Abstract
Finding better therapies for the treatment of brain tumors is hampered by the lack of consistently obtained molecular data in a large sample set, and ability to integrate biomedical data from disparate sources enabling translation of therapies from bench to bedside. Hence, a critical factor in the advancement of biomedical research and clinical translation is the ease with which data can be integrated, redistributed and analyzed both within and across functional domains. Novel biomedical informatics infrastructure and tools are essential for developing individualized patient treatment based on the specific genomic signatures in each patient’s tumor. Here we present Rembrandt, Repository of Molecular BRAin Neoplasia DaTa, a cancer clinical genomics database and a web-based data mining and analysis platform aimed at facilitating discovery by connecting the dots between clinical information and genomic characterization data. To date, Rembrandt contains data generated through the Glioma Molecular Diagnostic Initiative from 874 glioma specimens comprising nearly 566 gene expression arrays, 834 copy number arrays and 13,472 clinical phenotype data points. Data can be queried and visualized for a selected gene across all data platforms or for multiple genes in a selected platform. Additionally, gene sets can be limited to clinically important annotations including secreted, kinase, membrane, and known gene-anomaly pairs to facilitate the discovery of novel biomarkers and therapeutic targets. We believe that REMBRANDT represents a prototype of how high throughput genomic and clinical data can be integrated in a way that will allow expeditious and efficient translation of laboratory discoveries to the clinic.
Keywords: Rembrandt, personalized medicine, translational research, clinical genomics, data integration
Introduction
Primary brain tumors are a leading cause of cancer mortality in adults and children in the United States (1). The molecular and genetic heterogeneity of gliomas undoubtedly contributes to the varied and often suboptimal response to treatment that is usually predicated on standard pathological diagnoses. Improvement in the prognosis of patients with gliomas will likely come about through use of new targeted therapies, based in the biological knowledge of the tumors at a molecular level.
To identify glioma-specific targets, consistent molecular characterization of a large number of tumors is required. To date, all the studies published have limitations due to either incomplete coverage of whole-genome expression due to the usage of small or outdated, legacy, microarray platforms (2, 3), limited number of samples studied and/or incomplete inclusion of various different glioma subtypes and grades (4, 5) or the narrow scope of targets being investigated. Thus, we have put together a national, publicly funded effort that we call the Glioma Molecular Diagnostic Initiative (GMDI), which coupled with its bioinformatics counterpart, REMBRANDT, is designed to breach the gap of biological information related to primary brain tumors in order to help patients receive a better, biologically oriented therapy, tailored to their specific needs..
Rembrandt is a powerful and intuitive informatics system designed to integrate genetic and clinical information for improved research, disease diagnosis and treatment (as shown in Figure 1). The platform supports clinical genomic research and (as data is collected and analyzed) will create a knowledge base that allows physicians to predict clinical outcomes and therapeutic efficacy based on an individual’s clinical and genetic profiles thereby enabling personalized medicine.
Figure 1.
Data integration via the Rembrandt discovery platform
To support discovery, the Rembrandt platform also allows researchers to search, import and aggregate additional data from internal and external databases (such as GenBank, UCSC golden path datasets and Biocarta pathways), analyze the combined data sets to identify meaningful patterns (including specific chromosomal abnormalities) and share their research with other physicians and researchers within their own institution or in other physical locations. Each user is assigned a specific role that governs how much of the study data is accessible. A series of intuitive tools enable users to easily analyze and interact with the integrated data to achieve greater insight into molecular signatures that characterize each tumor and correlate with clinical outcome.
Unlike many biomedical database systems, Rembrandt is a fully integrated platform that supports multiple facets of clinical and molecular research, discovery, and hypothesis generation. This shared environment crosses many disciplines including genetic research and clinical care. As such, the platform should serve to foster cooperation and integration between research and clinical disciplines, and expedite the time and increase the depth to which molecular data becomes relevant to the clinical environment.
Materials and Methods
Glioma Molecular Diagnostic Initiative (GMDI)
Sample Acquisition and Diagnostic
To better understand the genetic pathogenesis of gliomas and begin to identify potential glioma-specific molecular therapeutic targets, consistent molecular characterization of a large number of tumors is required.
