Abstract
To determine the in vivo function of intestinal cytochrome P450 (P450) enzymes, we have generated an intestinal epithelium (IE)-specific P450 reductase gene (Cpr) knockout mouse model (designated IE-Cpr-null). In the IE-Cpr-null mouse, CPR expression was abolished in IE cells; however, CPR expression was not altered in other tissues examined. The loss of CPR expression in the small intestine (SI) led to increased expression of several P450 proteins examined, including CYP1A1, CYP2B, CYP2C, and CYP3A. It is interesting to note that the expression of CYP1A1 was also increased in the liver, kidney, and lung of the IE-Cpr-null mice compared with wild-type (WT) littermates, a result strongly supporting the notion that SI metabolism of putative dietary CYP1A1 inducers can influence the systemic bioavailability of these inducers. The rates of SI microsomal metabolism of nifedipine (NFP) in the IE-Cpr-null mice were ∼10% of the rates in WT littermates, despite the compensatory expression of multiple P450 enzymes in the SI. Furthermore, the area under the concentration-time curve (AUC) values for blood NFP (dosed at 10 mg/kg) levels were 1.6-fold higher in IE-Cpr-null mice than in WT littermates when NFP was given orally; in contrast, the AUC values were comparable for the two strains when NFP was given intravenously. This result directly showed that P450-catalyzed NFP metabolism in the SI plays an important role in the first-pass clearance of oral NFP. Our findings indicate that the IE-Cpr-null mouse model can be used to study the in vivo function of intestinal P450 enzymes in the clearance of oral drugs and other xenobiotics.
The small intestine (SI), the first site capable of metabolism of orally ingested xenobiotics, including nutrients, toxicants, and therapeutic drugs, is believed to play an important role in the first-pass metabolism of numerous chemical compounds (Thummel et al., 1997; Doherty and Charman, 2002; Kaminsky and Zhang, 2003). Among the many biotransformation enzymes expressed in the SI, the cytochrome P450 (P450) monooxygenases are the greatest contributors to the catalysis of the biotransformation reactions. P450 comprises a superfamily of heme-containing enzymes (Nelson et al., 2004) that are active in the bioactivation or detoxification of numerous toxic chemicals, carcinogens, and therapeutic drugs. Of the P450s expressed in humans, many are known to be expressed in the SI (Kaminsky and Fasco, 1991; Kaminsky and Zhang, 2003; Paine et al., 2006). Based on the expression levels and the activities of P450 enzymes in the SI, it has been proposed that these P450s directly affect the bioavailability of many drugs (Suzuki and Sugiyama, 2000; Doherty and Charman, 2002; Ding and Kaminsky, 2003); however, few studies have directly examined the relative contributions of liver and SI to systemic bioavailability of oral drugs, mainly because of the difficulties of distinguishing between hepatic and intestinal first-pass metabolism.
NADPH-P450 reductase (CPR) is the obligate redox partner for microsomal P450 enzymes; therefore, the deletion of the Cpr gene causes the inactivation of all the microsomal P450 enzymes in targeted cells. Several mouse models, in which the Cpr gene is deleted in a tissue-specific fashion, have been reported, including the “liver-specific Cpr-null” (LCN) mouse (Gu et al., 2003) and the similar “hepatic P450 reductase null” mouse (Henderson et al., 2003), the “lung-specific Cpr-null” mouse (Weng et al., 2007), and the “cardiomyocyte-specific Cpr-null” mouse (Fang et al., 2008). A transgenic mouse with a hypomorphic Cpr gene was also developed (designated as “Cpr-low” or CL mouse); in this mouse, CPR expression was globally down-regulated (Wu et al., 2005). In addition, a “Cpr-low and liver-Cpr-null” mouse (Gu et al., 2007) has been reported, as has a mouse model with inducible deletion of the Cpr gene primarily in the liver and intestine (Finn et al., 2007). However, although a combined use of some of these mouse models (e.g., CL, LCN, and Cpr-low and liver-Cpr-null), as exemplified by our recent study on nifedipine (NFP) clearance (Zhang et al., 2007), can be useful for a preliminary and/or indirect assessment of the relative roles of liver and extrahepatic tissues (including the SI) in drug metabolism, none of these models is able to directly test the specific roles of intestinal epithelial cells in the in vivo metabolism of xenobiotic or endobiotic compounds.
Here, we describe the development and initial characterization of an intestinal epithelium-specific Cpr-knockout (IE-Cpr-null) mouse model. The IE-Cpr-null mouse was generated by crossbreeding the Vil-Cre transgenic mouse (Madison et al., 2002) with the Cpr-lox mouse (Wu et al., 2003). The Vil-Cre mouse expresses the Cre recombinase under the control of the mouse villin 1 promoter (Madison et al., 2002; el Marjou et al., 2004). Villin, an actin binding protein, is expressed in every cell of the IE (Maunoury et al., 1992). The Cpr-lox mouse, which has two LoxP sequences inserted into introns 2 and 15 of the mouse Cpr gene, has normal CPR expression (Wu et al., 2003); this strain has been crossed to a number of Cre-expressing transgenic mice to achieve conditional deletion of the Cpr gene (e.g., Gu et al., 2003; Weng et al., 2007). In the IE-Cpr-null mouse, expression of the Cre protein in the enterocytes is expected to allow Cre-mediated recombination of the floxed Cpr gene, leading to IE-specific Cpr gene deletion.
For the initial characterization of the IE-Cpr-null mouse model, we determined the specificity and time course of Cpr gene deletion, in addition to routine phenotypic examinations of the viability, fertility, growth rates, and potential embryonic lethality. We have also monitored the occurrence of compensatory expression changes for selected P450 enzymes in the SI and in several extragut organs. Finally, we have used the IE-Cpr-null mouse model to study the role of SI in the first-pass metabolism of NFP, a CYP3A substrate. Our results show that the IE-Cpr-null mouse model can be used to study the in vivo function of intestinal P450 enzymes in the clearance of oral drugs and other xenobiotics.
