Abstract
Attempts to construct artificial systems from biological molecules such as DNA and RNA by self-assembly are compatible with the recent development of synthetic biology. Genetic mechanisms can be used to produce or control artificial structures made from DNA and RNA, and these structures can in turn be used as artificial gene regulatory elements, in vitro as well as in vivo. Artificial biochemical circuits can be incorporated into cell-like reaction compartments, which opens up the possibility to operate them permanently out of equilibrium. In small systems, stochastic effects become noticeable and will have to be accounted for in the design of future systems.
The phenomenon of biological self-organization has played an important role in shaping the vision of a future nanoscale technology. Nanotechnology aims at the control of matter and production of artificial structures on the nanometer scale. Among the many directions and disciplines of current nanoscience, bionanotechnology—and DNA nanotechnology in particular—is arguably closest in approach to biological systems. Bionanotechnology utilizes biomolecular self-assembly for the construction of artificial structures and devices, which is based on the molecular recognition properties of biological macromolecules such as DNA or proteins. However, self-assembly represents only one aspect of biological self-organization, as many phenomena can be only understood as a result of the complex interplay of many interacting molecules or subunits within a network.
Systems biology is devoted to the study of such networks with the goal to mathematically describe and predict the behavior of whole cells or even organisms. Due to the complexity of the systems and the lack of reliable and sufficient data, this is an extremely challenging task. This is part of the reason why in the last few years, systems biology has been increasingly complemented by a novel “bottom up” discipline termed “synthetic biology.”
Synthetic biology uses an engineering approach to design and construct biological systems, potentially with novel functions that do not exist in nature. It relies on tools from genetic engineering, bioengineering, systems biology and many other disciplines. Synthetic biology aims at the design and fabrication of biological components and systems that do not already exist in the natural world and the redesign and fabrication of existing biological systems.
In that sense, DNA nanotechnology can be regarded as one aspect of in vitro synthetic biology. As will be surveyed briefly below, DNA has been successfully used as a building material for supramolecular assemblies, but also for the realization of molecular machines and computers. Some of these structures are capable of integration within complex networks and even within living organisms. In this perspective we will discuss up and coming research on the construction of more complex systems based on DNA and RNA. In particular, we will deal with artificial biochemical interaction networks realized with DNA and RNA molecules, and also with their incorporation into artificial cells or living systems. We will also address issues related to the operation of such networks in small reaction volumes, in which stochastic effects are expected to play an important role.
The goals and benefits of a fusion of DNA nanotechnology with synthetic biology are diverse: On the one hand, DNA nanotechnology can deliver concepts and components for artificial structures to be implemented in biological systems. For example, RNA nanodevices may be used as artificial riboswitches, and certain DNA computing algorithms may be implemented in vivo. On the other hand, artificial or biological cells may be used for the production of RNA based nanostructures and for control of complex multistage nanoassembly.
NUCLEIC ACIDS AS COMPONENTS FOR SYNTHETIC BIOSYSTEMS
DNA and RNA nanostructures
DNA nanotechnology was established by Nadrian Seeman more than 25 years ago (Seeman, 1982). It is based on the precisely predictable molecular recognition events between DNA strands with complementary sequences. Since its first inception, DNA molecules have been used to construct increasingly complex supramolecular structures, ranging from three-dimensional objects (Chen and Seeman, 1991; Shih et al., 2004; Goodman et al., 2005; Douglas et al., 2007) over two-dimensional (2D) lattices (Winfree et al., 1998; Yan et al., 2003a) to arbitrary 2D patterns (Rothemund, 2006). For details the reader is referred to a number of excellent reviews on the subject (Seeman, 2004; Seeman and Lukeman, 2005; LaBean and Li, 2007). From the perspective of synthetic biology, it is particularly interesting to also consider RNA based nanostructures, as one can envision in vivo production of such structures by transcription. In nature, RNA only rarely is used as a structural element. One prominent exception here is pRNA (Guo et al., 1998; Shu et al., 2004), which is involved in the DNA packaging motor of phage phi29. Nevertheless, Jaeger and co-workers could demonstrate a variety of RNA assemblies comparable in complexity to those realized with DNA (Chworos et al., 2004; Jaeger and Chworos, 2006). One of the problems for in vivo RNA nanoconstruction is the rapid degradation of RNA. However, recent work suggests that degradation can be avoided by incorporating tRNA structures into the transcripts (Ponchon and Dardel, 2007).
Even though the accomplishments of DNA and RNA-based nanoassembly are extremely impressive—and today represent the most advanced “bottom up” nanotechnology—current self-assembly strategies capture only part of the complexity found in biological self-organization. In DNA nanoconstruction, one usually tries to find DNA sequences, which assemble into a desired thermal equilibrium structure. Careful annealing is used to direct the assembly process into this structure.
