B) NFAT activity is required for activation of reporter genes encoding CR-C. Luciferase reporter gene (pGL3) constructs containing conserved region B, B and C, or vector alone were transiently cotransfected into EL4 cells with a constitutive renilla luciferase expression vector. Transfected cells were then left untreated, treated with PMA/ionomycin (P/I) alone or in combination with CsA or VIVIT. Following overnight incubation, cells were lysed and assayed for luciferase activity as in Figure 4. These data represent an average of at least three independent experiments.
To further define the role of T cell stimulation and NFAT inhibitors on PD-1 expression, EL4 cells were transfected with luciferase promoter reporter vectors containing either CR-B or CR-B and C and the transcriptional response of these constructs to PMA/ionomycin, CsA, and VIVIT was assessed (Fig. 6b). While CR-B displayed little response to PMA/ionomycin, CsA, or VIVIT, a robust increase in transcriptional activation was observed with the reporter vector containing CR-B and CR-C upon cell treatment with PMA/ionomycin. Importantly, this increase was attenuated upon addition of CsA or VIVIT. Taken together, these results suggest that PD-1 may be an NFAT regulated gene.
NFATc1 binds to a PD-1 regulatory element in vitro and in vivo
The above data implicate NFAT as having a role in the regulation of PD-1 expression. However, which NFAT family member was responsible and whether this role was through direct binding to the locus or indirect by influencing the regulation of another trans-acting factor was unclear. Software analysis of potential transcription factor binding sites (Genomatix) identified five NFAT consensus-binding sites within the 5′ conserved region of the PD-1 gene. Notably, several of these sites fall within CR-B and CR-C. To provide a direct association between NFAT and PD-1 expression, ChIP assays for NFAT family members NFATc1 and NFATc2 were carried out across the upstream portion of the pdcd1 locus. Assays conducted using EL4 cells treated with PMA/ionomycin to ensure full NFAT induction demonstrated that NFATc1 (NFAT2) binds to CR-C in EL4 cells (Fig. 7a) and that NFATc2 (NFAT1) does not bind to any of the sequences tested (Fig. 7b and data not shown). The location of the ChIP amplicon that was bound by NFATc1 was closest to DHS-C. To determine if NFATc1 bound to DHS-C in primary CD8 T cells, ChIP assays were carried out on ex vivo stimulated primary CD8 T cells. As with EL4 cells, NFATc1 binding was limited to PD-1 expressing cells, occurred only within CR-C (Fig. 7a), and NFATc2 binding was not detected (Fig. 7b).
Figure 7. NFATc1 binds to the CR-C regulatory element in vivo and in vitro.