Materials and Methods
Animals. Vil-Cre(+/–) mice (on a B6 background) were purchased from The Jackson Laboratory (Bar Harbor, ME) (Madison et al., 2002). The Cprlox/lox mouse (Gu et al., 2003; Wu et al., 2003) and the Cpr-low mouse (Wu et al., 2005), which have been described recently, were available at the Wadsworth Center (Albany, NY). Vil-Cre(+/–) transgenic mice were first crossed with Cprlox/lox mice (congenic on B6 background) to generate Vil-Cre(+/–)/Cprlox/+ pups, which were crossed with Cprlox/lox mice again, yielding Vil-Cre(+/–)/Cprlox/lox (IE-Cpr-null) mice. Genotype analyses for the Cre transgene and the Cpr allele were performed as described previously (Gu et al., 2003; Wu et al., 2003). All the studies with mice were approved by the Wadsworth Center Institutional Animal Care and Use Committee.
Histopathology and Immunohistochemistry. SI from 2- to 3-month-old male mice was prepared as a so-called “Swiss roll,” so that the full length of the duodenum, jejunum, and ileum was represented on the same slides, an arrangement that facilitates histological analysis. Paraffin sections (4 μm) were hematoxylin and eosin-stained according to standard procedure. For immunohistochemical analysis of CPR expression in the SI, paraffin sections of SI were processed essentially according to a published protocol (Chen et al., 2003), with minor modifications. Endogenous peroxidase was blocked with 3% H2O2. All the tissue sections were subjected to antigen retrieval with the Citra solution, pH 6.0 (Biogenex, San Ramon, CA), and then incubated with a protein block (Dako North America, Inc., Carpinteria, CA) to prevent nonspecific binding. For detection of CPR protein, the tissue sections were incubated overnight, at 4°C, with a polyclonal rabbit anti-rat CPR antiserum (Assay Designs/Stressgen, Ann Arbor, MI) at a 1:1000 dilution. Antigenic sites were visualized with reagents from Invitrogen (Carlsbad, CA), including peroxidase-conjugated goat anti-rabbit secondary antibody (at a 1:100 dilution) and Alexa Fluor 594-conjugated tyramide as the peroxidase substrate. Sections were mounted with Prolong mounting medium with 4,6-diamidino-2-phenylindole (DAPI) (Invitrogen). Fluorescent signals were detected using a Nikon (Melville, NY) Eclipse 50i microscope, with either a tetramethylrhodamine isothiocyanate filter (for Alexa 594) or a DAPI filter (for DAPI). Negative control slides were incubated with normal goat serum (Biogenex) in place of the anti-CPR antibody.
Isolation of Intestinal Epithelial Cells and Preparation of Microsomes. Tissues from three to six mice were combined for each microsomal preparation. Epithelial cells from the SI or the colon were isolated, and microsomes were prepared as described previously (Zhang et al., 2003). Liver microsomes were prepared essentially as previously reported (Fasco et al., 1993), but with use of protease inhibitors (Sigma-Aldrich, St. Louis, MO), as described for the preparation of SI microsomes (Zhang et al., 2003). Microsomes were stored at –80°C until use. Microsomal protein concentrations were determined with the bicinchoninic acid protein assay kit (Pierce Chemical Co., Rockford, IL), with bovine serum albumin as the standard.
Immunoblot Analysis. For immunoblot analysis, microsomal proteins were separated on NuPAGE Bis-Tris gels (10%) (Invitrogen). Polyclonal antibodies to rat CYP1A1/2, CYP2B1, CYP2C6, and CYP3A2 were all purchased from BD Gentest (Woburn, MA). Polyclonal rabbit anti-rat CPR antibody was obtained from BD Gentest. Peroxidase-labeled rabbit anti-goat IgG or goat anti-rabbit IgG (Sigma-Aldrich) was detected with an enhanced chemiluminescence kit (GE Healthcare, Piscataway, NJ). The optical density of detected bands was determined with a Personal Densitometer SI (GE Healthcare).
In Vitro Metabolism of NFP. The NFP oxidase assay and the high-performance liquid chromatography (HPLC) analysis for detection of NFP and its metabolite were performed as described previously (Zhang et al., 2007). Control experiments were performed in which NADPH was omitted.
Pharmacokinetic Analysis. Mice (four to six in each group) were treated with NFP, either intravenously (2 mg/kg; NFP was injected in a mixture of dimethyl sulfoxide and phosphate-buffered saline, 1:4, v/v) or orally through gavage (10 or 20 mg/kg; NFP was administered in a mixture of ethanol, polysorbate 80, and phosphate-buffered saline, 20:20:60, v/v/v). Blood samples after NFP oral dosing were collected from the tail vein at various times after NFP administration as described recently (Zhang et al., 2007), whereas blood samples after intravenous dosing were collected from the saphenous vein, according to the protocol of Hem et al. (1998) at 10, 30, 45, 60, and 90 min. NFP was extracted from whole blood using conditions described by Jankowski and Lamparczyk (1994). NFP concentration was determined by HPLC as described previously (Zhang et al., 2007). Pharmacokinetic parameters were calculated using the WinNonlin software (version 5.0.1; Pharsight, Mountain View, CA). Statistical significance of differences between groups was examined with Student's t test.
Results
The IE-Cpr-null (Vil-Cre(+/–)/Cprlox/lox) mice are viable, fertile, normal in size, and do not display any gross physical or behavioral abnormalities. There is no significant difference in body weights between IE-Cpr-null and wild-type (WT) littermates for either male or female mice (data not shown). Genotype distribution analyzed among 119 available pups derived from crosses between Vil-Cre(+/–)/Cprlox/+ and Cprlox/lox mice indicates that the IE-Cpr-null genotype is not associated with in utero lethality (results not shown). In other experiments not shown, histological examination of the SI from the IE-Cpr-null mice did not reveal any abnormalities in the structure of the various intestinal segments.