This process was undertaken under a national prospective clinical trial that would eventually be IRB-approved both within the NCI intramural program as well as through both CTEP-sponsored adult brain tumor consortia (NABTT and NABTC protocol # 01-07). With the activation of this study, we collected matched tumor, blood and plasma from the 14 contributing institutions (National Institutes of Health, Henry Ford Hospital, Thomas Jefferson University, University of California San Francisco, H. Lee Moffitt Hospital, University of Wisconsin, University of Pittsburgh Medical Center, University of California Los Angeles, M.D. Anderson Cancer Center, Dana Farber Cancer Center, Duke University, Johns Hopkins University, Massachusetts General Hospital and Memorial Sloan Kettering Cancer Center). All tissue collected is sent to the Neuro-Oncology Branch laboratory for processing. The samples were provided as snap frozen sections of areas immediately adjacent to the region used for the histopathological diagnosis. Initial histopathological diagnosis is performed at the tissue collecting institution following the World Health Organization (WHO) standards(6). The initial diagnosis is reviewed by in-house neuropathologists to assure a measure of consistency across samples. To date, 874 complete frozen sample sets have been accrued, of those 389 are Glioblastoma Multiforme, 122 are Astrocytomas, 113 are Oligodendrogliomas, 33 are Mixed with the reminder still unclassified.
Clinical data on the patients is collected prospectively until the patient’s death through the NABTC Operations Office at M.D. Anderson Cancer Center, Houston, Texas and the NABTT Operations office at the Johns Hopkins University, Baltimore, MD. The clinical data collected is updated into the Rembrandt database on a quarterly basis.
In order to assure consistency in the collection, shipment, processing, assaying, storage, data retrieval and dissemination, we have put together a series of standard operating procedures (SOPs) that have resulted in a streamlined, high-throughput operation capable of handling large numbers of samples in a consistent, operator-independent fashion. Consistency of data over time is continuously monitored by looking for any signs of batch effect in the analyses.
mRNA extraction and gene expression data processing
Approximately 50–80 mg of tissue from each tumor was used to extract total RNA using the Trizol reagent (Invitrogen, Carlsbad, CA), following the manufacturer’s instructions. The quality of RNA obtained was verified with the Bioanalyzer System (7)(Agilent Technologies, Palo Alto, CA) using the RNA Pico Chips. Five μg of RNA extracted from the accrued samples have been processed using U133 2 Plus mRNA expression chips (Affymetrix, Inc., Santa Clara, CA), which contains over 54,000 probesets analyzing the expression level of over 47,000 transcripts and variants, including 38,500 well-characterized human genes.
All arrays were confirmed to be within an acceptable minimal quality control according to internal SOP parameters following these criteria: 1) A scaling factor of less than 5 when the expression values are scaled to a target mean signal intensity of 500. 2) Signal intensity ratios of the 3′ to 5′ end of the internal control genes of β-actin and GAPDH less than 3. 3) Affymetrix spike control (BioC, BioDN and CreX) are always present, and percentage present calls is above 35 % for brain tissue.
The .cel files and .txt files of all the arrays that passed the minimal quality control were input into dChip for normalization. The model-based expression index algorithm implemented in dChip selects an invariant set with a small within-subset rank difference to serve as basis for adjusting the brightness of the arrays to a comparable level. The normalization was done at the PM and MM probe level, then model-based expression levels were calculated using normalized probe level data. We choose the average difference model (PM > MM) to compute expression values; negative average differences were truncated to 1 or a log transformed values of zeros to flag negative signal intensities with no biological meaning.
For data preprocessing, probe-level data was consolidated into probe set data using the Affy MAS5 algorithm, with the target scaling value at 500. Probe-level data was also processed with custom CDF (1) (Chip Definition Files) that rearranged Affymetrix probes into gene-based probe sets. Probes mapped to alternatively spliced exons were grouped into distinct probe sets. Most 3′ probes were selected for processing. Nonspecific probes were masked before processing.
Single tumor samples were compared to the non-tumor pool and the sample average to the non-tumor pool. Samples were averaged based on tumor subtypes in six categories: Glioblastoma Multiforme, Oligodendroglioma, Astrocytoma, Mixed, Unclassified and Unknown tumors. Group comparisons were performed in R with two sample t-tests. Signal values were first transformed to logarithm (base 2). The averages of the log2-signals of tumor and non-tumor groups were computed. The magnitude of the differences between the geometric means of expression levels for each reporter from the two groups was computed. The significance of the differences between tumors (or each tumor subtype) and the non-tumor samples for each reporter was also evaluated.