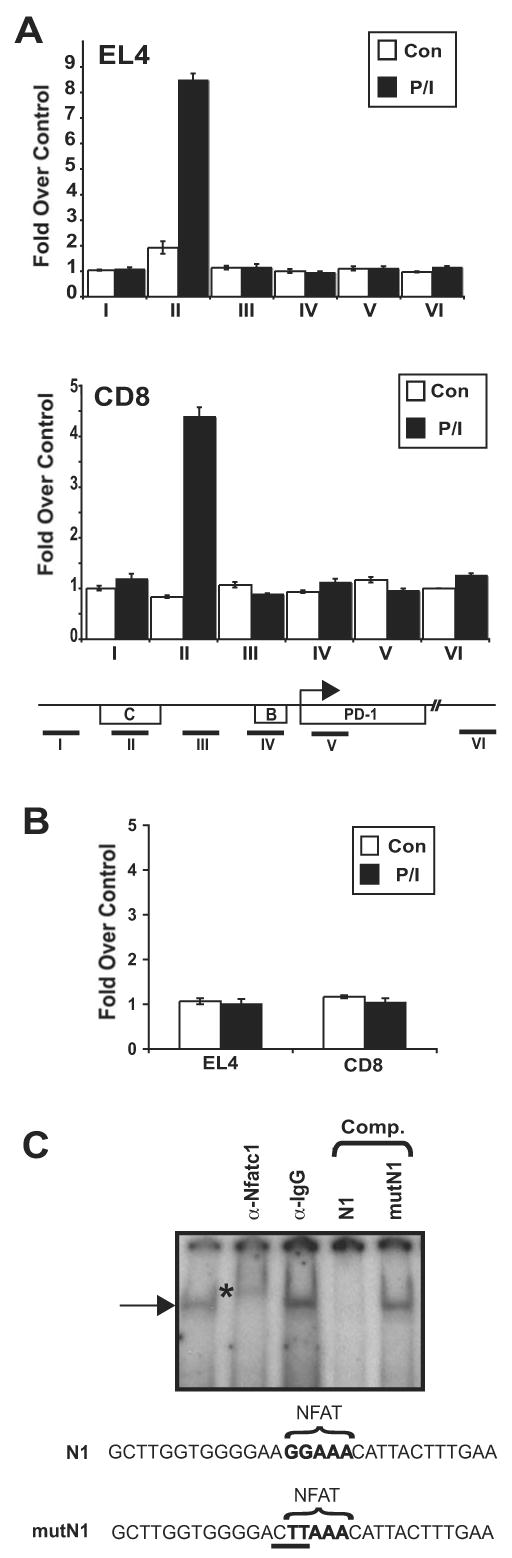
ChIP was performed on EL4 and CD8 T cells with antisera specific to (A) NFATc1, (B) NFATc2, or an irrelevant antiserum (anti-TCR). Cells were either untreated (con) or treated with PMA/Ionomycin (P/I) as indicated. Quantitative real-time PCR was used to determine the levels of NFATc1 binding at locations throughout the 5′ conserved region in each cell type. The locations of the PCR amplicons are shown on the schematic below the figures. PCR results were normalized to input DNA amounts and plotted as fold over an irrelevant antibody control (anti-TCR). The results shown represent an averaging of three independent experiments. In (B), data are shown for PCR amplicon II. (C) EL4 nuclear extracts from cells treated with PMA/ionomycin were incubated with the N1 NFAT-encoding binding site probe and antibodies where indicated. DNA binding competition assays were carried out with unlabeled N1 DNA or the same sequence containing a mutated NFAT binding site (mutN1). An asterisk marks the location of the anti-NFATc1 super-shifted complex.
To determine if NFATc1 binding to the above identified NFAT consensus binding site could be detected in vitro, electrophoretic mobility shift assays (EMSA) using nuclear extracts from EL4 cells stimulated with PMA/ionomycin were performed with a probe containing the portion of DHS-C encompassing the NFAT binding site (Fig 7c). A protein/DNA complex was observed using the NFAT binding site from CR-C (termed N1). This complex was supershifted using an NFATc1 antibody but not by an irrelevant antiserum (Fig 7c). Importantly, visualization of the N1 complex was abrogated by competition with unlabeled N1 DNA but not with a mutant N1 DNA sequence. These results demonstrate that PD-1 is regulated in part by the direct binding of NFATc1 to DHS-C located within conserved regulatory region C.
The N1 NFAT cis-element is necessary for PD-1 transcriptional regulation
To demonstrate that the N1 element is necessary for PD-1 promoter activity, we mutated the NFATc1 consensus binding site located in DHS-C and performed reporter assays in primary CD8 T cells. When compared to its wild-type counterpart, the mutated reporter showed a complete loss of promoter activity following cell stimulation (Fig. 8). Importantly, a reporter construct carrying a mutation of another consensus NFAT binding site within CR-C showed no loss of promoter activity. A construct carrying mutations at both sites showed a loss of transcriptional activation similar to the N1 mutation alone. These results suggest that the NFAT site within CR-C is critical to the expression of PD-1.
Figure 8. NFATc1 binding is necessary for PD-1 transcriptional regulation.
