Abstract
Biomolecules dynamically work in cells in which a variety of molecules assemble and interact in unique manner. The molecular mechanisms underlying several biological processes have been elucidated from the results obtained from the descriptions of cell function, from the snapshots of the structures of biomolecules involved in these processes, and from the biochemical properties of these reactions in vitro. Recently developed single molecule measurements have revealed the dynamic properties of the biomolecules that have been hidden in the data that have been averaged over large numbers of molecules in both ensemble measurement and in cells. Single molecule imaging and manipulation of single molecules have allowed the visualization of the dynamic operations of molecular motors, enzymatic reactions, structural dynamics of biomolecules, and cell signaling processes. The results have shown that the single molecule techniques are powerful tools to monitor the dynamic actions of biomolecules and their assemblies. This approach has been applied to a variety of fields within the life sciences. As new information emerges about the dynamic actions of biomolecules using methods of single molecule detection new views on how biological processes work will be revealed.
Single molecule measurements are based on two techniques: manipulation and imaging. Manipulation of biomolecules allows biomolecules to be arranged specifically as in their biological environment and their interactions to be accurately measured (Ishii et al., 2001; Mehta et al., 1999). Visualization of single biomolecules allows the motion of biomolecules and their association and dissociation events to be measured in biomolecules at the single molecule level in real time (Ishijima and Yanagida, 2001; Kulzer and Orrit, 2001; Weiss, 2000). These methods have been used in various areas of life science. Movement of molecular motors along biomolecular tracks along with the passive and diffusive motion of biomolecules can be tracked in cells. The enzymatic reactions involving the association and dissociation events can be visualized at the single molecule level. These single molecule techniques have been extended by combining them with fluorescence spectroscopic techniques. This has allowed structural changes of single biomolecules and the changes they undergo when interacting with other biomolecules to be measured in real time.
Single molecule measurements have many unique features that are distinct from other ensemble measurements including the following. (i) The dynamic changes of biomolecules can be detected without averaging over a large number of molecules. (ii) Reactions that occur repeatedly can be traced on the same molecule and characterized by statistically analyzing the data for individual biomolecules. Events of biomolecules can be affected by the previous events that have occurred in that molecule. (iii) Events that are made up of a series of reactions can be monitored without synchronizing large numbers of molecules. (iv) The input-output, stimulus-response, or structure-function relation can be directly determined, by simultaneously measuring two parameters representing these properties in the same biomolecules. (v) Events that occur rarely can be detected. Just because these events do not occur often does not mean they are not important. Minority events can be of great importance in biological systems. Such measurements provide new information that cannot be obtained in ensemble measurements. In this perspective review, we search for the route to describe the dynamic action of biomolecules in life using single molecule measurements. Some of the interesting topics and papers are not described because of limited space.
SINGLE MOLECULE IMAGING
Optical microscopy on which the single molecule measurement systems are built is the tool that enables active events in cells to be observed with minimal damage to the molecules. Individual biomolecules in the nanometer size range have been observed by attaching large or bright probes such as polystyrene beads, glass microneedles, gold particles, and fluorescent probes. In fluorescence measurements signals can be detected from biomolecules of interest without being disturbed by light and Raman scattering. This is possible because the observations are made at a wavelength different from that of the excitation light (Michalet et al., 2003). Weak fluorescence signals from single molecules can be monitored using local illumination from a laser, which reduces the background noise and increases the signal-to-noise (S/N) ratio. Single molecules were first visualized using near-field scanning microscopy (Betzig and Chichester, 1993). Total internal reflection fluorescence (TIRF) microscopy uses an evanescent field created in the vicinity of the interface between the sample solution and the slide glass when a laser is total internally reflected (Axelrod, 1989) [Fig. 1A]. It is possible to image the events occurring in a two-dimensional (2D) plane on the surface of the slide glass (Funatsu et al., 1995). In confocal microscopy, fluorescence signals from a small area in the order of the wavelength are detected through a pinhole in front of the detector. This allows only fluorescent light from a conjugated focal plane to be detected (Amos and White, 2003). The observation points are scanned in 3D for imaging (Trautman and Macklin, 1996) or fixed to monitor molecules passing through the small illumination area [fluorescence correlation spectroscopy (FCS)] (Rigler, 1995).
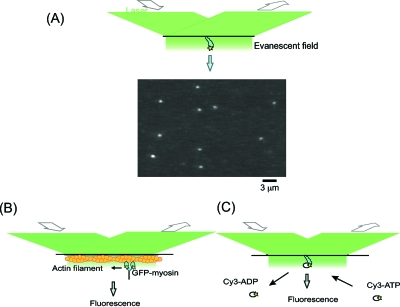
Figure 1. Single molecule imaging.
(A) Schematic drawing of a basic system of illuminating single fluorescent molecules on the glass surface in total internal reflection fluorescence microscopy (above) and typical images of single molecules. (B) Visualization of the sliding movement of a GFP-tagged myosin molecule along an actin filament immobilized on the glass surface. (C) Visualization of the turnover of ATP hydrolysis by a single myosin molecule on a glass surface.
Single fluorescence spots are an accumulation of photons successively emitted from single fluorescent molecules. The number of photons that emit fluorescence fluctuates randomly with time (Kulzer and Orrit, 2001). In addition, single molecules occasionally exhibit blinking in which the fluorescence is turned off and on due to photochemical reactions of the fluorophores (Dickson et al., 1997). Photobleaching is the event when a fluorescence probe suddenly ceases to emit fluorescence. Photobleaching has been used to confirm that the fluorescence arises from single molecules. However, because of photobleaching the measurement time is limited. Thus, fluorescence probes used for single molecule measurements must be photochemically stable against blinking and photobleaching and emit a strong fluorescence signal with a high atomic absorption, a high quantum yield, or a short lifetime. Organic fluorescent dyes can be attached to specific functional groups such as a thiol group of a cysteine residue of proteins or an amino group. Using mutagenesis techniques, cysteine residues can be introduced to specific positions that one wishes to label. Cyanine dyes and tetramethylrhodamine (TMR) were first used for single molecule imaging and since then many fluorescent dyes have been developed. YOYO families, which attach to the base of DNA molecules, are used to stain DNA. Small molecule, ATP and cAMP, which play an important role in biological activities, can be labeled chemically (Tokunaga et al., 1997; Oiwa et al., 2003). Fluorescent proteins that occur naturally such as green fluorescent protein (GFP) and their mutant forms have been used for labeling proteins both in vitro and in vivo (Tsien, 1998; Miyawaki, 2003). Recently, varieties of fluorescent proteins with a wide range of excitation and emission wavelength have been developed. The fluorescent proteins are fused to target proteins at the DNA level. Therefore, fluorescent proteins are suitable for imaging proteins in living cells. One drawback is that these probes are bulky and can only be attached to a limited number of positions. Quantum dots (QDs) are a new class of probes, which have a high intensity and a long lifetime against photobleaching (Smith et al., 2006). Once again there are problems with QDs affecting the kinetic properties of proteins and steric hindrances. Fluorescent labels having a large surface area can be used for a variety of functions, which will expand the numbr of applications they can be used for.
SINGLE MOLECULE MANIPULATION
Manipulation techniques allow biomolecules to interact with other molecules in an appropriate arrangement for effective measurements (Fig. 2). Laser and magnetic trap and glass microneedle methods have been used. The laser trap is a method used to manipulate small dielectrics by pushing the proteins towards the focal point of the laser using light pressure when a laser of suitable waverlength is irradiated (Ashkin, 1997). Biomolecules can be manipulated by attaching them to a trapped bead (Svoboda et al., 1993) or microneedle (Kishino and Yanagida, 1988). The biotin-streptavidin, antigen-hapten, and His-tag systems act as a molecule glue to attach the biomolecules or their assemblies to beads or microneedles.
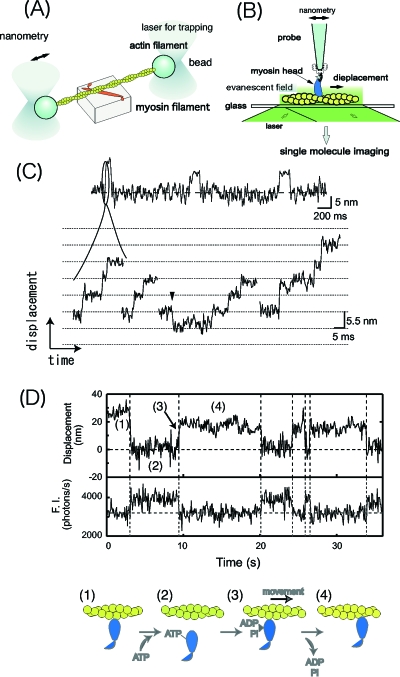
Figure 2. Single molecule manipulation and step movement of nonprocessive muscle myosin II.
(A) A single actin filament is manipulated through two beads attached at both ends using a laser trap. For mechanical measurements, the actin filament interacts with a myosin molecule immobilized on a pedestal in the presence of ATP. The movement caused by single myosin molecules can be measured by recording the displacement of one of the beads. (B) A single myosin molecule is attached to a tip of a microneedle and manipulated. The movement of a single myosin molecule is measured from displacement of the tip of the microneedle, when it interacts with actin filaments on the glass surface. (C) Displacement records of the movement of muscle myosin II on an actin filament using a scanning probe microscope. Cycles of the step movement of myosin and its return to the original position were monitored as in a laser trap measurement (top). The rising phase of the step movement was expanded and substeps of 5.5 nm were monitored. (D) Simultaneous measurement of the mechanical movement measured using a laser trap (top) and ATP hydrolysis turnover measured using single molecule imaging (bottom) with a fluorescent ATP analogue. Myosin strongly interacting with actin (1) dissociates upon binding of ATP to myosin. Myosin interacts weakly with actin (2) and generates mechanical motion when Pi and ADP dissociates from myosin (3) and then myosin interacts with actin strongly (4 and 1).