In contrast, many structures in biological systems are inherently nonequilibrium structures. For instance, the cytoskeleton is composed of dynamic protein filaments, which are in a constant process of assembly and disassembly, consuming ATP or GTP fuel molecules. Furthermore, for building complex structures it is sometimes necessary to control the spatial and temporal order of assembly. Biological examples for this are the complex intracellular rearrangements preceding cell division, or the development of multicellular organisms.
To achieve similar complexity also for nanoassembly, it may be necessary to incorporate self-assembly processes into nonequilibrium thermodynamic systems (Qian, 2005) and direct them using molecular control circuits. Other desirable, bioinspired features for self-assembling structures include adaptability and evolvability. In fact, sequence evolution experiments (Koltermann and Kettling, 1997; Schuster, 1997; Joyce, 2007) and in vitro selection of functional nucleic acids (Wilson and Szostak, 1999) indicate that evolutionary processes could be also exploited for the realization of artificial structures made from DNA or RNA.
A different concept, but clearly related to the spirit of synthetic biology, is the idea of a “translation machinery” for nanotechnology (Garibotti et al., 2007). According to the central dogma of molecular biology (and disregarding regulatory RNAs for a moment), genetic information stored on a DNA sequence is translated—via an RNA intermediate—to an amino acid sequence, which folds into a protein. A similar approach is highly desirable for the production of complex materials. One can envision the translation of assembly information stored on an informational molecule (again, e.g., DNA) into a programmed sequence of materials (see also Kauffman and Ellington, 1999; Li and Liu, 2004; Halpin and Harbury, 2004; Scheuermann et al., 2006). In contrast to conventional chemical approaches, with such machinery one could realize, e.g., block copolymers with arbitrary, pre-programmed sequences of blocks, or chains of nanoparticles with aperiodic order. While the artificial translation machinery developed by Garibotti et al. most probably cannot be used to produce artificial materials in vivo, this may be possible using the modified (“orthogonal”) ribosomal systems developed, e.g., by Chin and co-workers (Rackham and Chin, 2005; Chin, 2006).
Molecular machines and computers
Apart from nanoconstruction, in recent years DNA-based molecular recognition has been used to realize numerous machine-like assemblies with mechanical or information-processing properties (Seeman, 2005; Bath and Turberfield, 2007). These devices are composed of single or multiple strands of DNA, which fold into a particular, in most of the cases rationally designed, structure. The conformation of the devices can be switched between several structures by the addition of DNA or RNA strands, or by a change in buffer conditions. In this manner, the devices can act as simple sensors for the DNA sequences or environmental factors they react on. The conformational changes can also be utilized to induce mechanical motion, and structures displaying rotational (Mao et al., 1999; Yurke et al., 2000; Yan et al., 2002) and translational movement (Shin and Pierce, 2004; Sherman and Seeman, 2004; Bath et al., 2005; Venkataraman et al., 2007) have been demonstrated. Recently, the adaptation of functional nucleic acids for DNA nanodevices has considerably enhanced their versatility (Dittmer et al., 2004). Aptamers—oligonucleotides which bind specifically to other molecules—and ribozymes—nucleic acids with catalytic function—have been used to realize novel biosensors (Lu and Liu, 2006) and autonomous molecular computing devices (Stojanovic and Stefanovic, 2003; Penchovsky and Breaker, 2005).
Since Adleman’s original work on DNA-based computation (Adleman, 1994), a large variety of examples for DNA-based information processing have been demonstrated (Ezziane, 2006). DNA has been utilized for a molecular realization of finite state automata (Benenson et al., 2001; Benenson et al., 2004), and simple computer algorithms have been implemented in molecular self-assembly to produce supramolecular patterns (Yan et al., 2003b; Rothemund et al., 2004).
Even more than for supramolecular assemblies, it seems straightforward to incorporate nucleic acid based nanodevices and computers into synthetic biological systems. First, biological cells could be simply used to produce RNA nanodevices by transcription. Second, gene regulatory mechanisms can be used to control the time of production of the nanodevices and naturally occuring RNA—e.g., microRNA—or RNA transcribed from artificial control genes can be used to drive them. Finally, concepts from DNA nanotechnology and DNA computing can be adapted to devise novel strategies for the control of gene transcription and translation. Due to their comparatively simple and programmable structures, RNA-based devices and control circuits should also be of considerable interest as components for artificial cells (Szostak et al., 2001; Pohorille and Deamer, 2002; Forster and Church, 2006).