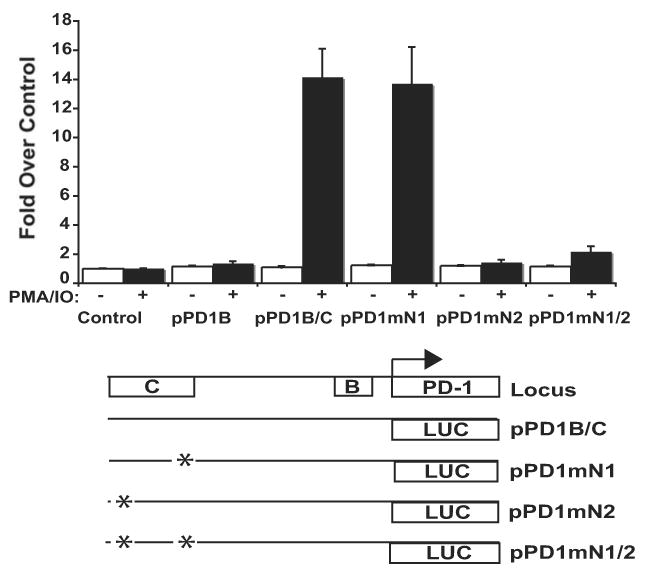
PD-1 promoter reporter constructs (pGL3) containing vector alone, CR-B, CR-B and -C, or CR-B and -C with the indicated NFAT site mutation were transiently transfected into primary CD8 T cells using nucleofection. Cells were left untreated or treated with PMA/ionomycin for six hours. Following incubation, cells were lysed and assayed for luciferase activity as above. The results presented represent the average of three independent transfection assays.
Discussion
The regulation and function of PD-1 has profound implications on how the mammalian immune system resolves infection and disease and at the same time governs tolerance. Disruption of the balance between these critical tasks can have consequences in the maintenance of a healthy and active immune system. Here, we have identified both cis- and trans-acting elements involved in the transcriptional regulation of PD-1 that are associated with CD8 T cell activation.
CR-B and CR-C, which were initially identified due to sequence homology are the centers of strong DNase I hypersensitive regions. The use of cross species homology searches has served as an outstanding tool for the initial identification of possible regulatory sequences (43-45). DNase I hypersensitivity assays provide an unbiased approach for the identification of potential regions. While the former approach is effective without respect to cell type, a positive identification of a site in the latter is completely dependent on the chromatin configuration of the region being assayed. Thus, it is critical to be able to compare expressing and non-expressing cells as well as to verify that the results observed in cell lines is portrayed in primary cells. This was the case for the DNase I hypersensitive sites analyzed. Each DNase hypersensitive site was observed only in PD-1 expressing cell lines and in primary CD8 T cells that were activated to express PD-1. Consistent with these findings was the fact that only in PD-1 expressing cells CR-B, CR-C, and the intervening sequence displayed histone acetylation marks that were indicative of activated genes (40). Additionally in PD-1 expressing cells, H3K4me3, a mark that is often associated with the transcription start site of actively transcribing genes displayed its highest levels at CR-B, the site closest to the promoter of PD-1 (40). Conversely, the absence of these activation marks in A20 cells indicates that epigenetic gene silencing is likely involved in PD-1 regulation. This could explain why a modest level of transcriptional activation was observed upon transfection of CR-B/CR-C containing reporter constructs despite the fact that A20 cells do not express PD-1.
CR-B was able to enhance expression and in combination with CR-C displayed a robust transcriptional activation function in EL4 cells. In contrast, when constructs containing both CR-B and CR-C were transfected into A20 cells, a decrease in transcriptional activation was observed. This finding suggests the possibility that both positive and negative regulatory elements are present in CR-C and that regulation of PD-1 expression by these elements occurs in a tissue-specific manner. Intriguingly, CR-C was not able to drive transcription independently of CR-B (data not shown), suggesting that the CR-C is not an independent element and requires additional interactions (such as those with CR-B) to be fully active. Thus, CR-C does not have the independent activity ascribed to enhancer regions. However, CR-C was essential for responses to T cell activation signals. Because these signals function through calcium mobilization pathways, it was logical that CsA and other NFAT inhibitors may function either directly or indirectly on PD-1 expression. The finding that NFATc1 binds to native PD-1 chromatin places a direct link between NFATc1 and PD-1 expression in response to T cell activating signals.