These manipulation methods are also used for mechanical measurements. When biomolecules attached to large probes are displaced from the equilibrium position, mechanical forces are applied to pull them back to their original position. The response of the biomolecules against such mechanical forces can be monitored. For example, the force vs extension relation of biomolecules and their assemblies can be determined, when both ends that have been attached to probes are extended. The structural changes or breakage of an assembly of biomolecules caused by the mechanical forces can also be monitored. The molecular motors move against the trapping force. The force generated by molecular motors can be measured from the displacement when the stiffness of the system is known. The displacement of the beads or tip of a microneedle can be measured with nm accuracy using a pair of photodiodes. Given that the spring constant is less than 1 pN∕nm, forces can be measured in the order of pNs. Trapped beads and the tips of a microneedle undergo thermal fluctuation depending on the stiffness of the system. The extent of thermal fluctuation can be greater than the size of the measurement that is being measured.
PROBES AND THEIR EFFECTS ON MEASUREMENTS
Improvements in the time and space resolution, stability of the measurements, length of the measurement time, and the configuration of the measurement systems will make new insights of the mechanisms underlying the behavior of biomolecules possible. The development of stable fluorescence probes is important. Rigid linkers between the biomolecules and the large probes improve the resolution of the mechanical measurements. In instances where the fine structure needs to be resolved on a high scale, it is critical to improve the resolution.
In single molecule measurements, the use of either fluorescent probes to visualize molecules or probes to monitor mechanical changes are essential. It is almost impossible to trace the changes of single molecules without the use of probes. However, the probes may alter the properties of the biomolecules themselves or their behavior. For accurate measurements it is important to minimize the effects of probes on the function of biomolecules to which the probes are attached. The effect of the fluorescent probes will depend on their size, physical and chemical properties, and the position where the probes attach. The probes restrict some degree of freedom of motion. The restricted motion may mimic the behavior in the biological environment. Biomolecules can also become restricted when they are assembled with large numbers of other biomolecules. In other cases probes may slow down reactions. This however, can be used as an advantage when rapid reactions are being measured that could not otherwise be measured without the probe. However, it is important to confirm that the essential mechanisms have been maintained.
NONPROCESSIVE MOVEMENT OF MUSCLE MYOSIN II
Basic techniques of single molecule measurements have been developed and used in the research of molecular motors (Dantzig et al., 2005). The single molecule technology was a prerequisite for understanding the mechanism of the molecular motors and in fact its development greatly contributed to the description of the operations of molecular motors. Muscle is a macroscopic machine in which myosin and actin assemble in a hierarchic structured manner. The sliding movement of myosin and actin filaments is thought to cause muscle contraction and this molecular event is amplified to macroscopic force and motion. However, the direct observation of the sliding movement using purified actin filaments and myosin molecules under microscopy was only possible once individual actin filaments could be visualized (Yanagida et al., 1984). Actin filaments fluorescently labeled with phalloidin move on myosin molecules immobilized on the slide glass when ATP was added as an energy source (Kron and Spudich, 1986). A fundamental question arising from the original motility assays was: how far do actin filaments move during hydrolysis of a single ATP molecule? Many assumptions were required to estimate this distance because many molecules are involved in the movement of a single actin filament. Single molecule measurements were essential to answer this question unambiguously (Toyoshima et al., 1990; Harada et al., 1990). The development of techniques to manipulate actin filaments using a laser trap or glass microneedle allowed the unitary step caused by myosin interacting with actin in the presence of single ATP molecules to be determined (Finer et al., 1994; Ishijima et al., 1994). The stepwise movements of the actin filaments were observed when they were attached to beads trapped by a laser at both their ends or to a glass microneedle at one end [Figs. 2A, 2C]. However, muscle myosin readily dissociates from actin filaments after each stepwise movement. The step size of the individual step movement has not been determined as it has only been possible to determine the average step size (Molloy et al., 1995). This is because the initial position of the step movement could not be determined due to the large Brownian motion just prior to the steps. The obtained average step size varies greatly (5–20 nm) between the different research groups (Mehta et al., 1999; Tyska et al., 1999; Molloy et al., 1995; Tanaka et al., 1998).
Manipulating single myosin molecules instead of the actin filaments allowed the measurements to be made with a greater resolution (Kitamura et al., 1999). A single myosin molecule attached to a scanning probe will interact with actin filaments on the surface of a glass slide [Fig. 2B]. It is possible to detect the rising phase of a step using this measurement system partly because the step is slowed down. Single steps have been found to contain several substeps 5.5 nm in size which corresponds to the size of an actin monomer [Fig. 2C]. The steps occur randomly and are independent of ATP. Occasionally steps occur in the backward direction which suggests that the movement of muscle myosin is driven by biased Brownian movement during hydrolysis of single ATP molecules. The velocity (inverse of the dwell time between the substeps) of myosin was measured as a function of load. The force-velocity relation obtained from single molecules was the same as that obtained in muscle, indicating that some of the characteristic properties of macroscopic machines originate from the basic units of the molecular machine. So far the substeps have not been observed with any of the other single molecule detection techniques. The difference in the results may be explained by the difference in the arrangement of proteins and probes in the different measurement system.
OBSERVATION OF SINGLE ATP TURNOVERS AND THE COUPLING WITH THE STEP MOVEMENT OF MYOSIN
The binding and dissociation of fluorescently labeled molecules can be visualized on partner molecules that have been immobilized on the surface of a glass slide [Fig. 1C] (Funatsu et al., 1995). Using such imaging methods the chemical reactions that involve the binding and dissociation of small molecules and large biomolecules have been visualized at the single molecule level. The hydrolysis of ATP by single myosin molecules was detected using fluorescently labeled ATP analogues. When fluorescently labeled ATP was bound to myosin molecules that have been fixed on a glass surface, fluorescence spots appeared where the myosin molecules were located and disappeared when fluorescent ADP dissociated after the hydrolysis of the fluorescent ATP. The average duration time between the binding and dissociation was almost the same as the turnover time of the ATP hydrolysis at high ATP concentration in ensemble measurement in solution.
A basic question regarding the mechanisms of molecular motors is: how is the chemical energy converted to mechanical work? Single molecule measurements allow the timing of the chemical reactions to be related to the mechanical events, because both parameters can be measured from the same biomolecule without averaging. The ATP turnover was measured using fluorescently labeled ATP. The step movement of muscle myosin was measured using a laser trap. The two measurements were combined to measure how the step movement of myosin is related to the hydrolysis of ATP (Ishijima et al., 1998). As expected from the biochemical studies, myosin dissociated from actin when ATP bound to myosin [Fig. 2D]. However, the timing of the generation of the step movements varied relative to the dissociation of ADP. In 50% of the events, the step movement was delayed after ADP had dissociated while the steps occurred at the same time as the dissociation of ADP for the remainder of the events. The thermodynamic and kinetic studies using ensemble measurements suggests that the free energy derived from ATP hydrolysis is released when Pi is released. This occurs at almost the same time as the release of ADP. Measurement systems need further improvement to obtain a clear answer.
Until single molecule measurements were possible, the measurements of the energy supply and their use were carried separately using different measurement systems. ATPase reactions were measured in solution in test tubes using purified proteins and the mechanical measurements were performed using muscle fibers. As both parameters were measured separately in different systems the only way to relate them was to assume a one-to-one coupling between the chemical reaction of ATP and the mechanical events. This assumption seemed reasonable in the framework of the measurement of average values. It is now possible to make both measurements under the same experimental conditions. The coupling between the two reactions can be experimentally determined.
Simultaneous measurements of two different parameters from the same molecules potentially have great importance. Given that structure of biomolecules can be measured at the single molecule level, the structure-function relation can be determined by measuring structure and function at the same time. In these types of measurements, the function must be defined and measured at the single molecule level. The ion currents through single ion channels can be monitored at the single molecule level using a patch clamp method. Single channel molecules and single molecules bound to channel molecules can now be visualized under the same conditions as the ion current is measured (Ide et al., 2002; Sonnleitner et al., 2002; Blunck et al., 2004; Iwamoto et al., 2006). Unveiling the structural-functional relationship of an ion channel is an intriguing project with realistic outcomes.
PROCESSIVE MOLECULAR MOTORS
Nonprocessive muscle myosin II readily dissociates from actin filaments in each cycle of the ATPase reaction. This makes single molecule measurements difficult. In contrast, processive molecular motors such as kinesin and unconventional myosin V and VI successively step for long distances without dissociating. Kinesin is a microtubule-based motor that transports cargo in cells. The step movement of kinesin is measured using a laser trap system in which kinesin attached to a bead interacts with microtubules immobilized on a glass surface [Fig. 3A] (Svoboda et al., 1993). The step size determined from individual steps is 8 nm. This is the same as the unit size of the microtubules. At low loads, the step movement is primarily directed towards the plus end of the microtubules. Steps in the backward direction occasionally occur. The population of backward steps relative to the forward steps increases with increasing load. Eventually the number of steps in the forward and backward direction is the same and this occurs at a stall force of 7 pN (Nishiyama et al., 2002). In superloaded conditions (forces greater than 7 pN), kinesin exhibits successive steps in the backward direction (Carter and Cross, 2004).