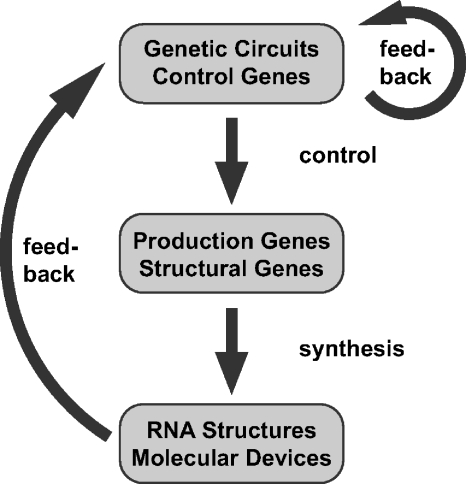
A conceptual overview of the relation between DNA nanotechnology and synthetic biology is depicted in Fig. 1. One can envision “production genes” which code for RNA-based nanostructures, which self-assemble upon transcription, and “control genes,” which control the temporal order of production and the behavior of molecular devices. The devices themselves may also have regulatory function and feed back into the control circuits. Artificial control and construction circuits may be incorporated into natural biological systems, or may be part of artificial cell-like reaction compartments.
Figure 1. Potential integration of DNA nanotechnology with synthetic biology.
As discussed in the text, artificial “production” genes may be used to produce nanostructures, whereas “control genes” may be used to decide or control, when or which nanostructure is produced or operated (cf. Fig. 3). Synthetic control networks may be produced with RNA regulators alone (see Fig. 2), and nucleic acid nanodevices may feed back on the control circuits. The circuit diagram may be implemented either in vitro or in vivo.
SYNTHETIC CIRCUITS IN VITRO
In live cells, signal transduction and information processing, needed, e.g., for survival and reproduction, are based on many species of interacting “modules” and molecules (Hartwell et al., 1999). There are different approaches to investigate the properties of these functional circuits. The reductionist systems biology approach attempts to explain the behavior of these circuits by trying to reveal the complex interplay of the circuit components. Although recent genetic and biochemical techniques allow for identification of many molecular components of biological organisms, one still cannot reliably predict more complex circuit behavior except for the simplest systems. In synthetic biology, as a complementary approach to study biological systems behavior, less complicated analogs of natural circuits are constructed. These artificially engineered circuits can be used to verify theoretical models, and thereby confirm and advance current understanding of biological complexity. In this way, synthetic biology complements insights already gained by systems biology. If simple, well-characterized artificial circuits can be arranged into more complex networks with behavior that can be predicted from that of the individual components, an understanding of regulatory processes from first principles seems possible. Furthermore, it should then be possible to construct novel functions and behaviors from well-characterized circuit modules, which has obvious and exciting implications for bionanotechnology.
A variety of synthetic networks with fascinating behavior have already been implemented in cells by rearranging known regulatory components, among them a bistable circuit (Gardner et al., 2000), a genetic oscillator (Elowitz and Leibler, 2000), a sender-receiver system (Weiss and Knight, 2000) and an artificial system for population control based on quorum sensing (You et al., 2004).
Analysis and modeling of these systems are challenging because of the many unknown parameters in the cellular host environment. In vitro reconstruction of genetic circuits with known components in an artificial cell-like environment, e.g., vesicles, is one possibility to overcome these limitations.
Cell-free genetic circuit assembly
In a first step towards in vitro genetic networks, Noireaux et al. (2003) constructed cell-free circuits using a commercial transcription∕translation system based on a cell extract (Noireaux et al., 2003). These circuits consisted of engineered transcriptional activation and repression cascades, in which protein products from each stage of the cascade were used as an input to activate or inhibit the following stage. Cell-free expression systems exhibit several advantages over in vivo protein synthesis because larger parameter ranges can be studied, gene and polymerase concentrations can be controlled and reporter measurements are quantitative. One-, two-, and three-stage gene expression cascades were constructed and used to study basic principles of cell-free genetic circuit assembly. Many applications of cell-free protein expression were optimized for maximal protein synthesis and thus focused on mRNA stability and reduction of nuclease activity (Jermutus et al., 1998). Noireaux et al. could show, that engineering in vitro genetic circuits using cell-free expression systems requires optimization of different parameters. Absence of a continuous supply with nutrients (even in ATP regenerating systems) and the accumulation of waste products limit the expression in batch mode.