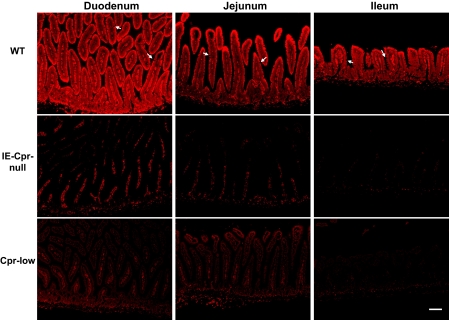
The sites of Cpr gene deletion in the SI were identified by immunohistochemical staining for the CPR protein (Fig. 1). In WT mice, CPR protein was found to be highly expressed in enterocytes of the duodenum, jejunum, and ileum. As expected, the signal intensity was stronger in the villi than in the crypts. Gaps visible within the epithelium represent SI goblet cells (secretory cells), which have little CPR expression and are thus not “stained” by the antibody (Fig. 1). In the IE-Cpr-null mice, CPR expression was not detected in the SI epithelial cells in any of the sections examined; this negative result confirmed Cpr gene deletion in all the enterocytes. It is noteworthy that relatively strong immunostaining was detected in the lamina propria of the WT mice and that of the IE-Cpr-null mice. This staining probably represents nonspecific reactivity of the antibody, given that cells in the lamina propria are not known to contain high levels of CPR protein. To further confirm the artifactual nature of the staining in the lamina propria, we analyzed the SI of the Cpr-low mice, which should have reduced CPR expression in all the cell types. As shown in Fig. 1, although signal intensity in the SI epithelial cells was drastically lower in the Cpr-low mice compared with WT mice, the staining in the lamina propria was not reduced, indicating that the signal detected there does not represent CPR expression.
Fig. 1.
Immunohistochemical analysis of CPR expression in the SI of IE-Cpr-null, Cpr-low, and WT (littermates) mice. Paraffin sections of SI from 2-month-old male mice were processed for immunohistochemistry as described under Materials and Methods. The tissue sections were incubated with a polyclonal rabbit anti-rat CPR antiserum. Antigenic sites were visualized with a peroxidase-conjugated goat anti-rabbit secondary antibody, with Alexa Fluor 594-conjugated tyramide as the peroxidase substrate. Sections were mounted with Prolong mounting medium with DAPI. Fluorescent signals were detected with a tetramethylrhodamine isothiocyanate filter (for Alexa 594) as shown here, or a DAPI filter (results not shown); scale bar, 100 μm. No signal was detected in negative control slides (data not shown), which were incubated with a normal goat serum in place of the anti-CPR antibody. Arrows indicate examples of goblet cells (with little CPR expression) in the SI epithelium. Results shown are typical of three mice per strain analyzed.
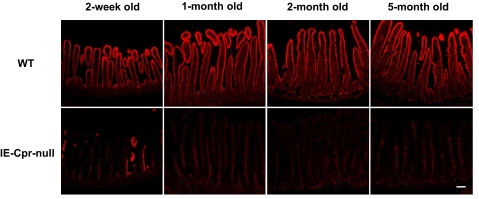
The time course of Cre-mediated Cpr deletion in the SI was also examined through comparison of the results of immunohistochemical staining of tissue sections obtained from mice of various ages (Fig. 2). In WT mice, CPR protein was readily detected in the SI of all the age groups, ranging from 2 weeks to 5 months old. In IE-Cpr-null mice, only a few CPR-positive epithelial cells were detected in the SI at 2 weeks of age, whereas no CPR-positive epithelial cells were detected at 1 month or at older ages. Therefore, the Cre-mediated Cpr deletion has occurred in all the enterocytes by 1 month after birth, and there was no indication of a recovery in CPR expression as late as 5 months after birth.
Fig. 2.
Time course of Cre-mediated Cpr deletion in the SI of the IE-Cpr-null mouse. Male mice of the indicated ages were studied. Immunohistochemical analysis of CPR expression was as described in the legend to Fig. 1. Sections shown are from the jejunum. A similar developmental pattern was observed for the duodenum and ileum (data not shown). Results shown are typical of three mice per group analyzed; scale bar, 100 μm.
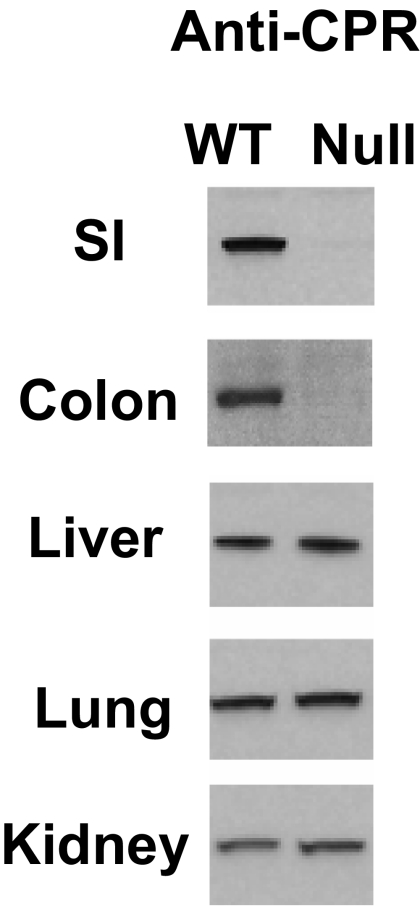
The extent of overall decrease of SI microsomal CPR protein levels in the IE-Cpr-null mice was determined by immunoblot analysis. As shown in Fig. 3, CPR protein was essentially undetectable in enterocyte microsomes from 3-month-old IE-Cpr-null mice, in contrast to the abundant CPR expression seen in enterocyte microsomes from WT littermates. Similar strain differences were seen for 1-month-old mice (data not shown). In 3-month-old mice, CPR protein could be detected in the blots for IE-Cpr-null mouse on prolonged exposure of the film; a quantitative analysis of CPR levels in IE-Cpr-null and WT enterocyte microsomes suggested that the residual amount of CPR protein in the IE-Cpr-null mouse was less than 5% of the level present in WT microsomes. CPR expression was also nearly abolished in colonic microsomes from 3-month-old IE-Cpr-null mice; however, CPR expression in other tissues examined, including liver, lung, and kidney, did not differ between the IE-Cpr-null and the WT mice (Fig. 3), indicating that Cpr deletion occurred specifically in the intestine of the IE-Cpr-null mice.