For each individual tumor sample, signals for each tumor and the ratio between each tumor and the average of normal (geometric means, computed the same way as described above) were computed. All processes were performed separately for various data groups (public data and institution-based data).
DNA extraction and Genomic Alteration analysis
Approximately 10 μg of tissue (as recommended by the manufacturer) from each tumor was used to extract high molecular weight, genomic DNA using QIAamp DNA Micro DNA extraction kit (Qiagen, Valencia, CA), following the manufacturer’s instructions. The quality of DNA was checked by electrophoresis run in a 2% agarose gel.
Two hundred and fifty ng of genomic DNA from samples received has been hybridized to 100K SNP chips (8) (Affymetrix, Inc., Santa Clara, CA), which covers 116,204 single-nucleotide polymorphism (SNP) loci in the human genome with a mean intermarker distance of 23.6 Kb. These arrays give two simultaneous data types: Allelic Calls and Signal intensity, allowing for the determination of both Copy Number alterations (CNAs) and regions of Allelic Imbalances (Loss of heterozygosity, LOH). Calls were determined by the GTYPE software version 3.0 with 25% level of confidence. Only samples with call rates > 90% were accepted for any analysis.
Clinical data processing
MD Anderson’s Clinical Center (MDACC) serves as the operating center for clinical data collection for the GMDI trial. Clinical data reports from the case report forms were accessed through the Data Management Initiative (DMI) Web portal at MDACC, parsed and uploaded to the REMBRANDT data warehouse after various preprocessing and data validations steps. The clinical data collected is updated into the Rembrandt database on a quarterly basis.
Results
A Rembrandt storyboard
To exemplify the powerful integration that Rembrandt provides to analyze a large dataset of both molecular and clinical data, we would like to show how one could come about to explore the validity of a scientific hypothesis using the system.
Suppose that one would have come across two publications on Glioma Tumor Stem Cells that mentioned the irregular expression of BMPR1B in such cells (9, 10).
A typical Rembrandt usage scenario might be to ask if BMPR1B is a potential therapeutic target as it has been recently been postulated to be involved in cell differentiation. To answer this question, a researcher can take a step-wise workflow approach in Rembrandt as shown in Figure 2 and 3.
Figure 2.
A) Gene expression box plot for BMPR1B. Samples are shown categorized by histological type. Different Affymetrix probesets are shown as different color bars. B) BMPR1B probeset in Affymetrix probeset viewer. Information for selected probeset can be displayed, allowing the user to decide on the quality of information retrieved. C) BMPR1B probeset of interest showing outliers in GBM samples. The ability to display expression graphs in different formats allow the use to gain a wealth of information without having to redo the queries.
Figure 3.
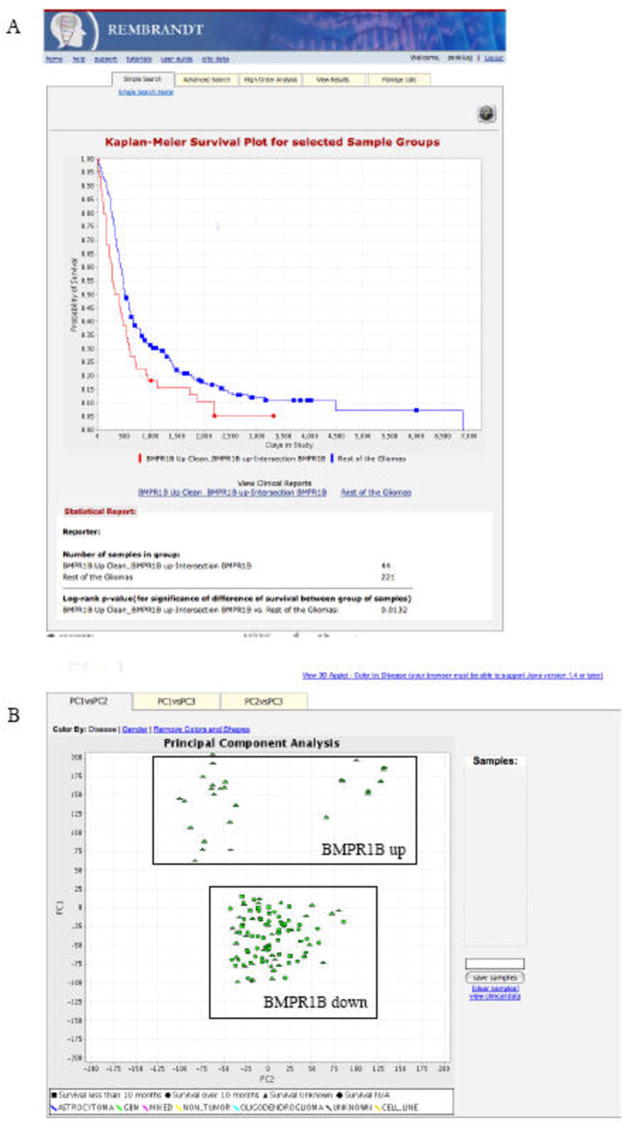