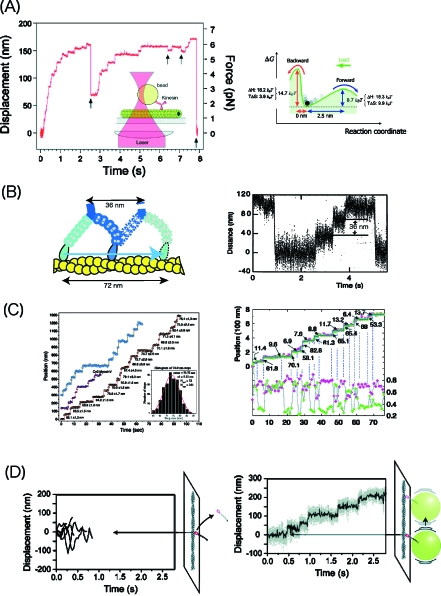
Figure 3. Step movement of processive molecular motors.
(A) The step movement of kinesin was measured by attaching to a bead trapped by a laser. Successive stepping movements were monitored as the external force increased up to 7 pN (left panel). Backward steps was observed (indicated by arrows). An energy landscape for the forward and backward steps was determined by statistically analyzing the step movement. (B) Step movement of myosin V. A cartoon of a step movement (left) and step movement in the presence of a load obtained from a laser trap measurement. (C) Step movement of myosin V measured in the absence of a load using single molecule imaging (left) and a histogram of a step size (insert). Changes in the orientation of a head coupled with the step movement determined by simultaneous measurement of the position (top two records) and polarization (bottom two records) of a fluorescent probe rigidly attached to a myosin head. Individual records for two sets of data were measured using two orthogonally polarized excitation lasers (green and red points). (D) Nonprocessive movement (left) and cargo-anchored processive movement of myosin VI (right). In the absence of a bead, single-headed myosin VI undergoes Brownian movement and readily dissociates from the actin filaments (left). When myosin VI is attached to a bead without a trap laser, step movement of 36 nm was monitored.
Kinesin has two motor domains (heads) connected at its tail domain. For processive movement, one head steps while the other head remains bound to the microtubules. It has been suggested that the ATPase reaction of the two heads is performed in a coordinated manner via interhead strain. Evidence that the affinity of ADP for kinesin changes, under a strain, has been obtained from experiments where kinesin is pulled by an external force through a trapped bead (Uemura and Ishiwata, 2003). The effect of the force was dependent on the direction: the binding affinity of ADP is enhanced when the force is applied in the same direction as the movement, and it is reduced when the force opposes the movement. During step movements, ADP may dissociate more readily at the leading head than at the tailing head due to a strain between the two heads. Direct evidence for the presence of this interhead strain is important for future research. Hand-over-hand processive movement requires two heads. However, some single-headed kinesin molecules have been found to move processively but in a diffusive manner when additional interaction with microtubules prevents them from dissociating. Unconventional KIF1A, a naturally single-headed motor in the kinesin family exhibits Brownian movement on microtubules in the presence of ATP (Okada et al., 2003). The step size is distributed around a multiple of 8 nm steps stochastically and the direction of the movement varies randomly while the overall movement is clearly directional toward the plus end of microtubules. The Brownian movement observed in this single-headed motor may be similar to a mechanism used in part in two-headed motors. Cytoplasmic dynein is a gigantic molecular motor that moves toward the minus end of the microtubules. Like kinesin, dynein moves with 8 nm steps processively (Reck-Peterson et al., 2006; Toba et al., 2006). Diffusive movements are involved in the step movement as the step size is variable and the movement is directional.
Unconventional myosin V and VI are actin-based processive motors. The step movement of myosin V has been intensively studied using a laser trap (Rief et al., 2000). The step size of the movement is large, 36 nm [Fig. 3B]. The step movement of myosin V can also be visualized using single molecule imaging [Fig. 3C]. In a fluorescence spot, the photons from single molecules are distributed around a real position and the movement can be tracked with the accuracy for several nm by fitting a photon distribution for each spot. The hand-over-hand mechanism was formulated after the two heads were differentially labeled and their movement monitored (Yildiz et al., 2003). The two heads of myosin V moved alternatively using 72 nm steps. In the meantime, fluorescence polarization measurement from probes, which had been rigidly fixed to calmodulin on the neck region connecting the heads and the tail, showed the change in the orientation of the neck between the two directions during the step movement [Fig. 3C] (Forkey et al., 2003). Furthermore, it was confirmed by simultaneously measuring the step movement and polarization that the change in the orientation is associated with the step movement (Syed et al., 2006). These data appear consistent with the widely accepted lever-arm model in which the step movement is driven by the rotation of the neck region. However, it is also possible that the movement causes the rotation of the neck domain, which then triggers the chemical reaction. In these measurements, only the states before and after the step movement can be monitored. It will be important to monitor the processes underlying the movement to understand the mechanism of the step movement. This process is very fast, but the details of the step can be monitored when it is slowed down.
The step size of two-headed myosin VI (36 nm) is the same as that of myosin V although the neck domain of myosin VI is short (Rock et al., 2001). Diffusion plays an important role in producing long steps and processive movement which is consistent with the broad distribution of the step size of myosin VI. In cells, however, myosin VI is single headed rather than double headed (Lister et al., 2004). Single-headed native myosin VI is nonprocessive but when the beads bind to the head the native myosin VI moves processively [Fig. 3D] (Iwaki et al., 2006). A myosin head will detach from the binding site and attach to the next binding site on the actin filaments before a bead attached to myosin VI diffuses away, because diffusion of large beads is slow. In cells, it is likely that cargoes make the movement of single-headed myosin VI processive. However, there has been a report that cargoes initiate the dimerization of myosin VI (Park et al., 2006).
The observations of individual steps has clearly shown that Brownian movement is involved in the step movement. In these molecular motors Brownian movement is biased to one direction, since overall movement occurs primarily in one direction. How does Brownian movement become biased is the next question that needs to be addressed. Conformational changes, strain sensor, and steric effects are possible mechanisms.
FROM SINGLE MOLECULES TO SYSTEMS
The basic nature of molecular motors has been studied using isolated single motors. In cells, however, numerous motors exist and the behavior of single motors may be influenced by other motors. Therefore, it is important to monitor the behavior of individual biomolecules in the systems and cells where multiple molecules are present. These results can then be compared to results obtained from isolated single molecules. Take, for example, the in vitro gliding assay of multiple numbers of molecular motors. When the number of kinesin motors attached to the fluorescent bead increased in the in vitro gliding assay in the presence of ATP, the long range cooperativity of the binding of kinesin was indicated (Muto et al., 2005). In these measurements, once a kinesin motor was bound to the microtubules, the binding of the other motors was attracted to neighboring motors. These results should be confirmed and the role of the cooperativity should be discussed.
The final goal of the studies of the mechanism of molecular motors is to understand the movement of molecular motors in living cells. The movement of microtubules-based motors in living cells was monitored by tracking cargo fluorescently labeled with GFP (Kural et al., 2005) or introducing QDs via endocytosis (Nan et al., 2005). High resolution measurements and analysis showed step movements of 8 nm and multiple 8 nm steps along microtubules. Many questions were raised as a result of these measurements. Do the two microtubules-based motors, kinesin and dynein, who transport cargo in opposite directions, compete or share their work load? How many motors are involved in the transport of the cargo? Is the speed of the molecular motors different in living cells? It will be possible to answer such questions in the near future.
Nonprocessive motors, muscle myosin II, appear to be designed such that many motors work together. These motors may have special strategies which will be revealed as the single molecule measurements are being extended to systems that contain multiple numbers of motors.
DIFFUSION OF BIOMOLECULES IN BIOSYSTEMS
Thermal Brownian movement has been implicated in the step movement of molecular motors. One-dimension diffusion along long molecules or the linear assemblies of biomolecules has been observed as the biomolecules search for their target positions. RNA polymerase uses diffusion to search for promoter sites along DNA (Harada et al., 1999). Unconventional kinesin that depolymerizes the end of microtubules, diffuses to search for its binding site along the microtubules (Helenius et al., 2006). The diffusion motion is not fueled by ATP. Two-dimension diffusion has been observed in biomembranes. Phospholipids bilayer membranes that separate cells and organelles from the extracellular environment are fluid. The Brownian movement of fluorescently labeled lipid and protein molecules in the biomembranes has been monitored on bilayer membranes artificially formed on the glass surface or on the tip of a pipette (Ide and Yanagida, 1999; Schütz et al., 2000; Schwille et al., 1999). The mean square displacement is proportional to the time and the diffusion constant is determined as a proportional constant. The diffusion of lipid and protein molecules is restricted in plasma membranes as the transmembrane proteins are anchored by the actin cytoskeleton inside the cells (Kusumi et al., 2005). In agreement with the compartmentalized structure of the membrane, the protein and lipid molecules diffuse freely within a compartment and then hop between other compartments. In the biomembranes of living cells, it is thought that raft domains exist in which specific lipids such as cholesterol and sphingolipids cluster and play an important role in cell signaling. Single molecule imaging has also been used to characterize the structure of this raft domain (Kahya, 2006).