Fig. 3.
Immunoblot analysis of CPR protein expression. SI epithelial cells of IE-Cpr-null and WT mice at the age of 3 months were studied. For each mouse strain, epithelial cells from the SI (three male mice used) or the colon (five male mice used) were pooled for microsomal preparation. Microsomal proteins (10 μg) were analyzed on immunoblots with a rabbit anti-rat CPR antibody. Microsomes from the liver (2 μg), lung (10 μg), and kidney (10 μg) were also analyzed.
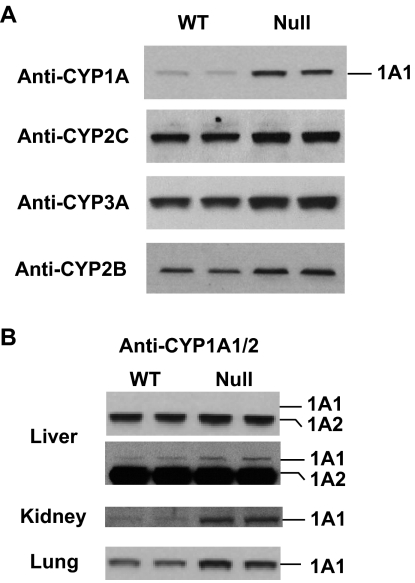
Immunoblot analysis also indicated up-regulation of CYP1A1, CYP2B, CYP2C, and CYP3A expression in the SI of the IE-Cpr-null mouse (Fig. 4A); these results are similar to those seen previously in the SI of the Cpr-low mice (Zhang et al., 2007). The apparently compensatory up-regulation of CYP2B, CYP2C, and CYP3A proteins occurred only in the SI, and not in the liver (data not shown), of the IE-Cpr-null mice. However, the up-regulation of CYP1A1 was seen not only in the SI but also in extragut organs examined, including the lung, kidney, and liver, from the IE-Cpr-null mouse (Fig. 4B). It is noteworthy that although the antibodies to rat CYP1A react with both mouse CYP1A1 and CYP1A2, the two isoforms can be resolved by gel electrophoresis, thus permitting specific detection of CYP1A1. In the liver, the main CYP1A protein detected was CYP1A2; level of this isoform also seemed to be at slightly higher levels (∼2-fold) in the IE-Cpr-null liver than in the WT liver (Fig. 4B, liver-top). Detection of hepatic and renal CYP1A1 required longer exposures of the film (Fig. 4B); nevertheless, it is clear that CYP1A1 was up-regulated in both organs, as well as in the lung. A densitometric analysis indicated that the level of CYP1A1 protein was increased approximately 3-, 6-, and 3-fold in liver, kidney, and lung, respectively. The up-regulation of CYP1A1 in extragut organs was similar to the recently reported (Ito et al., 2007) CYP1A1 induction in extragut organs of the IE-specific aryl hydrocarbon receptor nuclear translocator (Arnt)-knockout mouse model, an induction attributed by these authors to altered SI metabolism of putative dietary CYP1A1 inducers.
Fig. 4.
Immunoblot analysis of CYP1A, CYP2B, CYP2C, and CYP3A protein expression. A, microsomal proteins from the enterocytes (10 μg) of WT or IE-Cpr-null (Null) mice were loaded in duplicate and analyzed on immunoblots with use of anti-CYP1A, anti-CYP2B, anti-CYP2C, or anti-CYP3A antibodies (as indicated). B, microsomes from the liver (2 μg), lung (10 μg), and kidney (10 μg) were analyzed for CYP1A expression. The positions of CYP1A1 and CYP1A2 proteins are indicated for the liver: two films, exposed either briefly (top) or for an extended time (bottom) to the same blot, are shown to illustrate the induction of CYP1A2 (top) and CYP1A1 (bottom).
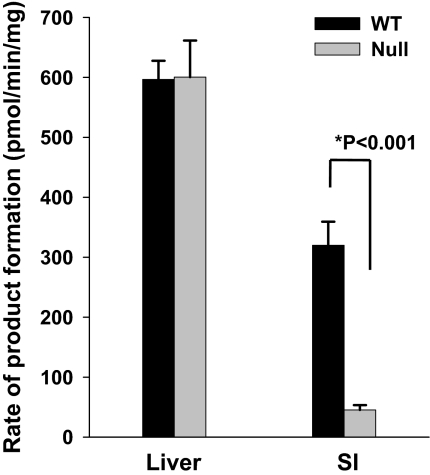
We determined the impact, with respect to NFP, of the loss of CPR expression on microsomal P450 activities in IE cells by comparing activities between the IE-Cpr-null and the WT mice (Fig. 5). Despite the compensatory increases in P450 expression in enterocytes, the rates of NFP oxide formation from NFP in SI epithelial cell microsomes from the IE-Cpr-null mice were ∼10% of the rates for WT mice. In contrast, hepatic microsomal metabolism of NFP did not differ between the two mouse strains. These findings confirm that targeted deletion of enterocyte Cpr does ablate IE microsomal P450 activity, and thus the IE-Cpr-null mouse can be used to study the role of IE P450s in the clearance of oral drugs.
Fig. 5.
In vitro metabolism of NFP by liver and SI microsomes from IE-Cpr-null and WT mice. Rates of formation of NFP oxide from NFP were determined as described under Materials and Methods. Reaction mixtures contained 200 mM phosphate buffer, pH 7.4, 25 μM NFP, and 0.5 mg/ml microsomal protein in a final volume of 0.5 ml. Reactions were performed at 37°C for 10 min in the presence or absence of 1.0 mM NADPH. Metabolites were extracted and analyzed by HPLC. Each microsomal preparation was obtained from pooled tissues from three mice. The values reported represent mean ± S.D. for quadruplicate determinations.