A) K-M survival plot showing survival comparing BMRP1-upregulating samples and rest of the gliomas in the database. This plot allows you to identify putative clinically relevant genes to explore as new targets for therapy. Users can query gene expression and graph changes in survival rate at each time point on the study. Kaplan-Meier (K-M) estimates are calculated based on the last follow-up time and the censor status (0=alive, 1=dead) from the samples of interest. The Kaplan-Meier estimates are then plotted against the survival time. Users can dynamically modify the fold change (up and down regulation) thresholds and redraw the plot. A log-rank p-value is provided as an indication of significance of the difference in survival between any two groups of samples segregated based on gene expression of the gene of interest. B) Performing principal component analysis and correlating with clinical data. This figure shows an example Principal Component Analysis report from the REMBRANDT application. This two-dimensional (a) and three-dimensional graph plots (b) the various principal components from the gene expression PCA analysis. Various analysis options are provided to the user to select from an array of gene/reporter filtering and sample selection settings. Users can select samples in the 2-dimensional plot to retrieve related clinical information on the selected patients.
Explore the expression levels of BMPR1B in different subtypes of Glioma. Analysis of the box plots in Rembrandt (Figure 2A) indicate that probeset 210523_at is differentially expressed in GBMs when compared to non-tumors (borderline significance: pvalue < 0.04)
Where does this probe map onto the transcripts of BMPR1B? Review of probe mapping in Affymetrix probe viewer integrated into Rembrandt (figure 2B) shows that this probe maps to coding region
Are there 2 sub-populations of BMPR1B regulating samples? Review of the “box and whisker” plot in figure 2C indicates that GBMs have low end outliers for BMPR1B expression.
Now, can we identify samples that show high (up >2) and low (down<1.5) expression of BMPR1B? Advanced queries can be set up in the Rembrandt application to create sample sets with separate up and down regulation criteria for BMPR1 expression.
Does BMPR1 upregulation impact survival? Can this sample group be compared to the rest of the gliomas? Figure 3A shows the difference in probability of survival between BMRP1 upregulating group and the rest of the gliomas. Results indicate that BMPR1B upregulation is bad as a prognostic factor, and could be a good target for therapy
How different are these sample groups beyond BMPR1B expression? By analyzing the whole gene expression patterns in both groups using the high order analysis tool of Principal Component Analysis (Figure 3B), it is possible to see that BMPR1B over and under expressors are indeed quite different at a global expression level, suggesting that this gene may hold a key to glioma diversity.
The storyboard here presented indicates that Rembrandt can effectively be used to test in silico a scientific hypothesis and allow for additional experimentation to occur. In fact, this has been the case with the scenario here presented and we have shown that BMPR1B is able in fact to modulate the tumorigenic potential of glioma cells (11). Additionally, a Rembrandt search of newly identified (NF1) and well-known (IGFBP2) targets of deregulation in gliomas show that the result produced by our dataset are concordant with the current knowledge of clinical features (Supplemental Figure 1).