In cells biomolecules diffuse or are transported by molecular motors. Being able to track signals from molecules in living cells is crucial to understand the signal transduction processes. The signal molecules from the external environment bind to receptors on the biomembranes. The movement of individual receptors has been monitored using fluorescently labeled receptors or ligands that bind to the receptors (Lidke et al., 2004; Jacquier et al., 2006). The ability of receptors to move laterally for the binding of the neurotransmitter plays a central role in the development and plasticity of synapses. The movement of individual glycine receptors to which the neurotransmitter binds has been tracked using QD-tagged receptors in the time scale ranging from milliseconds to minutes (Dahan et al., 2003). Multiple diffusion domains have been characterized in relation to synaptic, perisynaptic, and extrasynaptic receptor localization. Diffusion has been used to distinguish between different states of biomolecules and their locations within the systems.
Inside cells, the performance of DNA and RNA is crucial in the complex events of gene expression. DNA and RNA can be visualized in cells by fusing fluorescent proteins to their binding proteins. The movement of individual mRNA molecules that have been transcribed has been tracked after it had been released from the transcription site (Shav-Tal et al., 2004). The movement of this complex is not directed but is governed by Brownian movement in the nucleus in living cells. Many movements are constrained to chromatin-poor domains and excluded by newly formed chromatin barriers.
ROTARY MOTORS F0F1 AND THE SYNTHESIS OF ATP
It has been proposed that the proton-flow driven ATP synthase F0F1 is a rotary motor (Boyer, 1993) and its crystalline structure supported this proposed model (Abrahams et al., 1994). Direct evidence of the rotation of F1lecule measurements (Noji et al., 1997). The rotational motion of a central rotor γ subunit around a peripheral stator (αβ)3 of F1 was first visualized by attaching a fluorescently labeled actin filament several μm in length to the γ subunit, while the stator was immobilized to the glass surface. The rotation was unidirectional. A unit step of 120° rotation was clearly identified when the rotation was slowed at low ATP concentrations (Yasuda et al., 1998). The kinetic analysis of the stepping movement at various ATP concentrations showed that the 120° step is coupled to the hydrolysis of single ATP molecules. The rotational movement of F1 at zero load was observed by attaching small and bright gold beads instead of actin filaments (Yasuda et al., 2001). The bead image was taken in dark field on a time scale of submilliseconds and the full speed rotation of about 130 revolutions per second was consistent with the ATPase rate. The measurement at high resolution also made it possible to resolve the substeps of the rotation.
Together with F0, the F1 rotary motors synthesize ATP from ADP and Pi, using the rotational movement resulting from the proton flow through the F0 section of the cells (Boyer, 1997). Single molecule measurements have shown that ATP is synthesized when the rotation of F1 is mechanically induced (Ito et al., 2004). The rotation of F1 is induced in the presence of ADP and Pi. This was determined by monitoring magnetic beads attached to the γ subunits on the F1 complex. The amount of ATP synthesized by F1 immobilized on the glass surface in a small droplet has been measured using a luciferase-luciferin reaction. ATP is synthesized only when the γ subunits are rotated in the opposite direction to which F1 rotates in the presence of ATP. Thus, these studies show that unlike linear motors the F0F1 rotary motors are reversible.
DNA-BASED MOTORS
The duplication and transcription of DNA is controlled by a variety of DNA binding proteins. The proteins that processively move along a DNA strand by hydrolysis of NTP are also classified as molecular motors and their movement has been monitored using single molecule measurements (Bustamante et al., 2004). The force generated by DNA-based motors (25–35 pN) is greater than that generated by myosin or kinesin and the velocity (5 nm∕s) is slower (Wang et al., 1998; Wuite et al., 2000). However, the underlying mechanical processes are not as well known as motors such as myosin and kinesin.
RNA polymerase is the enzyme that copies RNA from DNA and DNA polymerase is the enzyme that copies DNA to make more DNA. A single RNA polymerase molecule moves along a DNA molecule that is attached at both ends to trapped beads in a dumbbell-type arrangement. The experimental system has been recently improved and the movement generated by RNA polymerase has been measured with ångström accuracy. The stepwise movement was 3.7 Å, which is a distance equivalent to a base pair of DNA (Abbondanzieri et al., 2005). Thus, RNA polymerase takes a single base pair per nucleotide in addition to the nascent RNA. RNA polymerase also exhibits backward movements, which may be related to the proofreading of the transcription (Shaevitz et al., 2003). RNA polymerase moves along a strand of DNA, which wraps around the main axis, while it advances. When RNA polymerase molecules are immobilized on a glass surface, the rotation of DNA molecules can be detected using magnetic trapping (Harada et al., 2001). The magnetic beads attached to one end of the DNA molecule are pulled upward and allowed to rotate freely. The rotation of small fluorescent beads attached to the magnetic bead is detected during the movement of RNA polymerase.
Unlike actin and microtubules-based motors, the conformation and topology of the changes in the track upon the interaction with the motors. The changes have been used as a tool for detecting the translocation of the DNA-based motors. The movement of DNA polymerase is accompanied by the opening of double stranded DNA. Using the difference in the force-extension curve between double stranded and single stranded DNA, the movement of DNA polymerase has been monitored (Wuite et al., 2000). Topoisomerase II is an enzyme that relaxes the supercoil of DNA. The activity of this enzyme has been measured by monitoring the change in the topology of DNA (Strick et al., 2000). One end of a DNA molecule was attached to the glass surface and the other end to a magnetic bead, through which the DNA molecule was twisted by applying a magnetic field. The translocation of helicases that unwind DNA or RNA or destabilize the secondary structure of RNA were measured by monitoring the extension of DNA or RNA using manipulation techniques and the FRET imaging technique (Dumont et al., 2006; Ha et al., 2002). These DNA-based motors comprise multiple proteins. Dynamic assembly and disassembly of these molecular motors are important in relation to their functions. The formation of RecA filaments was directly observed on single DNA molecules using fluorescently labeled RecA (Galletto et al., 2006). Images were taken in succession with a short time interval allowing the nucleation and filament growth to be measured.
STOCHASTIC ENZYME REACTION
Single events of the reactions caused by single biomolecules can be measured. However, the reactions cannot be measured by monitoring just one single event because the reaction is stochastic. The reactions of numbers of events must be measured repeatedly and the data then analyzed statistically. Single molecule measurements allow reactions occurring on the same molecules to be detected in succession. Synchronization is not necessary in these measurements unlike in the reactions with ensemble measurements. An example of the analysis of an enzymatic reaction is the reaction underlying the step movement of kinesin [Fig. 3A]. The dwell time between the steps and the direction of individual steps, either in the forward or backward directions have been measured as a function of load, ATP concentration, and temperature (Taniguchi et al., 2005). These data have been interpreted by constructing a kinetic model which includes the rate constants. The energy landscape for the forward and backward step movement was determined using the rate constants obtained. The difference in the activation free energy between the forward and backward movement, which determines the directionality, was ∼6kBT. Furthermore, the difference in free energy between the backward and forward steps could be broken down into the contributions of enthalpy and entropy from the dependence of temperature. From these results it was concluded that the directional step movement was driven entropically. Thus, the behavior of biomolecules can be described thermodynamically by statistically analyzing the data.
Thermal effects are considerably large on biomolecules of nanometer size when water molecules collide randomly. In fact, thermal fluctuations are comparable in size to the biomolecules themselves. Thermal fluctuations are very fast since water molecules collide in the order of femtoseconds (10−15 s) to picoseconds (10−12). The enzymatic reaction and step movement of molecular motors commence when biomolecules acquire energy beyond the activation energy level for the reactions. The rate for this step is the order of submillisecond, millisecond to seconds.
When the turnover of the enzymes occurs repeatedly on the same molecule, each turnover occurs independently or dependent on the previous events. Memory effects have been reported. Cholesterol oxidase, a flavoenzyme catalyzes the oxidation of cholesterol by oxygen. The enzymatic turnover of cholesterol can be monitored at the single molecule level using an active fluorescent site flavin adenine dimucleotide (FAD), which is naturally fluorescent in an oxidized form (FAD) but not in a reduced form (FADH2) (Lu et al., 1998). The fluorescence of single FAD molecules turns on and off within a time interval in the order of several hundreds of milliseconds which corresponds to the turnover of single enzyme molecules. The reaction obeys Michaelis-Menten kinetics. The rate of the turnover fluctuates slowly. The turnover events are influenced by previous turnovers. The memory effect lasts for several turnovers. This memory effect was explained by slow conformational fluctuations, which were detected by single molecule spectroscopy.
FOLDING OF BIOMOLECULES
Structures of biomolecules have been determined in accordance with a thermodynamic principle. In their native structure, biomolecule chains connect hundreds to thousands of unit structures which are folded into unique structures corresponding to a free energy minimum. Biomolecules fold through an ensemble of pathways on a rugged energy landscape (Onuchic and Wolynes, 2004). In single molecule measurements, individual pathways can be distinguished, while only average routes can be detected using ensemble measurements. The folding processes of biomolecules have been studied at the single molecule level using imaging and mechanical methods (Weiss, 2000; Fisher et al., 2000). Single molecule FRET measurements showed the distribution of the structure when the protein unfolding was chemically induced (Schuler, 2005; Haas, 2005; Lipman et al., 2003). Technically it is important to measure the time trajectory of structure from biomolecules when they are not immobilized for long periods of time. The folding and unfolding processes have been reported using single protein molecules contained in tiny lipid vesicles tethered to the surface of the glass slide (Rhoades et al., 2003).