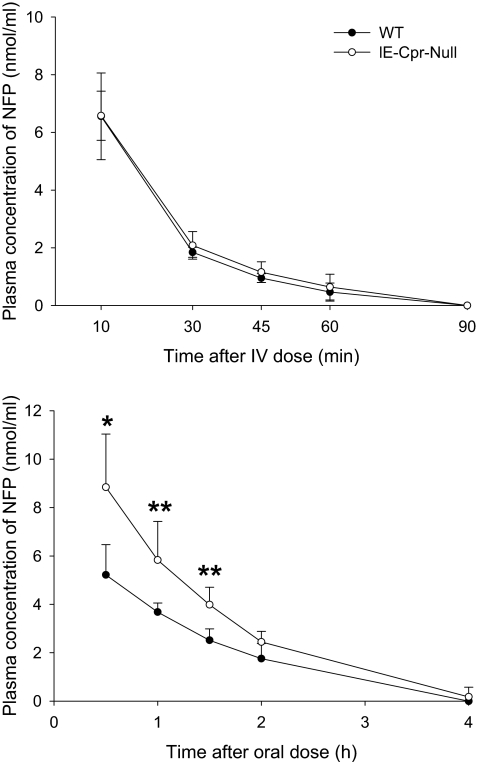
To investigate the impact, with respect to NFP bioavailability, of the loss of CPR expression and the consequent reduction in microsomal NFP oxidase activity, we performed a pharmacokinetics study on the IE-Cpr-null mice and WT mice. The concentration-time curves for blood NFP after a single-dose oral or intravenous administration of NFP are shown in Fig. 6. The calculated pharmacokinetic parameters are shown in Table 1. Male and female mice of a given genotype were combined for use in this experiment; no noticeable gender-related differences in NFP clearance were seen. After NFP oral administration at 10 mg/kg, the absorption of NFP was rapid in both IE-Cpr-null and WT mice. However, blood NFP levels were significantly higher in the IE-Cpr-null mice than in the WT mice at the first three time points monitored. Furthermore, the Cmax and area under the concentration-time curve values were, respectively, 1.8- and 1.6-fold higher in the IE-Cpr-null mice than in the WT mice, indicating that NFP disposition was much slower in the IE-Cpr-null mice than in WT mice, when administration was by the oral route. Similar differences in pharmacokinetic parameters between the WT and IE-Cpr-null mice were observed when NFP was orally given at a 20-mg/kg dose (data not shown). In contrast, after a single intravenous administration of NFP at 2 mg/kg, there was no difference in blood NFP levels at any of the time points monitored, or in any of the pharmacokinetic parameters determined, between the two strains of mice. Taken together, these data indicate that SI P450-catalyzed NFP metabolism plays an important role in controlling systemic bioavailability of NFP administered orally but not intravenously.
Fig. 6.
Blood concentrations of NFP as a function of time after a single dose of NFP. Two- to 4-month-old WT and IE-Cpr-null mice were given a single oral (10 mg/kg) or intravenous (2 mg/kg) dose of NFP. Blood samples were collected from individual mice at 0 to 4 h after oral administration or 0 to 90 min after intravenous injection. NFP was determined by HPLC as described under Materials and Methods. Values represent the mean ± S.D. for six (oral) or four (intravenous) mice in each group. Student's t test was used for statistical analysis. *, p < 0.01; **, p ≤ 0.02; significant difference between IE-Cpr-null and WT mice at the indicated time points.
TABLE 1.
Pharmacokinetic parameters for nifedipine clearance
Blood NFP levels (from Fig. 6) were used to calculate pharmacokinetic parameters, including AUC, Tmax (the time at which maximum concentration occurs), Cmax (the maximum concentration), and t1/2 (the elimination half-life). Values presented are mean ± S.D. (n = 6 for oral and n = 4 for intravenous administration). Student's t test was used for statistical analysis of differences between WT and IE-Cpr-null strains.
| Strain | Treatment Route | Tmax | Cmax | t1/2 | AUC0-∞ |
|---|---|---|---|---|---|
| h | nmol/ml | h | nmol · h/ml | ||
| WT | Oral | 0.50 | 4.9 ± 1.2 | 1.38 ± 0.74 | 8.0 ± 0.5 |
| IE-Cpr-null | Oral | 0.50 | 9.0 ± 2.5a | 0.91 ± 0.26 | 12.8 ± 2.3a |
| WT | Intravenous | 0.17 | 6.6 ± 1.5 | 0.23 ± 0.02 | 3.6 ± 0.8 |
| IE-Cpr-null | Intravenous | 0.17 | 6.6 ± 0.9 | 0.26 ± 0.05 | 3.8 ± 0.6 |
AUC, area under the concentration-time curve.
p < 0.01, as compared with similarly treated WT mice.
Discussion
Oral bioavailability of a drug is determined by multiple factors, including the extent of SI absorption, the extent of SI metabolism, and the extent of hepatic metabolism (Paine and Oberlies, 2007). Thus, the effects of SI drug metabolism on the bioavailability of some drugs can be masked by variations in other factors. NFP, known to have high solubility and high permeability, is expected to be minimally influenced by efflux or uptake transporters, unlike other drugs that can either be regulated by both P450-mediated metabolism and efflux transport, or else are eliminated (mainly through excretion into bile and urine) without undergoing metabolism (Wu and Benet, 2005). Therefore, the IE-Cpr-null mouse model will be valuable for determination of the extent to which SI P450-mediated metabolism can influence bioavailability of a given drug.
The tissue specificity of the Cpr gene deletion in the IE-Cpr-null mouse is dictated by the tissue specificity of the Vil-Cre transgene expression. Because villin is predominantly expressed in the epithelial cells of the SI, the Vil-Cre transgenic mice express Cre almost exclusively in the IE (Madison et al., 2002; el Marjou et al., 2004). In the intestine, Cre protein and activity are expressed in both differentiated epithelial cells and immature (progenitor) crypt cells throughout the crypt-villus axis, but not in goblet cells (also located in the epithelium) or in the nonepithelial cells of the gut. The endogenous villin gene is expressed in the kidney cortex, in addition to the SI; however, in Vil-Cre mice, the Cre recombinase expression and activity were much lower in the kidney than in the IE (El Marjou et al., 2004). In our IE-Cpr-null mice, Cpr deletion was detected only in the intestine and not in the kidney. Furthermore, Cre-mediated Cpr deletion was continuous over the entire length of SI with no signs of mosaicism.