Key features in Rembrandt
Integrating genome characterization data with clinical outcomes
Users can query gene expression or copy number data and graph changes in survival rate at each time point in the study. Kaplan-Meier (K-M) estimates are calculated based on the last follow-up time and the censor status (0=alive, 1=dead) from the samples of interest. K-M estimates are then plotted against survival time (Figure 3A). The points that correspond to the events with a censor status of 0 are indicated on the graph. Users can dynamically modify the fold change (up and down regulation) thresholds and redraw the plot. A log-rank p-value is provided as an indication of significance of the difference in survival between any two groups of samples segregated based on gene expression of the gene of interest. The log rank p-value is calculated using the Mantel-Haenszel procedure (12). The p-values are recalculated every time a new threshold is selected. Users can toggle to a unified gene expression view with lesser reporters to get a gene-based view of the expression data. To obtain the unified gene expression values, the probe-level data is processed with custom CDF (Chip Definition Files) that rearranges Affymetrix probes into gene-based probe sets. Probes mapped to alternatively spliced exons are grouped into distinct probe sets. Most 3′ probes are selected for processing. Nonspecific probes are masked before processing. Similar to K-M plots for differential fold change analysis, K-M plots can be drawn for copy number data where genes are mapped to SNP probe sets by aligning the probe’s physical position to aligned mRNA sequences plus 50 kb up and down steam for maximum coverage. Also K-M plots can be drawn by selecting 2 patient groups of interest. These groups can be user-defined or pre-defined lists of patients. These groups can be user-defined or pre-defined lists of patients.
Performing higher-order statistical analysis on genomic and clinical data set
Rembrandt supports computer-intensive, high-memory utilizing tasks such as higher-order gene expression analyses (such as class comparison, clustering and principal component analysis), where the data sets could be as large as 4 GB with an analytic cluster to allow for several simultaneous analytic jobs.
Figure 3B shows an example of a Principal Component Analysis (PCA) report from the REMBRANDT application. This two-dimensional graph plots the various principal components from the gene expression PCA analysis. Various analysis options are provided from which users can select gene/reporter filtering and sample selection settings. Users can click on the three tabs at the top of the graph to display either PC1 vs. PC2, or PC1 vs. PC3, or PC2 vs. PC3. Each point on the graph represents a sample. The samples are colored by disease type. Users can click on the link on the upper left-hand corner of the graph to color by gender. Patients with different survival ranges are indicated by different shapes on the graph. Users can select samples of interest by clicking on the graph and drawing a rectangle around samples to save them for future use.
GenePattern link
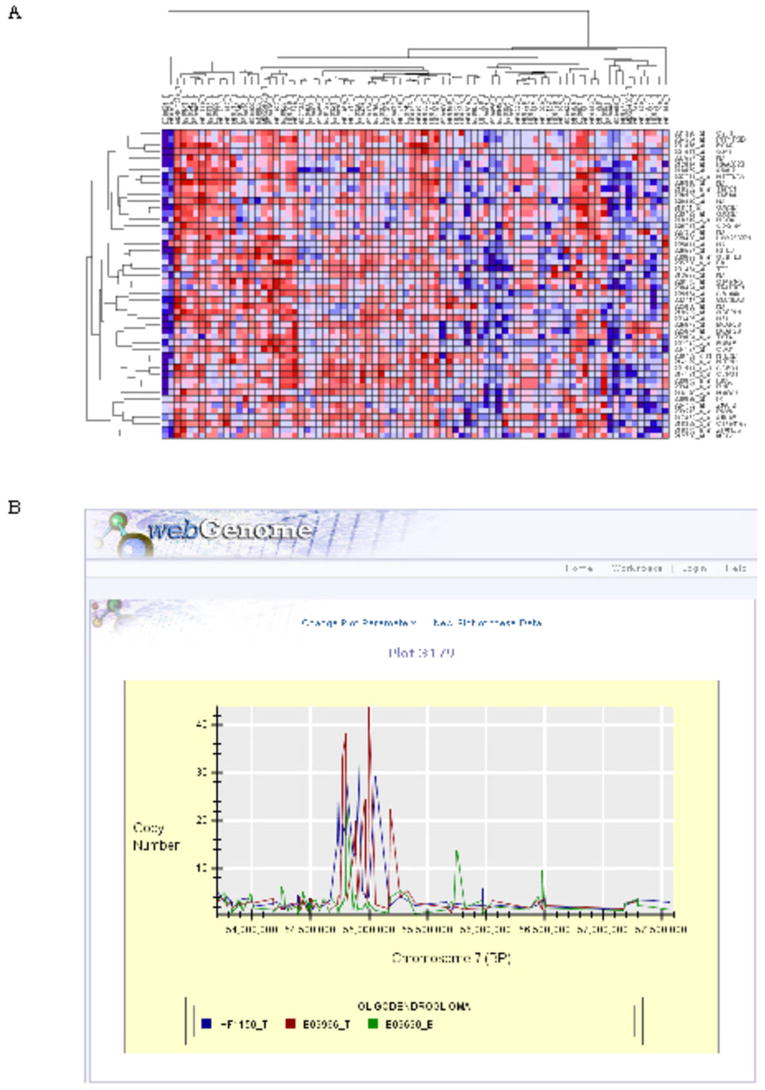
Broad’s GenePattern (13) combines a powerful scientific workflow platform with more than 90 computational and visualization tools for the analysis of genomic data. In order to expand a researcher’s ability to analyze the glioma data sets, Rembrandt has been seamlessly integrated with GenePattern. Shown in Figure 4A is an expression heatmap of 50 additional genes that have expression patterns related to SCF (14) in GBM.