NFAT typically interacts with other factors to control transcription of its target genes (46-49). Genes regulated by this mechanism include the NFAT mediated activation and repression of the IL-2 gene, as well as the activation of CTLA4. By choosing different partner proteins, NFAT can repress as well as activate transcription (47). For example, the T regulatory cell transcription factor Foxp3 has been shown to partner with NFAT to repress IL-2 transcription, while at the same time promoting CTLA4 expression. An initial screen using the ChIP assay to find NFAT partners, including AP-1 and NF-κB family members at CR-C did not yield a positive result (data not shown). Thus, a role for these factors in regulating PD-1 is inconclusive at this point. Additionally, it is possible that a trans-acting factor(s) working with or independently of NFAT is responsible for the transcription activity of DHS-B seen in EL4 cells.
PD-1 expression is transiently induced following the initial encounter between T cell and antigen. The data presented here demonstrate that NFATc1 is a critical factor in promoting the initial induction of PD-1 expression following T cell activation. Thus, the question of whether NFATc1 and/or CR-C are required for PD-1 expression in the exhausted state is of interest. We suggest that the NFATc1 mediated pathway is likely involved in the expression of PD-1 in the exhausted cell setting, as continued exposure to antigen and repetitive cell stimulation is required to maintain the exhausted phenotype and PD-1 expression. (12). These same stimuli should lead to continual nuclear translocation of NFAT and subsequent NFAT-mediated gene transcription. In support of this argument, EL4 cells have been identified as having a mutant calcineurin gene that results in the constitutive dephosphorylation and activation of NFAT (50). Thus, in a way this mutation may mimic a persistent cell activation state that results in T lymphocyte exhaustion. Furthermore, comparative cDNA microarray analysis between naïve, effector, and exhausted CD8 T cells identified NFATc1 as an upregulated gene, suggesting that it plays a role in this phenotype (32).
Here we have identified cis- and trans-acting factors responsible for promoting the induction of PD-1 expression. The identification of two novel regulatory regions provides an excellent platform to further define these known and other unknown cis-acting elements involved in PD-1 regulation. Additionally, the association of NFATc1 hints at a number of other possible trans-acting factors necessary for governing PD-1 expression. These questions not withstanding, the identification of the cis-acting elements and trans-acting factors presented here are the first steps in understanding the mechanism of PD-1 regulation in disease. Ultimately, the illumination of the mechanisms of action governing PD-1 gene regulation will provide insight into novel pathways of disease pathogenesis and tumor invasion, potentially identifying therapeutic strategies to treat a variety of disorders, including chronic viral infection, cancer, autoimmune disease, and graft-versus-host disease.
Supplementary Material
Acknowledgments
We thank Drs. Benjamin Youngblood and Melinda Horne for assistance and comments on this work.
Abbreviations
- ChIP
Chromatin Immunoprecipitation
- CR
Conserved Region
- CsA
Cyclosporine A
- DHS
DNase I Hypersensitive Site
- PD-1
Programmed Death-1
Footnotes
This work was supported by a grant from the Concerned Parents for AIDS Research. KJO was supported by a Ruth Kirschstein Institutional Postdoctoral Training Award from The NIH T32 AI007610.
Disclosures The authors have no financial conflict of interest.