The measurements of the mechanically induced unfolding of a protein at the single molecule level was first carried out with a gigantic protein titin, which is composed of a repeating unit of immunoglobulin (Ig) domain (Tskhovrebova et al., 1997; Rief et al., 1997; Kellermayer et al., 1997). The force-extension relation of titin showed a characteristic sawtooth pattern when the molecule fixed at one end was stretched through a trapped bead or a cantilever. This protein is extensible when it is stretched and an abrupt drop of the force occurs due to the unfolding of the Ig domain in an all-or-none fashion. The force required for the unfolding of the Ig domain depends on the speed at which the filament is stretched.
The folding and unfolding trajectories of individual protein molecules can be detected using force microscopy in the force clamp mode [Fig. 4A] (Fernandez and Li, 2004). A single polyubiquitin molecule, in which a small protein ubiquitin is polymerized, is attached to a cantilever. Upon a step increase in the stretching force, ubiquitin is stretched to unfold to the unfolded structure in a discrete step. Furthermore, when the stretching force is rapidly quenched after stretching, ubiquitin folds with a continuous trajectory with distinct stages. The folding of ubiquitin does not occur by transitions between well defined structural states. The kinetics of the folding from different unfolded states may be different. Mechanically induced folding and unfolding is essentially different from the chemical and thermal unfolding traditionally used in biochemical measurements. Mechanically induced unfolded conformation is the conformation of which end-to-end distance is fixed and long. In chemically or thermally induced unfolding conformation the end-to-end distance is not fixed and is thought to be relatively short. In cells biomolecules fold in a diverse range of environments, many of which are not known. The folding of GFP was monitored at the single molecule level in the presence of chaperonins, molecular machines that assist the folding of proteins (Taguchi et al., 2001).
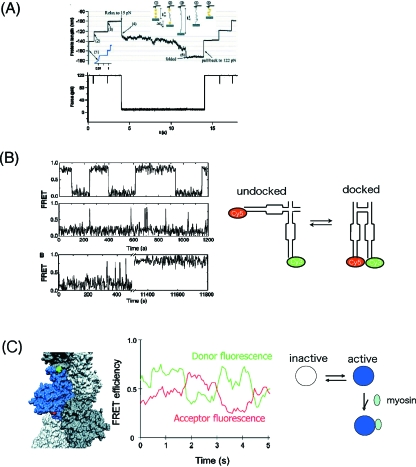
Figure 4. Dynamic structures of biomolecules revealed by single molecule measurements.
(A) Folding processes monitored using force clamp measurement. Polyubiquitin was attached to a probe and constant forces were applied to measure the protein length (top panel) in response to rapid changes in force (bottom panel). When the force was applied, stepwise expansion of the polyprotein was observed due to the unfolding of a unit of ubiquitin. When the force was released suddenly, continuous folding of ubiquitin was observed. (B) Spontaneous structural fluctuations of single hairpin ribozyme molecules. The ribozyme switched its structure between docked and undocked structures, which could be monitored using FRET between two probes on a molecule. Two kinetic states were observed with a long time duration. (C) Dynamic structure of actin. Actin molecules were specifically labeled with two different probes (left) and the donor and acceptor fluorescence showed the FRET varied between two conformational states (middle). The data were interpreted as actin dynamically changes the structure between active and inactive states and preferential binding of myosin to an active state of actin activates myosin motility (right).
DYNAMIC STRUCTURES OF BIOMOLECULES
Dynamic structures of biomolecules play an important role in the dynamic action of biomolecules. Multiple conformational states and the spontaneous transition between them are observed for RNA enzymes (ribozymes) as well as proteins. Using single molecule FRET from two probes in a hairpin ribozyme, the dynamic changes in the structure of individual molecules can be monitored (Zhuang et al., 2002). The time trace of the FRET efficiency shows transitions between active docking states (high FRET) and inactive nondocking states (low FRET) [Fig. 4B]. The existence of multiple docking states has been shown by heterogeneous undocking kinetics. These can be explained by the slow interconversion due to cleavage and ligation reactions which are not able to be measured in the FRET change (Nahas et al., 2004). The slow interconversion may result in the memory effect on the docking and undocking reactions.
Multiple conformations of the protein, actin, and the transitions between them have been observed [Fig. 4C] (Kozuka et al., 2006). Actin serves as a track for the movement of myosin when in filamentous form. The conformational dynamics of actin is important to understanding the mechanism of the activation of myosin motility. Single molecule FRET of actin monomers in the filament has revealed that actin spontaneously switches conformation between two states. The conformational states relate to the activation and inhibition of myosin motility. The active and inactive conformation states preexist and the binding of myosin preferentially selects the active conformation states. Thus actin filaments play an active role in the activation of the myosin motility through their conformational changes. Similar conformational dynamics has been observed in the signaling protein, Ras (Arai et al., 2006). Ras changes conformation between multiple states. The binding of the Ras binding protein preferentially selects one of the conformations. The idea of self-regulation of proteins using the structural dynamics is not new but is important to understand the function of these proteins in the systems and cells, in which biomolecules self-assemble, self-activate, and self-control. The single molecule detection techniques that can measure these structural dynamics will be powerful tools for measurements in both the systems and cells as well.
CHANGE IN THE STATE OF BIOMOLECULES IN LIVING CELLS
In in vitro single molecule measurements, the diverse conformational, kinetic, and behavioral states of biomolecules can be observed over time and molecules. In cells, biomolecules dynamically assemble and disassemble. Their location in the cells and the environments within the cells are also factors that affect the diversity of biomolecules. Biomolecules are localized spontaneously or in response to an external stimulus. The structures, biochemical states, physical and chemical properties, interaction states, or other parameters of the biomolecules may change between several states associated with the localization of the biomolecules and changes in the environments.
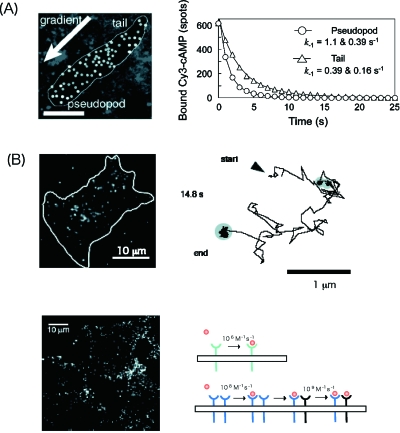
Heterogeneity in the kinetic states of signaling molecules occurs when a ligand binds to a receptor during chemotaxis in Dictyostelium cells, which exhibit chemotaxis to a chemical gradient of cyclic adenosine 3′, 5′-monophosphate (cAMP) (Ueda et al., 2001). Using a fluorescently labeled cAMP the binding of cAMP to the receptor on the cell surface was observed at the single molecule level [Fig. 5A]. Although the binding spots were distributed homogeneously on the cell surface, the dissociation rate of cAMP was faster in those cells forming the pseudopods than in the tail half of chemotaxing cells by a factor of approximately 3. The differences in the receptor states reflect a coupling with molecules downstream, i.e., a heterotrimeric G protein. It is likely that the receptors can transduce signals at the pseudopod regions more efficiently than at the tail regions of chemotaxing cells.
Figure 5. Imaging of activation processes of signaling in living cells at the single molecule level.
(A) Imaging of the binding of cAMP to dictyostelium. Fluorescently labeled cAMP was used for imaging the binding to living cells moving in the presence of a gradient of cAMP. The duration time of the binding measured was plotted for cells moving in two locations. The faster dissociation of cAMP was related to the polarization of the cell for chemotaxis. (B) The imaging of a signal receptor molecule on the cell membrane. Lateral movement of a receptor was measured by tracking a complex of NGF receptors with fluorescently labeled NGF. An activation occurs when the diffusive movement is paused. (C) Imaging of the binding of signaling molecules to a dimer of the receptor. Fluorescently labeled EGF was used for imaging. From the fluorescence intensity, the mechanism of the binding to EGF receptor was determined by fitting to a model (right). The binding affinity was enhanced in the order of the monomer, the first and second binding sites of the dimer.
A receptor molecule to which a signal molecule binds changes state between active and inactive states over time. The state changes occurring in nerve growth factor (NGF) receptors attached to fluorescently labeled NGF were detected by monitoring the changes in the diffusion on the cell surface [Fig. 5B]. The diffusion of the NGF receptor stopped and formed clusters in the active state. A monomer receptor separated from the cluster and resumed the diffusion in the inactive state (Shibata et al., 2006). Dimer formation is an important step for inducing activation in the binding of epidermal growth factor (EGF). Preferential binding of EGF to its receptor in the dimer form has been detected at the single molecule level (Teramura et al., 2006). The dimer preexisted even though the population of receptors was low. Fluorescently labeled EGF bound to the dimer more strongly than to a monomer. The binding of EGF to one of the two sites of the dimer enhanced the binding to the second site [Fig. 5C]. As small populations of the dimer, consistent with the small number of signal molecules (300), is sufficient for the activation of cells (Uyemura et al., 2005). Thus, a small number of molecules is all that is required to control the activation of cells. Single molecule detection is important to measure such processes in cells (Cai et al., 2006).
Difficulties can arise when interpreting the data of single molecule imaging in cells. Most of the molecules are hidden. In many cases, simulations are needed to test an idea used to interpret the data from single molecule measurements. Simulation thus plays an important role in single molecule techniques.