The loss of CPR expression in the SI resulted in remarkable reduction of microsomal P450-mediated metabolism of NFP; the ∼10% residual activity detected in SI microsomes from the IE-Cpr-null mice can be explained by the presence of low levels of CPR expressed in nonenterocytes, including contaminating submucosal cells, in the microsome preparations. It should be noted that the extent of reduction in P450 activity toward NFP, in IE microsomes from the IE-Cpr-null mouse, was much greater than the ∼50% reduction that was observed earlier in the Cpr-low mouse (Zhang et al., 2007). This finding is consistent with the immunohistochemical data, which indicated significant residual expression of CPR protein in the IE cells of the Cpr-low mouse, but not in those IE cells of the IE-Cpr-null mouse. The differences in SI CPR expression between the Cpr-low and the IE-Cpr-null mouse dictate that whereas compensatory increases in IE P450 expression in the Cpr-low mouse (Zhang et al., 2007) will result in greater-than-expected residual P450 activity in the enterocytes, a cell type in which CPR is present at reduced levels, similar increases in P450 expression in the IE-Cpr-null mouse will not lead to residual P450 activity in the enterocytes because CPR is absent in these cells in the latter strain.
In the SI of the Cpr-low mouse, we previously found (Zhang et al., 2007) that the suppression of CPR expression in the SI and in extragut organs causes induction of multiple P450 enzymes; similarly, we find in the present study that the loss of CPR expression in the enterocytes, in the SI of the IE-Cpr-null mouse, leads to increases in the expression of several P450 forms examined, including CYP1A1, CYP2B, CYP2C, and CYP3A. It is interesting to note that the expression of CYP1A1, but not that of CYP2B, CYP2C, or CYP3A, was also increased in several other organs of the IE-Cpr-null mouse examined, namely, liver, lung, and kidney. In the liver, the level of CYP1A2, a liver-specific P450, also appeared to be increased. Thus, the CYP1A induction seen previously in the extragut organs of the Cpr-low mouse (Weng et al., 2005; Wu et al., 2005; Zhang et al., 2007) could be partly because of the specific loss of SI CPR expression and partly because of the loss of CPR expression in situ in those extragut organs. The extragut organ CYP1A induction in the IE-Cpr-null mouse can perhaps be explained by a greater accumulation in the SI of the IE-Cpr-null mouse, compared with the WT mouse, of dietary (i.e., laboratory chow) aryl hydrocarbon receptor ligands that are normally degraded by P450 enzymes in the gut. In that context, it was recently reported (Ito et al., 2007) that CYP1A1 mRNA expression and enzymatic activities were markedly elevated in almost all the extragut tissues in an IE-specific Arnt knockout mouse model, in which SI CYP1A1 expression was suppressed. Furthermore, the extragut organ CYP1A1 induction was abolished when those mice were fed purified diet instead of a normal chow; the latter contains phytochemicals capable of activating the aryl hydrocarbon receptor. The above results strongly suggested that SI CYP1A1 can regulate the bioavailability of dietary CYP1A1 inducers and, consequently, systemic expression of CYP1A1, and our present findings in the IE-Cpr-null mouse lend further support to this notion.
The induction of CYP1A1 in the liver and other extragut organs of the IE-Cpr-null mouse could alter systemic clearance of xenobiotic compounds that are CYP1A1 substrates. This potential confounder can be eliminated by feeding animals with a synthetic diet, as described by Ito et al. (2007) for the Arnt-knockout mice. For the present study, the systemic induction of CYP1A1 and the hepatic induction of CYP1A2 in the IE-Cpr-null mouse were not expected to influence the clearance of NFP, a compound that is thought to be mainly metabolized by CYP3A (Guengerich et al., 1986); this expectation was confirmed by the results of our pharmacokinetic studies of intravenous NFP, which showed no difference between the WT and the IE-Cpr-null mice.
A significant role of the SI in the first-pass extraction of oral NFP has been suggested by previous studies (e.g., Bailey et al., 1991; Holtbecker et al., 1996; Grundy et al., 1997; Yoshisue et al., 2001; Zhang et al., 2007). In the present study, we have directly shown the in vivo contribution of SI CPR/P450-mediated metabolism to the clearance of oral, but not systemic, NFP. Indeed, the oral bioavailability (Foral) of NFP, calculated based on the area under the concentration-time curve values shown in Table 1, was 44% for WT mice and 67% for the IE-Cpr-null mice, an increase of 50%. It is noteworthy that although several non-P450 enzymes, including heme oxygenases, also depend on CPR for function (Gu et al., 2003), none of these enzymes are directly involved in drug metabolism. Therefore, the impact of a reduced CPR expression on microsomal drug metabolism can be unambiguously attributed to the associated decreases in microsomal P450 activity.
The capacity of the SI to extract oral drugs is not unlimited. The biotransformation enzymes and transporters can become saturated when a sufficiently large dose is given; as a result, the relative contribution of intestinal metabolism to the overall first-pass clearance of the drug will be decreased. In the current study, the pharmacokinetic parameters for oral NFP were determined using a 10-mg/kg dose. Similar differences in pharmacokinetic parameters between the WT and IE-Cpr-null mice were observed when we administered NFP at a dosage of 20 mg/kg; the agreement suggests that the capacity of the SI first-pass clearance was not yet saturated at 20 mg/kg. A more complete dose-response study was not conducted, given the difficulties encountered in the detection and quantification of blood NFP, when NFP was given orally at 5 mg/kg or lower doses; at the other end of the scale, NFP, when given at doses higher than 20 mg/kg, could potentially cause toxicity.