References
- 1.Agata Y, Kawasaki A, Nishimura H, Ishida Y, Tsubata T, Yagita H, Honjo T. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int Immunol. 1996;8:765–772. doi: 10.1093/intimm/8.5.765. [DOI] [PubMed] [Google Scholar]
- 2.Sharpe AH, Wherry EJ, Ahmed R, Freeman GJ. The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nat Immunol. 2007;8:239–245. doi: 10.1038/ni1443. [DOI] [PubMed] [Google Scholar]
- 3.Okazaki T, Honjo T. The PD-1-PD-L pathway in immunological tolerance. Trends Immunol. 2006;27:195–201. doi: 10.1016/j.it.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Wherry EJ, Barber DL, Kaech SM, Blattman JN, Ahmed R. Antigen-independent memory CD8 T cells do not develop during chronic viral infection. Proc Natl Acad Sci U S A. 2004;101:16004–16009. doi: 10.1073/pnas.0407192101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wherry EJ, Blattman JN, Ahmed R. Low CD8 T-cell proliferative potential and high viral load limit the effectiveness of therapeutic vaccination. J Virol. 2005;79:8960–8968. doi: 10.1128/JVI.79.14.8960-8968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klenerman P, Hill A. T cells and viral persistence: lessons from diverse infections. Nat Immunol. 2005;6:873–879. doi: 10.1038/ni1241. [DOI] [PubMed] [Google Scholar]
- 7.Trabattoni D, Saresella M, Biasin M, Boasso A, Piacentini L, Ferrante P, Dong H, Maserati R, Shearer GM, Chen L, Clerici M. B7-H1 is up-regulated in HIV infection and is a novel surrogate marker of disease progression. Blood. 2003;101:2514–2520. doi: 10.1182/blood-2002-10-3065. [DOI] [PubMed] [Google Scholar]
- 8.Trautmann L, Janbazian L, Chomont N, Said EA, Gimmig S, Bessette B, Boulassel MR, Delwart E, Sepulveda H, Balderas RS, Routy JP, Haddad EK, Sekaly RP. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat Med. 2006;12:1198–1202. doi: 10.1038/nm1482. [DOI] [PubMed] [Google Scholar]
- 9.Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, Mncube Z, Duraiswamy J, Zhu B, Eichbaum Q, Altfeld M, Wherry EJ, Coovadia HM, Goulder PJ, Klenerman P, Ahmed R, Freeman GJ, Walker BD. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 10.Petrovas C, Casazza JP, Brenchley JM, Price DA, Gostick E, Adams WC, Precopio ML, Schacker T, Roederer M, Douek DC, Koup RA. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. J Exp Med. 2006;203:2281–2292. doi: 10.1084/jem.20061496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freeman GJ, Wherry EJ, Ahmed R, Sharpe AH. Reinvigorating exhausted HIV-specific T cells via PD-1-PD-1 ligand blockade. J Exp Med. 2006;203:2223–2227. doi: 10.1084/jem.20061800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 13.Nishimura H, Minato N, Nakano T, Honjo T. Immunological studies on PD-1 deficient mice: implication of PD-1 as a negative regulator for B cell responses. Int Immunol. 1998;10:1563–1572. doi: 10.1093/intimm/10.10.1563. [DOI] [PubMed] [Google Scholar]
- 14.Nishimura H, Nose M, Hiai H, Minato N, Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 1999;11:141–151. doi: 10.1016/s1074-7613(00)80089-8. [DOI] [PubMed] [Google Scholar]
- 15.Nishimura H, Okazaki T, Tanaka Y, Nakatani K, Hara M, Matsumori A, Sasayama S, Mizoguchi A, Hiai H, Minato N, Honjo T. Autoimmune dilated cardiomyopathy in PD-1 receptor-deficient mice. Science. 2001;291:319–322. doi: 10.1126/science.291.5502.319. [DOI] [PubMed] [Google Scholar]