CONCLUSION
Great advances have been made with single molecule measurements and these techniques have proven to be a powerful tool in various fields of the life sciences. The impact of the direct visualization of the behavior of individual biomolecules made a huge impact. The single molecule measurements provide quantitative measurements about the mechanisms underlying biomolecules. What the single molecule measurements have revealed is not only something we had speculated for some time but did not observe directly, but also information that was not previously known. We believe that the single molecule measurements provide us with entirely new views in the field of life sciences rather than just a technique to answer individual questions. Single molecule techniques have shown that biomolecules function under the influence of thermal motion. At the beginning of 18th century thermal Brownian motion was discovered in pollen moving in an aqueous solution. At that time it was thought that Brownian motion provided a vital power. The original idea is not true because metal particles also undergo Brownian movement. The kinetic theory of gas was then established. Single molecule measurements have directly demonstrated that thermal Brownian motion is involved in the actions of biomolecules. Biomolecules move diffusively. The thermal Brownian motion is also involved in the movement of molecular motors. The structures and states of biomolecules change spontaneously. Enzymatic and biomolecular reactions proceed using thermal energy. Individual events caused in single biomolecules occur in a stochastic manner because of the thermal effects. Single molecule measurements also reveal that in many systems the biomolecules have mechanism to bias reactions toward one direction when they interact with other biomolecules. The movement involving Brownian movement is biased to directional motion when the molecular motors interact with the protein tracks. The fluctuations of the states of biomolecules is preferentially shifted to an active or inactive state upon interaction with other biomolecules. These mechanisms are further modulated in the system where large numbers of biomolecules participate in a specific arrangement. Single molecule measurements will be used in a variety of stages to provide new breakthroughs to the basic processes of life.
References
- Abbondanzieri, E A, Greenleaf, W J, Shaevits, J W, Landick, R, and Block, S M (2005). “Direct observation of base-pair stepping by RNA polymerase.” Nature (London) 10.1038/nature04268 438, 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahams, U M, Leslie, A GW, Lutter, R, and Walker, J E (1994). “Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria.” Nature (London) 10.1038/370621a0 370, 621–628. [DOI] [PubMed] [Google Scholar]
- Amos, W B, and White, J G (2003). “How the confocal laser scanning microscope entered biological research.” Biol. Cell 95, 335–342. [DOI] [PubMed] [Google Scholar]
- Arai, Y, Iwane, A H, Wazawa, T, Yokota, H, Ishii, Y, Kataoka, T, and Yanagida, T (2006). “Dynamic polymorphism of Ras observed by single molecule FRET is the basis for molecular recognition.” Biochem. Biophys. Res. Commun. 343, 809–815. [DOI] [PubMed] [Google Scholar]
- Ashkin, A (1997). “Optical trapping and manipulation of neutral particles using lasers.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.94.10.4853 94, 4853–4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod, D (1989). “Total internal reflection fluorescence microscopy.” Methods Cell Biol. 30, 245–270. [DOI] [PubMed] [Google Scholar]
- Betzig, E, and Chichester, R J (1993). “Single molecules observed by near-field scanning optical microscopy.” Science 10.1126/science.262.5138.1422 262, 1422–1424. [DOI] [PubMed] [Google Scholar]
- Blunck, R, Starace, D M, Correa, A M, and Bezanilla, F (2004). “Detecting rearrangements of shaker and NaChBac in real-time with fluorescence spectroscopy in patch-clamped mammalian cells.” Biophys. J. 10.1529/biophysj.103.034512 86, 3966–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer, P D (1993). “The binding change mechanism for ATP synthase—some probabilities and possibilities.” Biochim. Biophys. Acta 10.1016/0005-2728(93)90063-L 1140, 215–250. [DOI] [PubMed] [Google Scholar]
- Boyer, P D (1997). “The ATP synthase—a splendid molecular machine.” Annu. Rev. Biochem. 10.1146/annurev.biochem.66.1.717 66, 717–749. [DOI] [PubMed] [Google Scholar]
- Bustamante, C, Chemla, Y R, Forde, N R, and Izhaky, D (2004). “Mechanical processes in biochemistry.” Annu. Rev. Biochem. 10.1146/annurev.biochem.72.121801.161542 73, 705–748. [DOI] [PubMed] [Google Scholar]
- Cai, L, Friedman, N, and Xie, X S (2006). “Stochastic protein expression in individual cells at the single molecule level.” Nature (London) 10.1038/nature04599 440, 258–262. [DOI] [PubMed] [Google Scholar]
- Carter, N J, and Cross, R A (2005). “Mechanics of the kinesin step.” Nature (London) 10.1038/nature03528 435, 308–318. [DOI] [PubMed] [Google Scholar]
- Dahan, M, Levi, S, Luccardini, C, Rostaing, P, Rivearu, B, and Triller, A (2003). “Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking.” Science 10.1126/science.1088525 302, 442–445. [DOI] [PubMed] [Google Scholar]
- Dantzig, J A, Liu, T Y, and Goldman, Y E (2005). “Functional studies of individual myosin molecules.” Ann. N.Y. Acad. Sci. 1080, 1–18. [DOI] [PubMed] [Google Scholar]
- Dickson, R M, Cubitt, A B, Tsien, R Y, and Moerner, W E (1997). “On/off blinking and switching behaviour of single molecules of green fluorescent protein.” Nature (London) 10.1038/41048 388, 355–358. [DOI] [PubMed] [Google Scholar]
- Dumont, S, Cheng, W, Beran, R, TinocoJr., I, Pyle, A M, and Bustamante, C (2006). “RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP.” Nature (London) 10.1038/nature04331 439, 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, J M, and Li, H (2004). “Force-clamp spectroscopy monitors the folding trajectory of a single protein.” Science 10.1126/science.1092497 303, 1674–1678. [DOI] [PubMed] [Google Scholar]
- Finer, J T, Simmons, R M, and Spudich, J A (1994). “Single myosin molecule mechanics; piconewton forces and nanometre steps.” Nature (London) 10.1038/368113a0 368, 113–119. [DOI] [PubMed] [Google Scholar]
- Fisher, T R, Marszalek, P E, and Fernandez, J M (2000). “Stretching single molecules into novel conformations using the atomic force microscope.” Nat. Struct. Biol. 10.1038/78936 7, 719–724. [DOI] [PubMed] [Google Scholar]
- Forkey, J N, Quinlan, M E, Shaw, M A, Corrie, E T, and Goldman, Y E (2003). “Three-dimensional structural dynamics of myosin V by single-molecule fluorescence polarization.” Nature (London) 10.1038/nature01529 422, 399–404. [DOI] [PubMed] [Google Scholar]
- Funatsu, T, Harada, Y, Tokunaga, M, Saito, K, and Yanagida, T (1995). “Imaging of single fluorescent molecules and individual ATP turnovers by single myosin molecules in aqueous solution.” Nature (London) 10.1038/374555a0 374, 555–559. [DOI] [PubMed] [Google Scholar]
- Galletto, R, Amitani, I, Baskin, R J, and Kowalczykowski, S C (2006). “Direct observation of individual RecA filaments assembling on single DNA molecules.” Nature (London) 10.1038/nature05197 443, 875–877. [DOI] [PubMed] [Google Scholar]
- Ha, T, Ransnik, I, Cheng, W, Babcook, H P, Gauss, G H, Lohman, T M, and Chu, S (2002). “Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase.” Nature (London) 10.1038/nature01083 419, 638–641. [DOI] [PubMed] [Google Scholar]
- Haas, E (2005). “The study of protein folding and dynamics by determination of intramolecular distance distributions and their fluctuations using ensemble and single-molecule FRET measurements.” ChemPhysChem 10.1002/cphc.200400617 6, 858–870. [DOI] [PubMed] [Google Scholar]
- Harada, Y, Funatsu, T, Murakami, K, Nonomura, Y, Ishihama, A, and Yanagida, T (1999). “Single molecule imaging of RNA polymerase-DNA interactions in real time.” Biophys. J. 76, 709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada, Y, Ohara, O, Takatsuki, A, Ito, H, Shimamoto, and N, KinositaJr., K (2001). “Direct observation of DNA rotation during transcription by Escherichia coli RNA polymerase.” Nature (London) 10.1038/35051126 409, 113–115. [DOI] [PubMed] [Google Scholar]
- Harada, Y, Sakurada, K, Aoki, T, Thomas, D D, and Yanagida, T (1990). “Mechanochemical coupling in actomyosin energy transduction studied by in vitro movement assay.” J. Mol. Biol. 10.1016/0022-2836(90)90010-J 216, 49–68. [DOI] [PubMed] [Google Scholar]
- Helenius, J, Brouhard, G, Kaladzidis, Y, Diez, S, and Howard, J (2006). “The depolimerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends.” Nature (London) 10.1038/nature04736 441, 115–119. [DOI] [PubMed] [Google Scholar]