Figure 4.
A) Heatmap view in GenePattern. Subsets of data from Rembrandt can be transferred to GenePattern using standard interfaces to invoke a number of run-time data analysis capabilities. A heatmap for 50 neighbors of SCF is shown for astrocytoma and mixed glioma samples in Rembrandt. B) Scatter plot for copy number data across physical genomic locations. Scatter plots (shown above) display measured copy number against physical genome location in an application called webGenome, which has been integrated with Rembrandt via standard data interfaces. These plots are context-sensitive to the copy number reports generated from the copy number queries in the caIntegrator application. You can view data at arbitrary resolutions from the entire genome on down.
Plotting Copy Number Data from patient DNA samples against genomic location
Scatter plots (shown in Figure 4B) display measured copy number against the physical genome location in an application called webGenome, which has been integrated with Rembrandt. These plots are context-sensitive to the copy number reports generated from the copy number queries in the Rembrandt application. Users can view data at arbitrary resolutions from the entire genome on down. When users move the mouse over specific probes, the system provides mouse-over probe names. Clicking on the name of an experiment or bioassay in the plot legend will highlight the corresponding data.
Advanced Query and Report Interfaces
Biomedical researchers struggle to meaningfully integrate their findings across multiple data types. Cancer is a complex disease requiring genomic, proteomic, pathology, imaging, and clinical data for a true understanding of the scope of the problem (The Scientist reference). Advanced query interfaces (as shown in Figure 5) in Rembrandt enable this meaningful integration across data types. It allows users to mine the REMBRANDT database using various genomic and clinical criteria. These queries can be combined to arrive at reports (shown in Figure 6) that integrate data from various data domains, such as gene expression, copy number analysis and clinical trials. A number of filtering and data download options are presented in Rembrandt reports.
Figure 5.
User-friendly data query interface. Query pages enable users to restrict their searches in the database to specific genomic and/or clinical criteria.
Figure 6.
Gene Expression Fold Report. All reports in Rembrandt are fully customizable at the report window, making unnecessary to re-run queries to refine the results.
Rembrandt system architecture
Rembrandt was developed using an n-tier architecture. The system was developed using Java 2 Enterprise Edition (J2EE), a hybrid star datawarehouse schema and various open source technologies. The back end consists of an Oracle 10g database for storing precomputed microarray differential expression, computed copy number, clinical data and user security information. For performance reasons, normalized gene expression data used by the real-time analysis module is stored as R-binary files. The middle tier, which handles application logic and core functionality, was developed using Java and caBIG™ software development and compatibility guidelines (15). Rembrandt application consists of standard interfaces that enable integration with 3rd party tools such as caArray, webGenome and GenePattern. Rembrandt has an Analytical Server that provides on-the-fly computational analysis capability. The Analytical Server communicates asynchronously with Rembrandt’s middle tier via the Java Messaging Service (JMS). JMS allows Rembrandt to abstract the statistical packages being utilized for heavy computational tasks.