- 16.Ansari MJ, Salama AD, Chitnis T, Smith RN, Yagita H, Akiba H, Yamazaki T, Azuma M, Iwai H, Khoury SJ, Auchincloss H, Jr, Sayegh MH. The programmed death-1 (PD-1) pathway regulates autoimmune diabetes in nonobese diabetic (NOD) mice. J Exp Med. 2003;198:63–69. doi: 10.1084/jem.20022125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salama AD, Chitnis T, Imitola J, Ansari MJ, Akiba H, Tushima F, Azuma M, Yagita H, Sayegh MH, Khoury SJ. Critical role of the programmed death-1 (PD-1) pathway in regulation of experimental autoimmune encephalomyelitis. J Exp Med. 2003;198:71–78. doi: 10.1084/jem.20022119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kroner A, Mehling M, Hemmer B, Rieckmann P, Toyka KV, Maurer M, Wiendl H. A PD-1 polymorphism is associated with disease progression in multiple sclerosis. Ann Neurol. 2005;58:50–57. doi: 10.1002/ana.20514. [DOI] [PubMed] [Google Scholar]
- 19.Hatachi S, Iwai Y, Kawano S, Morinobu S, Kobayashi M, Koshiba M, Saura R, Kurosaka M, Honjo T, Kumagai S. CD4+ PD-1+ T cells accumulate as unique anergic cells in rheumatoid arthritis synovial fluid. J Rheumatol. 2003;30:1410–1419. [PubMed] [Google Scholar]
- 20.Nielsen C, Hansen D, Husby S, Jacobsen BB, Lillevang ST. Association of a putative regulatory polymorphism in the PD-1 gene with susceptibility to type 1 diabetes. Tissue Antigens. 2003;62:492–497. doi: 10.1046/j.1399-0039.2003.00136.x. [DOI] [PubMed] [Google Scholar]
- 21.Prokunina L, Castillejo-Lopez C, Oberg F, Gunnarsson I, Berg L, Magnusson V, Brookes AJ, Tentler D, Kristjansdottir H, Grondal G, Bolstad AI, Svenungsson E, Lundberg I, Sturfelt G, Jonssen A, Truedsson L, Lima G, Alcocer-Varela J, Jonsson R, Gyllensten UB, Harley JB, Alarcon-Segovia D, Steinsson K, Alarcon-Riquelme ME. A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nat Genet. 2002;32:666–669. doi: 10.1038/ng1020. [DOI] [PubMed] [Google Scholar]
- 22.Dong H, Chen L. B7-H1 pathway and its role in the evasion of tumor immunity. J Mol Med. 2003;81:281–287. doi: 10.1007/s00109-003-0430-2. [DOI] [PubMed] [Google Scholar]
- 23.Iwai Y, Ishida M, Tanaka Y, Okazaki T, Honjo T, Minato N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc Natl Acad Sci U S A. 2002;99:12293–12297. doi: 10.1073/pnas.192461099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohigashi Y, Sho M, Yamada Y, Tsurui Y, Hamada K, Ikeda N, Mizuno T, Yoriki R, Kashizuka H, Yane K, Tsushima F, Otsuki N, Yagita H, Azuma M, Nakajima Y. Clinical significance of programmed death-1 ligand-1 and programmed death-1 ligand-2 expression in human esophageal cancer. Clin Cancer Res. 2005;11:2947–2953. doi: 10.1158/1078-0432.CCR-04-1469. [DOI] [PubMed] [Google Scholar]
- 25.Thompson RH, Gillett MD, Cheville JC, Lohse CM, Dong H, Webster WS, Krejci KG, Lobo JR, Sengupta S, Chen L, Zincke H, Blute ML, Strome SE, Leibovich BC, Kwon ED. Costimulatory B7-H1 in renal cell carcinoma patients: Indicator of tumor aggressiveness and potential therapeutic target. Proc Natl Acad Sci U S A. 2004;101:17174–17179. doi: 10.1073/pnas.0406351101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao R, Wang L, Han R, Wang T, Ye Q, Honjo T, Murphy TL, Murphy KM, Hancock WW. Differential effects of B and T lymphocyte attenuator and programmed death-1 on acceptance of partially versus fully MHC-mismatched cardiac allografts. J Immunol. 2005;175:5774–5782. doi: 10.4049/jimmunol.175.9.5774. [DOI] [PubMed] [Google Scholar]
- 27.Ishida Y, Agata Y, Shibahara K, Honjo T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. Embo J. 1992;11:3887–3895. doi: 10.1002/j.1460-2075.1992.tb05481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 29.Emmel EA, Verweij CL, Durand DB, Higgins KM, Lacy E, Crabtree GR. Cyclosporin A specifically inhibits function of nuclear proteins involved in T cell activation. Science. 1989;246:1617–1620. doi: 10.1126/science.2595372. [DOI] [PubMed] [Google Scholar]
- 30.Flanagan WM, Corthesy B, Bram RJ, Crabtree GR. Nuclear association of a T-cell transcription factor blocked by FK-506 and cyclosporin A. Nature. 1991;352:803–807. doi: 10.1038/352803a0. [DOI] [PubMed] [Google Scholar]