- Ide, T, Takeuchi, Y, and Yanagida, T (2002). “Development of an experimental apparatus for simultaneous observation of optical and electrical signals from single channels.” Single Mol. 3, 33–42. [DOI] [Google Scholar]
- Ide, T, and Yanagida, T (1999). “An artificial lipid bilayer formed on an agarose-coated glass for simultaneous electrical and optical measurement of single ion channels.” Biochem. Biophys. Res. Commun. 10.1006/bbrc.1999.1720 265, 595–599. [DOI] [PubMed] [Google Scholar]
- Ishii, Y, Ishijima, A, and Yanagida, T (2001). “Single molecule nanomanipulation of biomolecules.” Trends Biotechnol. 10.1016/S0167-7799(01)01635-3 19, 211–216. [DOI] [PubMed] [Google Scholar]
- Ishijima, A, Harada, Y, Kojima, H, Funatsu, T, Higuchi, H, and Yanagida, T (1994). “Single-molecule analysis of the actomyosin motor using nano-manipulation.” Biochem. Biophys. Res. Commun. 10.1006/bbrc.1994.1336 199, 1057–1063. [DOI] [PubMed] [Google Scholar]
- Ishijima, A, Kojima, H, Higuchi, H, Harada, Y, Funatsu, T, and Yanagida, T (1998). “Simultaneous measurement of chemical and mechanical reaction.” Cell 10.1016/S0092-8674(00)80911-3 70, 161–171. [DOI] [PubMed] [Google Scholar]
- Ishijima, A, and Yanagida, T (2001). “Single molecule nanobioscience.” Trends Biotechnol. 26, 438–444. [DOI] [PubMed] [Google Scholar]
- Itoh, H, Takahashi, A, Adachi, K, Nojji, H, Yasuda, R, Yoshida, M, and KinositaJr., K (2004). “Mechanically driven ATP synthesis by F1-ATPase.” Nature (London) 10.1038/nature02212 427, 465–468. [DOI] [PubMed] [Google Scholar]
- Iwaki, M, Tanaka, H, Iwane, A H, Katayama, E, Ikebe, M, and Yanagida, T (2006). “Cargo binding makes a wild-type single-headed myosin-VI move processively.” Biophys. J. 10.1529/biophysj.105.075721 90, 3643–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto, M, Shimmizu, H, Inoue, F, Kono, T, Sasaki, Y C, and Oiki, S (2006). “Surface structure and its dynamic rearrangements of the KcsA potassium channel upon gating and tetrabutylammonium blocking.” J. Biol. Chem. 10.1074/jbc.M602018200 281, 28379–28386. [DOI] [PubMed] [Google Scholar]
- Jacquier, V, Prummer, M, Segure, J-M, Pick, H, and Vogel, H (2006). “Visualizing odorant receptor trafficking in living cells down to the single-molecule level.” 103, 14325–14330. [DOI] [PMC free article] [PubMed]
- Kahya, N (2006). “Targeting membrane proteins to liquid-ordered phases: molecular self-organization explored by fluorescence correlation spectroscopy.” Chem. Phys. Lipids 10.1016/j.chemphyslip.2006.02.026 141, 158–168. [DOI] [PubMed] [Google Scholar]
- Kellermayer, M S, Smith, S B, Granzier, H L, and Bustamante, C (1997). “Folding-unfolding transitions in single titin molecules characterized with laser tweezers.” Science 10.1126/science.276.5315.1112 276, 1112–1116. [DOI] [PubMed] [Google Scholar]
- Kishino, A, and Yanagida, T (1988). “Force measurement of micromanipulation of a single actin filament by glass needles.” Nature (London) 10.1038/334074a0 334, 74–76. [DOI] [PubMed] [Google Scholar]
- Kitamura, K, Tokunaga, M, Iwane, A H, and Yanagida, and T (1999). “A single myosin head moves along an actin filament with regular steps of 5.3 nanometres.” Nature (London) 10.1038/16403 397, 129–134. [DOI] [PubMed] [Google Scholar]
- Kozuka, J, Yokota, H, Arai, Y, Ishii, Y, and Yanagida, T (2006). “Dynamic polymorphism of single actin molecules in the actin filament.” Nat. Chem. Biol. 10.1038/nchembio763 2, 83–86. [DOI] [PubMed] [Google Scholar]
- Kron, S J, and Spudich, J A (1986). “Fluorescent actin filaments move on myosin fixed to a glass surface.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.83.17.6272 83, 6272–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulzer, F, and Orrit, M (2001). “Single-molecule optics.” Annu. Rev. Phys. Chem. 10.1146/annurev.physchem.54.011002.103816 55, 585–617. [DOI] [PubMed] [Google Scholar]
- Kural, C, Kim, H, Syed, S, Goshima, G, Gelfand, V I, and Selvin, P R (2005). “Kinesin and dynein move a peroxisome in vivo: a tug-of-war or coordinated movement?” Science 10.1126/science.1108408 308, 1469–1472. [DOI] [PubMed] [Google Scholar]
- Kusumi, A, Ike, H, Nakada, C, Murase, K, and Fujiwara, T (2005). “Single-molecule tracking of membrane mmolecules: plasma membrane compartmentalization and dynamic assembly of raft-philic signaling molecules.” Semin Immunol. 17, 3–21. [DOI] [PubMed] [Google Scholar]
- Lidke, D S, Heinzmann, R, Arndt-Jovin, D, Post, J N, Grecco, H E, Jares-Erijiman, E A, and Jovin, T M (2004). “Quantum dot ligands provide new insights into erbB/HER receptor-mediated signal transduction.” Nat. Biotechnol. 10.1038/nbt929 22, 198–203. [DOI] [PubMed] [Google Scholar]
- Lipman, E A, Schler, B, Bakajin, O, and Eaton, W A (2003). “Single-molecule measurement of protein folding kinetics.” Science 10.1126/science.1085399 301, 1233–1235. [DOI] [PubMed] [Google Scholar]
- Lister, I, Schnitz, S, Walker, M, Trinick, J, Buss, F, Veigel, C, and Kendrick-Jones, J (2004). “A monomeric myosin VI with a large working stroke.” EMBO J. 10.1038/sj.emboj.7600180 23, 1729–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H P, Xun, L, and Xie, X S (1998). “Single-molecule enzymatic dynamics.” Science 10.1126/science.282.5395.1877 282, 1877–1882. [DOI] [PubMed] [Google Scholar]
- Mehta, A D, Rief, M, and Spudich, J A (1999). “Single-molecule biomechanics with optical methods.” Science 10.1126/science.283.5408.1689 283, 1689–1695. [DOI] [PubMed] [Google Scholar]
- Michalet, X, Kapanids, A N, Laurence, T, Pinaud, F, Doose, S, Pflughoefft, M, and Weiss, S (2003). “The power and prospects of fluorescence microscopies and spectroscopies.” Annu. Rev. Biophys. Biomol. Struct. 10.1146/annurev.biophys.32.110601.142525 32, 161–182. [DOI] [PubMed] [Google Scholar]
- Miyawaki, A (2003). “Fluorescence imaging of physiological activity in complex systems using GFP-based probes.” Curr. Opin. Neurobiol. 10.1016/j.conb.2003.09.005 13, 591–596. [DOI] [PubMed] [Google Scholar]
- Molloy, J E, Burns, J E, Kendrick-Jones, J, Tregear, R T, and White, D C (1995). “Movement and force produced by a single myosin head.” Nature (London) 10.1038/378209a0 378, 209–212. [DOI] [PubMed] [Google Scholar]
- Muto, E, Sakai, H, and Kaseda, K (2005). “Long-range cooperative binding of kinesin to a microtubule in the presence of ATP.” J. Cell Biol. 10.1083/jcb.200409035 168, 691–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahas, M K, Wilson, T J, Hohng, S, Jarvie, K, Liller, D M, and Ha, T (2004). “Observation of internal cleavage and ligation reactions of a ribozyme.” Nat. Struct. Mol. Biol. 10.1038/nsmb842 11, 1107–1113. [DOI] [PubMed] [Google Scholar]
- Nan, X, Sims, P A, Chen, P, and Xie, X S (2005). “Observation of fundamental microtubule motor steps in living cells with endocytosed quantum dots.” J. Phys. Chem. B 10.1021/jp056360w 109, 24220–24224. [DOI] [PubMed] [Google Scholar]
- Nishiyama, M, Higuchi, H, and Yanagida, T (2002). “Chemomechanical coupling of the forward and backward steps of single kinesin molecules.” Nat. Cell Biol. 10.1038/ncb857 4, 790–797. [DOI] [PubMed] [Google Scholar]
- Noji, H, Yasuda, R, Yoshida, M, and KinositaJr, K (1997). “Direct observation of the rotation of F1-ATPase.” Nature (London) 10.1038/386299a0 386, 299–302. [DOI] [PubMed] [Google Scholar]
- Oiwa, K, Jameson, D M, Croney, J C, Davis, C T, Eccleston, J F, and Anson, M (2003). “The 2′-O- and 3′-O-Cy3-EDA-ATP(ADP) complexes with myosin subfragment-1 are spectroscopically distinct.” Biophys. J. 84, 634–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, Y, Higuchi, H, and Hirokawa, N (2003). “Processivity of the single-headed kinesin KIF1A through biased binding to tubulin.” Nature (London) 10.1038/nature01804 424, 574–577. [DOI] [PubMed] [Google Scholar]
- Onuchic, J N, and Wolynes, P G (2004). “Theory of protein folding.” Curr. Opin. Struct. Biol. 10.1016/j.sbi.2004.01.009 14, 70–75. [DOI] [PubMed] [Google Scholar]
- Park, H, Ramamurthy, B, Tavaglia, M, Safer, D, Chen, L, Franzini-Armstrong, C, and Sweeney, H L (2006). “Full-length myosin VI dimerizes and moves processively along actin filaments upon monomer clustering.” Mol. Cell 21, 331–336. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson, S L, Yildiz, A, Carter, A P, Gennerich, A, Zhang, N, and Vale, R D (2006). “Single-molecule analysis of dynein processivity and stepping behavior.” Cell 10.1016/j.cell.2006.05.046 126, 335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, E, Gussakovsky, E, and Haran, G (2003). “Watching proteins fold one molecule at a time.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.2628068100 100, 3197–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rief, M, Gautel, M, Oesterheilt, F, Ferrandez, J M, and Gaub, H E (1997). “Reversible unfolding of individual titin immunoglobulin domains by AFM.” Science 10.1126/science.276.5315.1109 276 1109–1112. [DOI] [PubMed] [Google Scholar]