Rembrandt caBIG™ Grid service
Basic and clinical research has increasingly become dependent on advanced information technologies for management, exchange and analysis of diverse biomedical data. Although a wealth of information is collected by the cancer research community, any one given researcher is faced with challenges in discovering, extracting and analyzing the information relevant to his/her research. To address this need the National Cancer Institute (NCI) has initiated a national-scale effort, called the cancer Biomedical Informatics Grid (caBIG™), to develop a federation of interoperable research information systems. At the heart of the caBIG approach to federated interoperability effort is a Grid middleware infrastructure, called caGrid (16). caGrid Data Services provide the means to share data via the caGrid federated infrastructure. One of the major goals of the current release of Rembrandt was to create a clinical genomic object model (CG-OM) and expose the domain model through a caGrid data service. The purpose of the object model is to help capture the relationships between the clinical study and its associated experimental observations. The Rembrandt caGrid service can be used to obtain programmatic access to public data in Rembrandt in a federated fashion and can be found at http://caintegrator.nci.nih.gov/wsrf-rbt/services/cagrid/RembrandtGridService
Conclusion
Large-scale data sets from genomics, proteomics, population genetics and imaging are driving research at a previously unprecedented pace. Bioinformatics data management providers must serve these datasets in a usable way that helps find the needle in a haystack effectively and accurately. The goal of the “omic” sciences is not to generate numbers but rather “insight”. The web interfaces are burdened with displaying terabytes of data in ways that physician scientists can comprehend and use the results to develop hypothesis for their next study or trial. Ultimately, we feel that information must be standardized, integrated and made available at the point of care to help patients and physicians make optimal decisions.
Tools like Rembrandt have primarily focused on the usability aspect of high throughout heterogeneous data and yet enabling power users and bioinformaticians to tap into runtime analysis tools such as gene pattern or use the programmatic interfaces that are provided via the caGrid service. From a technical standpoint, the Rembrandt platform provides developer tools for a highly scalable system to include new data types (as shown in figure 1) and connect with existing ones to present integrated data views to users. This flexible discovery informatics platform has aided in implementing data portals to host a number of other cancer clinical datasets including those from the I-SPY stage 3 Breast cancer study and the Cancer Genome Atlas (TCGA)(17) project data included in the Cancer Molecular Analysis Portal. In this respect it is worth to point out that the new Cancer Molecular Data portal has reutilized many of the features available in Rembrandt to suit a more general set of tumor sample analysis. At the sample level, GMDI and TCGA are complementary in many levels. GMDI is a prospective study wherein 14 institutions recruited patients with any type of glioma giving a wide spectrum of demographical sampling due to the geographical dispersion of the sites. TCGA’s samples collection pipeline included 2 centers that had retrospective sample collections of Glioblastoma Multiforme. Thus, TCGA focused its analysis on high grade GBMs, while the samples in GMDI represent all glioma grades and subtypes described in the WHO classification, allowing for studies on the differences of gliomas as they progress. The clinical data obtained by the GMDI project is comprehensive, since the study was conceived as a prospective, natural history clinical trial, thus allowing for the collection of a wide range of clinical datapoints. On the other hand, the TCGA project, in virtue of its more focused nature, has produced more molecular data types (methylation, sequencing, miRNA expression) than GMDI. However, the GMDI samples are being used to acquire those datatypes, and they will be incorporated to Rembrandt as sufficient number of samples are processed.
The ultimate beneficiaries of Rembrandt are the brain tumor patients themselves. Rembrandt is designed to bridge the gap between biological and clinical information in order to help patients receive a better, biologically oriented therapy, tailored to their specific needs. As such, we plan to incorporate new and useful capabilities in future releases that are not available at present time; such as the ability for researchers to incorporate their own data to the system to compare with the large dataset already in the database. It is hoped that the GMDI and Rembrandt will provide a much needed resource for scientists and physicians combating brain cancer, and ultimately other forms of cancer, for providing the data and bioinformatics tool set that may allow the development of a biologically and clinically significant pathological classification of brain tumors and help elucidate novel molecular targets for therapy.