The tissue specificity and the ablation of all the microsomal P450 activities in targeted cells represent unique advantages of the IE-Cpr-null mouse model. Yet, there are situations where it would also be desirable to determine the specific contributions of a given P450 enzyme to SI drug metabolism in vivo. In that context, a mouse model with gut-specific CYP3A4-transgene expression on a whole-body Cyp3a-knockout background was described recently (van Herwaarden et al., 2007). Using that mouse model, the authors convincingly showed that transgenic expression of CYP3A4 in the mouse intestine can dramatically decrease systemic bioavailability of oral docetaxel, an anticancer agent. Despite potential confounding by the compensatory increase in the expression of CYP2C enzymes in the livers of the Cyp3a-null mice (van Waterschoot et al., 2008), the “Cyp3a-knockout/CYP3A4-transgenic” mouse model developed by that group should be very useful for assessing the ability of human CYP3A4 to metabolize drugs in the SI.
In summary, we have generated an IE-Cpr-null mouse model for determination of the in vivo function of intestinal P450 enzymes. We have shown that deletion of the Cpr gene is IE-specific and that, despite compensatory increases in the expression of multiple P450s in the SI, the model can be used to show the role of SI P450 enzymes in the first-pass clearance of a model drug, NFP. We further provide evidence that strongly supports the notion that SI metabolism of putative dietary CYP1A1 inducers can influence the systemic bioavailability of these inducers. Our findings indicate that the IE-Cpr-null mouse model can be broadly applied as a much-needed in vivo tool in studies on the function of intestinal P450 enzymes in the clearance of numerous oral drugs and other xenobiotics.
Acknowledgments
We thank the Wadsworth Center for the use of the services of the Advanced Light Microscopy and the Molecular Genetics Core Facilities. We also thank Dr. Adriana Verschoor for reading the manuscript and Weizhu Yang for assistance with mouse breeding.
This work was supported in part by the National Institutes of Health National Institute of General Medical Sciences [Grant GM082978]; and the National Institutes of Health National Institute of Environmental Health Sciences [Grant ES07462].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.108.025429.
ABBREVIATIONS: SI, small intestine; P450, cytochrome P450; CPR, NADPH-cytochrome P450 reductase; LCN, liver-specific Cpr-null; NFP, nifedipine; IE-Cpr-null, intestinal epithelium-specific Cpr-knockout; DAPI, 4,6-diamidino-2-phenylindole; HPLC, high-performance liquid chromatography; WT, wild-type; Arnt, aryl hydrocarbon receptor nuclear translocator.
References
- Bailey DG, Spence JD, Munoz C, and Arnold JM (1991) Interaction of citrus juices with felodipine and nifedipine. Lancet 337 268–269. [DOI] [PubMed] [Google Scholar]
- Chen Y, Liu YQ, Su T, Ren X, Shi L, Liu D, Gu J, Zhang QY, and Ding X (2003) Immunoblot analysis and immunohistochemical characterization of CYP2A expression in human olfactory mucosa. Biochem Pharmacol 66 1245–1251. [DOI] [PubMed] [Google Scholar]
- Ding X and Kaminsky LS (2003) Human extrahepatic cytochromes P450: function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu Rev Pharmacol Toxicol 43 149–173. [DOI] [PubMed] [Google Scholar]
- Doherty MM and Charman WN (2002) The mucosa of the small intestine: how clinically relevant as an organ of drug metabolism? Clin Pharmacokinet 41 235–253. [DOI] [PubMed] [Google Scholar]
- el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, and Robine S (2004) Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39 186–193. [DOI] [PubMed] [Google Scholar]
- Fang C, Gu J, Xie F, Behr M, Yang W, Abel ED, and Ding X (2008) Deletion of the NADPH-cytochrome P450 reductase gene in cardiomyocytes does not protect mice against doxorubicin-mediated acute cardiac toxicity. Drug Metab Dispos 36 1722–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasco MJ, Silkworth JB, Dunbar DA, and Kaminsky LS (1993) Rat small intestinal cytochromes P450 probed by warfarin metabolism. Mol Pharmacol 43 226–233. [PubMed] [Google Scholar]
- Finn RD, McLaren AW, Carrie D, Henderson CJ, and Wolf CR (2007) Conditional deletion of cytochrome P450 oxidoreductase in the liver and gastrointestinal tract: a new model for studying the functions of the P450 system. J Pharmacol Exp Ther 322 40–47. [DOI] [PubMed] [Google Scholar]
- Grundy JS, Eliot LA, and Foster RT (1997) Extrahepatic first-pass metabolism of nifedipine in the rat. Biopharm Drug Dispos 18 509–522. [DOI] [PubMed] [Google Scholar]
- Gu J, Chen CS, Wei Y, Fang C, Xie F, Kannan K, Yang W, Waxman DJ, and Ding X (2007) A mouse model with liver-specific deletion and global suppression of the NADPH-cytochrome P450 reductase gene: characterization and utility for in vivo studies of cyclophosphamide disposition. J Pharmacol Exp Ther 321 9–17. [DOI] [PubMed] [Google Scholar]
- Gu J, Weng Y, Zhang QY, Cui H, Behr M, Wu L, Yang W, Zhang L, and Ding X (2003) Liver-specific deletion of the NADPH-cytochrome P450 reductase gene: impact on plasma cholesterol homeostasis and the function and regulation of microsomal cytochrome P450 and heme oxygenase. J Biol Chem 278 25895–25901. [DOI] [PubMed] [Google Scholar]
- Guengerich FP, Martin MV, Beaune PH, Kremers P, Wolff T, and Waxman DJ (1986) Characterization of rat and human liver microsomal cytochrome P-450 forms involved in nifedipine oxidation, a prototype for genetic polymorphism in oxidative drug metabolism. J Biol Chem 261 5051–5060. [PubMed] [Google Scholar]