- Rief, M, Rock, R S, Mehta, A D, Moosker, M S, Cheney, R E, and Spudich, J A (2000). “Myosin-V stepping kinetics: a molecular model for processivity.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.97.17.9482 97, 9482–9486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigler, R (1995). “Fluorescence correlations, single molecule detection and large number screening. Applications in biotechnology.” J. Biotechnol. 10.1016/0168-1656(95)00054-T 41, 177–186. [DOI] [PubMed] [Google Scholar]
- Rock, R S, Rice, S E, Wells, A L, Purcell, T J, Spudich, J A, and Sweeney, H L (2001). “Myosin VI is a processive motor with a large step size.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.191512398 98, 13655–13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler, B (2005). “Single-molecule fluorescence spectroscopy of protein folding.” ChemPhysChem 10.1002/cphc.200400609 6, 1206–1220. [DOI] [PubMed] [Google Scholar]
- Schütz, G J, Kada, G, Pastushenko, V Ph, and Schindler, H (2000). “Properties of lipid microdomains in a muscle cell membrane visualized by single molecule microscopy.” EMBO J. 10.1093/emboj/19.5.892 19, 892–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwille, P, Korlach, J, and Webb, W W (1999). “Fluorescence correlation spectroscopy with single-molecule sensitivity on cell and model membranes.” Cytometry 36, 76–82. [DOI] [PubMed] [Google Scholar]
- Shaevitz, J W, Abbondanzierl, E A, Landick, R, and Block, S M (2003). “Backtracking by single RNA polymerase molecules observed at near-base-pair resolution.” Nature (London) 10.1038/nature02191 426, 684–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal, Y, Darzacq, X, Shenoy, S M, Fusco, D, Janckl, S M, Spector, D L, and Singer, R H (2004). “Dynamics of single mRNPs in nuclei of living cells.” Science 10.1126/science.1099754 304, 1797–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, S C, Hibino, K, Mashimo, T, Yanagida, T, and Sako, Y (2006). “Formation of signal transduction complexes during immobile phase of NGFR movements.” Biochem. Biophys. Res. Commun. 342, 316–322. [DOI] [PubMed] [Google Scholar]
- Smith, A M, Ruan, G, Rhyner, M N, and Nie, S (2006). “Engineering luminescent quantum dots for in vivo molecular and cellular imaging.” Ann. Biomed. Eng. 10.1007/s10439-005-9000-9 34, 3–14. [DOI] [PubMed] [Google Scholar]
- Sonnleitner, A, Mannuzzu, L M, Terakawa, S, and Isacoff, E Y (2002). “Structural rearrangements in single ion channels detected optically in living cells.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.192261499 99, 12759–12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strick, T R, Croquette, V, and Bensimmon, D (2000). “Single-molecule analysis of DNA uncoiling by a type II topoisomerase.” Nature (London) 10.1038/35009144 404, 901–904. [DOI] [PubMed] [Google Scholar]
- Svoboda, K, Schmidt, C F, Schnapp, B J, and Block, S M (1993). “Direct observation of kinesin stepping by optical trapping interferometry.” Nature (London) 10.1038/365721a0 365, 721–727. [DOI] [PubMed] [Google Scholar]
- Syed, S, Snyder, G E, Franzini-Armstrong, C, Selvin, P R, and Goldman, Y E (2006). “Adaptability of myosin V studied by simultaneous detection of position and orientation.” EMBO J. 10.1038/sj.emboj.7601060 25, 1795–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi, H, Ueno, T, Tadakuma, H, Yoshida, M, and Funatsu, T (2001). “Single-molecule observation of protein-protein interactions in the chaperonin system.” Nat. Biotechnol. 10.1038/nbt0901-861 19, 861–865. [DOI] [PubMed] [Google Scholar]
- Tanaka, H, Ishijima, A, Honda, M, Saito, K, and Yanagida, T (1998). “Orientation dependence of displacements by a single one-headed myosin relative to the actin filament.” Biophys. J. 75, 1888–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi, Y, Nishiyama, M, Ishii, Y, and Yanagida, T (2005). “Entropy rectifies the Brownian steps of kinesin.” Nat. Chem. Biol. 10.1038/nchembio741 1, 342–347. [DOI] [PubMed] [Google Scholar]
- Teramura, Y, Ichinose, J, Takagi, H, Nishida, K, Yanagida, T, and Sako, Y (2006). “Single-molecule analysis of epidermal growth factor binding on the surface of living cells.” EMBO J. 10.1038/sj.emboj.7601308 25, 4216–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba, S, Watanabe, T M, Yamaguchi-Okamoto, I, Toyoshima, Y Y, and Higuchi, H (2006). “Overlapped hand-over-hand mechanism of single molecular motility of cytoplasmic dynein.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0508511103 102, 5741–5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga, M, Kitamura, K, Saito, K, Iwane, A H, and Yanagida, T (1997). “Single molecule imaging of fluorophores and enzymatic reactions achieved by objective type total internal reflection fluorescence microscopy.” Biochem. Biophys. Res. Commun. 10.1006/bbrc.1997.6732 235, 47–53. [DOI] [PubMed] [Google Scholar]
- Toyoshima, Y Y, Kron, S J, and Spudich, J A (1990). “The myosin step size: measurement of the unit displacement per ATP hydrolyzed in an in vitro assay.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.87.18.7130 87, 7130–7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautman, J K, and Macklin, J J (1996). “Time-resolved spectroscopy of single molecules using near-field and far-field optics.” Chem. Phys. 10.1016/0301-0104(95)00391-6 205, 221–229. [DOI] [Google Scholar]
- Tsien, R Y (1998). “The green fluorescent proteins.” Annu. Rev. Biochem. 10.1146/annurev.biochem.67.1.509 67, 509–544. [DOI] [PubMed] [Google Scholar]
- Tskhovrebova, L, Trinick, J, Sleep, J A, and Simmons, F M (1997). “Elasticity and unfolding of single molecules of the giant muscle protein titin.” Nature (London) 10.1038/387308a0 387, 308–312. [DOI] [PubMed] [Google Scholar]
- Tyska, M J, Dupuis, D E, Guilford, W H, Patlak, J B, and Warshaw, D M (1999). “Two heads of myosin are better than one for generating force and motion.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.96.8.4402 96, 4402–4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, M, Sako, Y, Tanaka, T, Devreotes, P, and Yanagida, T (2001). “Single-molecule analysis of chemotactic signaling in Dictyostelium cells.” Science 10.1126/science.1063951 294, 864–867. [DOI] [PubMed] [Google Scholar]
- Uemura, S, and Ishiwata, S (2003). “Loading direction regulates the affinity of ADP for kinesin.” Nat. Struct. Biol. 10.1038/nsb911 10, 308–311. [DOI] [PubMed] [Google Scholar]
- Uyemura, T, Takagi, H, Yanagida, T, and Sako, Y (2005). “Single-molecule analysis of epidermal growth factor signaling that leads to ultrasensitive calsium response.” Biophys. J. 10.1529/biophysj.104.053330 88, 3720–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M D, Schnitzer, M J, Yin, H, Landick, R, Gelles, J, and Block, S M (1998). “Force and velocity measured for single molecules of RNA polymerase.” Science 10.1126/science.282.5390.902 282, 902–907. [DOI] [PubMed] [Google Scholar]
- Weiss, S (2000). “Measuring conformational dynamics of biomolecules by single molecule fluorescence spectroscopy nat.” Nat. Struct. Biol. 10.1038/78941 7, 724–729. [DOI] [PubMed] [Google Scholar]
- Wuite, G J L, Smith, S B, Young, M, Keller, D, and Bustamante, C (2000). “Single-molecule studies of the effect of template tension of T7 DNA polymerase activity.” Nature (London) 10.1038/35003614 404, 103–106. [DOI] [PubMed] [Google Scholar]
- Yanagida, T, Nakase, M, Nishiyama, K, and Oosawa, F (1984). “Direct observation of motion of single F-actin filaments in the presence of myosin.” Nature (London) 10.1038/307058a0 307, 58–60. [DOI] [PubMed] [Google Scholar]
- Yasuda, R, Noji, H, KinositaJr., K, and Yoshida, M (1998). “F1-ATPase is a highly efficient molecular motor that rotates with discrete 120° steps.” Cell 10.1016/S0092-8674(00)81456-7 93, 1117–1124. [DOI] [PubMed] [Google Scholar]
- Yasuda, R, Noji, H, Yoshida, M, KinositaJr., K, and Itoh, H (2001). “Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase.” Nature (London) 10.1038/35073513 410, 898–904. [DOI] [PubMed] [Google Scholar]
- Yildiz, A, Firjey, J N, McKinney, S A, Ha, T, Goldman, Y E, and Selvin, P R (2003). “Myosin V walks hand-over-hand: single fluorophores imaging with 1.5 nm localization.” Science 10.1126/science.1084398 300, 2061–2065. [DOI] [PubMed] [Google Scholar]
- Zhuang, X, Kim, H, Pereira, M J B, Babcock, H P, Walter, N G, and Chu, S (2002). “Correlating structural dynamics and function in single ribozyme molecules.” Science 10.1126/science.1069013 296, 1473–1476. [DOI] [PubMed] [Google Scholar]