- 31.Fruman DA, Klee CB, Bierer BE, Burakoff SJ. Calcineurin phosphatase activity in T lymphocytes is inhibited by FK 506 and cyclosporin A. Proc Natl Acad Sci U S A. 1992;89:3686–3690. doi: 10.1073/pnas.89.9.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 33.Lu Q, Richardson B. Methods for Analyzing the Role of DNA Methylation and Chromatin Structure in Regulating T Lymphocyte Gene Expression. Biological procedures online. 2004;6:189–203. doi: 10.1251/bpo89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyannopoulos JA, Wilson CB. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beresford GW, Boss JM. CIITA coordinates multiple histone acetylation modifications at the HLA-DRA promoter. Nat Immunol. 2001;2:652–657. doi: 10.1038/89810. [DOI] [PubMed] [Google Scholar]
- 36.Louis-Plence P, Moreno CS, Boss JM. Formation of a regulatory factor X/X2 box-binding protein/nuclear factor-Y multiprotein complex on the conserved regulatory regions of HLA class II genes. J Immunol. 1997;159:3899–3909. [PubMed] [Google Scholar]
- 37.Hasegawa SL, Boss JM. Two B cell factors bind the HLA-DRA X box region and recognize different subsets of HLA class II promoters. Nucleic Acids Res. 1991;19:6269–6276. doi: 10.1093/nar/19.22.6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Majumder P, Gomez JA, Boss JM. The human major histocompatibility complex class II HLA-DRB1 and HLA-DQA1 genes are separated by a CTCF-binding enhancer-blocking element. J Biol Chem. 2006;281:18435–18443. doi: 10.1074/jbc.M601298200. [DOI] [PubMed] [Google Scholar]
- 39.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 40.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 41.Ozkaynak E, Wang L, Goodearl A, McDonald K, Qin S, O'Keefe T, Duong T, Smith T, Gutierrez-Ramos JC, Rottman JB, Coyle AJ, Hancock WW. Programmed death-1 targeting can promote allograft survival. J Immunol. 2002;169:6546–6553. doi: 10.4049/jimmunol.169.11.6546. [DOI] [PubMed] [Google Scholar]
- 42.Aramburu J, Garcia-Cozar F, Raghavan A, Okamura H, Rao A, Hogan PG. Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Mol Cell. 1998;1:627–637. doi: 10.1016/s1097-2765(00)80063-5. [DOI] [PubMed] [Google Scholar]
- 43.Singh H, Sen R, Baltimore D, Sharp PA. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature. 1986;319:154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- 44.Church GM, Ephrussi A, Gilbert W, Tonegawa S. Cell-type-specific contacts to immunoglobulin enhancers in nuclei. Nature. 1985;313:798–801. doi: 10.1038/313798a0. [DOI] [PubMed] [Google Scholar]
- 45.Nardone J, Lee DU, Ansel KM, Rao A. Bioinformatics for the ‘bench biologist’: how to find regulatory regions in genomic DNA. Nat Immunol. 2004;5:768–774. doi: 10.1038/ni0804-768. [DOI] [PubMed] [Google Scholar]
- 46.Serfling E, Klein-Hessling S, Palmetshofer A, Bopp T, Stassen M, Schmitt E. NFAT transcription factors in control of peripheral T cell tolerance. Eur J Immunol. 2006;36:2837–2843. doi: 10.1002/eji.200536618. [DOI] [PubMed] [Google Scholar]
- 47.Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, Bates DL, Guo L, Han A, Ziegler SF, Mathis D, Benoist C, Chen L, Rao A. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 48.Hu H, Djuretic I, Sundrud MS, Rao A. Transcriptional partners in regulatory T cells: Foxp3, Runx and NFAT. Trends Immunol. 2007;28:329–332. doi: 10.1016/j.it.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 49.Jain J, McCaffrey PG, Miner Z, Kerppola TK, Lambert JN, Verdine GL, Curran T, Rao A. The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature. 1993;365:352–355. doi: 10.1038/365352a0. [DOI] [PubMed] [Google Scholar]
- 50.Fruman DA, Pai SY, Burakoff SJ, Bierer BE. Characterization of a mutant calcineurin A alpha gene expressed by EL4 lymphoma cells. Mol Cell Biol. 1995;15:3857–3863. doi: 10.1128/mcb.15.7.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