Availability
Rembrandt is freely available to all users at https://caintergator.nci.nih.gov/rembrandt. The source code for Rembrandt is also available under a non-viral caBIG™ license at https://gforge.nci.nih.gov/frs/download.php/1489/rembrandt_1_0.zip. The Rembrandt caGrid service is accessible at http://caintegrator.nci.nih.gov/wsrf-rbt/services/cagrid/RembrandtGridService
Web resources
Rembrandt clinical genomics object model:
http://Rembrandt.nci.nih.gov/content/Rembrandtlfs/RembrandtEA1.0docs/index.htm
Rembrandt clinical genomics data model:
http://Rembrandt.nci.nih.gov/developers/images/db_model2.jpg
REMBRANDT application:
http://rembrandt-db.nci.nih.gov
REMBRANDT information site:
Web Genome:
http://webgenome.nci.nih.gov/webgenome/home.do
GenePattern:
http://www.broad.mit.edu/cancer/software/genepattern/
caArray:
https://array.nci.nih.gov/caarray/home.action
I-SPY trial:
http://ncicb.nci.nih.gov/tools/translation_research/ispy
The Cancer Genome Atlas:
Cancer molecular analysis portal (Access to TCGA datasets)
Acknowledgments
We thank Anand Basu, Shine Jacobs, Alex Jiang, Huaitian Liu, Ram Bhattaru, Michael Harris, Kevin Rosso, Ryan Landy, Hangjiong Chen and Ying Long for their contributions to Rembrandt software development and data loading; George Komatsoulis for reviewing data sharing policies and contributing to the Rembrandt domain information model; Carl Schaefer and Tracy Lively for reviewing usecases and interim releases of the software; Juli Klemm for helping with the integration of Rembrandt with caArray data repository; Jill Hadfield for technical documentation; David Hall, Dean Jackman, Vessalina Bakalov for their efforts on webGenome. We also thank M.D. Anderson Cancer Center’s DMI team for making the clinical reports available from the NABTC GMDI study for populating the Rembrandt database. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and the National Institute of Neurological disorders and Stroke.
References
- 1.Cancer Statistics Branch N, NIH. Cancer Survival rates. In: Harras A, editor. Cancer: Rates & Risks. Washington, DC: US Dept of Health & Human Services, National Institutes of Health; 1996. pp. 28–34. [Google Scholar]
- 2.Nutt CL, Mani DR, Betensky RA, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63:1602–7. [PubMed] [Google Scholar]
- 3.Mischel PS, Shai R, Shi T, et al. Identification of molecular subtypes of glioblastoma by gene expression profiling. Oncogene. 2003;22:2361–73. doi: 10.1038/sj.onc.1206344. [DOI] [PubMed] [Google Scholar]
- 4.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 5.Nigro JM, Misra A, Zhang L, et al. Integrated array-comparative genomic hybridization and expression array profiles identify clinically relevant molecular subtypes of glioblastoma. Cancer Res. 2005;65:1678–86. doi: 10.1158/0008-5472.CAN-04-2921. [DOI] [PubMed] [Google Scholar]
- 6.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta neuropathologica. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller CL, Diglisic S, Leister F, Webster M, Yolken RH. Evaluating RNA status for RT-PCR in extracts of postmortem human brain tissue. Biotechniques. 2004;36:628–33. doi: 10.2144/04364ST03. [DOI] [PubMed] [Google Scholar]
- 8.Matsuzaki H, Dong S, Loi H, et al. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat Methods. 2004;1:109–11. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- 9.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 10.Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–21. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 11.Lee J, Son MJ, Woolard K, et al. Epigenetic-mediated dysfunction of the bone morphogenetic protein pathway inhibits differentiation of glioblastoma-initiating cells. Cancer Cell. 2008;13:69–80. doi: 10.1016/j.ccr.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–81. [Google Scholar]
- 13.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 14.Sun L, Hui AM, Su Q, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Phillips J, Chilukuri R, Fragoso G, Warzel D, Covitz PA. The caCORE Software Development Kit: streamlining construction of interoperable biomedical information services. BMC medical informatics and decision making. 2006;6:2. doi: 10.1186/1472-6947-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oster S, Langella S, Hastings S, et al. caGrid 1.0: an enterprise Grid infrastructure for biomedical research. J Am Med Inform Assoc. 2008;15:138–49. doi: 10.1197/jamia.M2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLendon R, Friedman A, Bigner D, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008 doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]