- Hem A, Smith AJ, and Solberg P (1998) Saphenous vein puncture for blood sampling of the mouse, rat, hamster, gerbil, guinea pig, ferret and mink. Lab Anim 32 364–368. [DOI] [PubMed] [Google Scholar]
- Henderson CJ, Otto DM, Carrie D, Magnuson MA, McLaren AW, Rosewell I, and Wolf CR (2003) Inactivation of the hepatic cytochrome P450 system by conditional deletion of hepatic cytochrome P450 reductase. J Biol Chem 278 13480–13486. [DOI] [PubMed] [Google Scholar]
- Holtbecker N, Fromm MF, Kroemer HK, Ohnhaus EE, and Heidemann H (1996) The nifedipinerifampin interaction. Evidence for induction of gut wall metabolism. Drug Metab Dispos 24 1121–1123. [PubMed] [Google Scholar]
- Ito S, Chen C, Satoh J, Yim S, and Gonzalez FJ (2007) Dietary phytochemicals regulate whole-body CYP1A1 expression through an arylhydrocarbon receptor nuclear translocator-dependent system in gut. J Clin Invest 117 1940–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowski A and Lamparczyk H (1994) Evaluation of chromatographic methods for the determination of nifedipine in human serum. J Chromatogr A 668 469–473. [DOI] [PubMed] [Google Scholar]
- Kaminsky LS and Fasco MJ (1991) Small intestinal cytochromes P450. Crit Rev Toxicol 21 407–422. [DOI] [PubMed] [Google Scholar]
- Kaminsky LS and Zhang QY (2003) The small intestine as a xenobiotic-metabolizing organ. Drug Metab Dispos 31 1520–1525. [DOI] [PubMed] [Google Scholar]
- Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, and Gumucio DL (2002) Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem 277 33275–33283. [DOI] [PubMed] [Google Scholar]
- Maunoury R, Robine S, Pringault E, Léonard N, Gaillard JA, and Louvard D (1992) Developmental regulation of villin gene expression in the epithelial cell lineages of mouse digestive and urogenital tracts. Development 115 717–728. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, and Nebert DW (2004) Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics 14 1–18. [DOI] [PubMed] [Google Scholar]
- Paine MF, Hart HL, Ludington SS, Haining RL, Rettie AE, and Zeldin DC (2006) The human intestinal cytochrome P450 “pie.” Drug Metab Dispos 34 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine MF and Oberlies NH (2007) Clinical relevance of the small intestine as an organ of drug elimination: drug-fruit juice interactions. Expert Opin Drug Metab Toxicol 3 67–80. [DOI] [PubMed] [Google Scholar]
- Suzuki H and Sugiyama Y (2000) Role of metabolic enzymes and efflux transporters in the absorption of drugs from the small intestine. Eur J Pharm Sci 12 3–12. [DOI] [PubMed] [Google Scholar]
- Thummel KE, Kunze KL, and Shen DD (1997) Enzyme-catalyzed processes of first-pass hepatic and intestinal drug extraction. Adv Drug Deliv Rev 27 99–127. [DOI] [PubMed] [Google Scholar]
- van Herwaarden AE, Wagenaar E, van der Kruijssen CM, van Waterschoot RA, Smit JW, Song JY, van der Valk MA, van Tellingen O, van der Hoorn JW, Rosing H, et al. (2007) Knockout of cytochrome P450 3A yields new mouse models for understanding xenobiotic metabolism. J Clin Invest 117 3583–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Waterschoot RA, van Herwaarden AE, Lagas JS, Sparidans RW, Wagenaar E, van der Kruijssen CM, Goldstein JA, Zeldin DC, Beijnen JH, and Schinkel AH (2008) Midazolam metabolism in cytochrome P450 3A knockout mice can be attributed to up-regulated CYP2C enzymes. Mol Pharmacol 73 1029–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, DiRusso CC, Reilly AA, Black PN, and Ding X (2005) Hepatic gene expression changes in mouse models with liver-specific deletion or global suppression of the NADPH-cytochrome P450 reductase gene. Mechanistic implications for the regulation of microsomal cytochrome P450 and the fatty liver phenotype. J Biol Chem 280 31686–31698. [DOI] [PubMed] [Google Scholar]
- Weng Y, Fang C, Turesky RJ, Behr M, Kaminsky LS, and Ding X (2007) Determination of the role of target tissue metabolism in lung carcinogenesis using conditional cytochrome P450 reductase-null mice. Cancer Res 67 7825–7832. [DOI] [PubMed] [Google Scholar]
- Wu CY and Benet LZ (2005) Predicting drug disposition via application of BCS: transport/absorption/elimination interplay and development of a biopharmaceutics drug disposition classification system. Pharm Res 22 11–23. [DOI] [PubMed] [Google Scholar]
- Wu L, Gu J, Cui H, Zhang QY, Behr M, Fang C, Weng Y, Kluetzman K, Swiatek PJ, Yang W, et al. (2005) Transgenic mice with a hypomorphic NADPH-cytochrome P450 reductase gene: effects on development, reproduction, and microsomal cytochrome P450. J Pharmacol Exp Ther 312 35–43. [DOI] [PubMed] [Google Scholar]
- Wu L, Gu J, Weng Y, Kluetzman K, Swiatek P, Behr M, Zhang QY, Zhuo X, Xie Q, and Ding X (2003) Conditional knockout of the mouse NADPH-cytochrome P450 reductase gene. Genesis 36 177–181. [DOI] [PubMed] [Google Scholar]
- Yoshisue K, Nagayama S, Shindo T, and Kawaguchi Y (2001) Effects of 5-fluorouracil on the drug-metabolizing enzymes of the small intestine and the consequent drug interaction with nifedipine in rats. J Pharmacol Exp Ther 297 1166–1175. [PubMed] [Google Scholar]
- Zhang QY, Dunbar D, and Kaminsky LS (2003) Characterization of mouse small intestinal cytochrome P450 expression. Drug Metab Dispos 31 1346–1351. [DOI] [PubMed] [Google Scholar]
- Zhang QY, Kaminsky LS, Dunbar D, Zhang J, and Ding X (2007) Role of small intestinal cytochrome P450 in the bioavailability of oral nifedipine. Drug Metab Dispos 35 1617–1623. [DOI] [PubMed] [Google Scholar]