In principle, continuous expression systems could solve this problem (Spirin et al., 1988). Bar-Ziv and co-workers (Buxboim et al., 2007) recently developed an elegant approach to such a continuous system, which also provided the solution to another problem of current cell-free gene expression methods: in experiments conducted in bulk solution or microcompartments, reactions are not “localized.” This is in contrast to the situation found in highly structured biological cells, where several stages of information-processing cascades are often co-localized. Therefore, natural gene circuit behavior is also influenced by reaction-diffusion effects. To be able to place several reaction sites into immediate vicinity, Buxboim et al. developed a technology for controlled cell-free gene expression on a microchip. To this end, a novel photoactivatable hybrid molecule was designed that forms a biocompatible lithographic interface on SiO2. This interface is used to immobilize long DNA molecules (i.e., gene templates) with sub-micrometer resolution and high densities. With this technique, a two-stage gene cascade was built, in which proteins are synthesized at one location, and then diffuse to regulate the synthesis of another protein at a second site. Cell-free transcription∕translation reactions based on localized gene templates can be coordinated and cascaded in place and time.
Although this approach allows for cell-free gene expression in a more controlled manner, the in vitro synthetic gene circuits realized so far rely on protein-based regulation and thus the comparatively complex translation machinery. The use of poorly characterized cell extracts makes it difficult to describe these artificial systems using a theoretical, component-oriented model, and quantitative predictions of circuit behavior have not been possible so far.
One possibility to overcome this obstacle for a quantitative treatment is to further reduce the number of components of the synthetic biochemical systems. As will be described in the next section, it is possible to construct simple gene regulatory circuits almost exclusively based on DNA and RNA, in which RNA itself is used as a transcriptional regulator. These circuits function on a transcriptional level and therefore do not require translation of RNA into proteins.
In principle, it should be possible to use these systems as simple models of biological control circuits, integrating elements acting as molecular sensors, signal transducers, genetic regulators, and also mechanical and chemical actuators. In an RNA-based system, the sensors and transducers could be RNA aptamers, allosteric ribozymes, and rationally designed molecular logic gates; the genetic regulators could be transcriptional variants of riboswitches and riboregulators; the chemical actuators could be ribozymes, and the mechanical actuators could be RNA nanodevices and self-assembling molecules. RNA components can be synthesized from DNA “construction genes” by an RNA polymerase, while “control genes” can determine which components are expressed as a function of molecular inputs and RNA regulators - thus, the general scheme of Fig. 1 could be employed using DNA, RNA molecules and transcriptional regulation alone. Due to the reduced number of components in such systems, accurate computational prediction of their behavior should be feasible and may also be used to improve their design.
An RNA-based bistable circuit in vitro
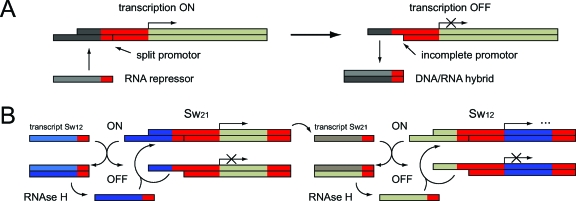
Theoretical work on transcriptional circuits has shown that in vitro systems containing only DNA, RNA, RNA polymerase, and ribonucleases can in principle be used to implement arbitrary circuit functions using RNA transcripts directly as regulators (Kim et al., 2004). Thus a full transcription∕translation system, which contains roughly 100 proteins, may not always be needed. Recent experiments have indeed shown that rationally designed transcriptional elements can regulate each other (Kim et al., 2006). The basic switching principle employed by Kim et al. is depicted in Fig. 2A: double-stranded “gene” templates are constructed, in which one of the DNA strands contains a nick in the promotor region. Transcription from such a split promotor is slightly reduced, but still efficient. Hybridization of an RNA repressor molecule with one of the promotor strands results in a gene template with an incomplete promotor region. Hence, in this situation transcription is switched off. The RNA part of the resulting DNA‐RNA hybrid duplex formed by the promotor and repressor strands can be degraded by ribonuclease H (RNAse H). The promotor region can be completed again and transcription is restored. This very simple synthetic transcriptional system therefore allows for enzyme-mediated controlled production of RNA molecules (by the gene templates and RNA polymerase), and also controlled degradation (by RNAse H).
Figure 2. Simple gene transcriptional circuits may be realized on the basis of DNA, RNA, RNA polymerase and RNAse H alone (Kim et al., 2006).
A: The basic switching principle is based on a promotor region, in which one of the gene strands contains a nick in the promotor region. Removal of one part of the promotor using strand displacement by an RNA regulator molecule leaves the gene with an incomplete promotor region. In this state, transcription is turned OFF. B: Using feedback, this switching principle can be used to realize a simple bistable reaction network: The RNA molecules transcribed from gene Sw21 can switch off gene Sw12, and RNA transcribed from Sw12 can switch off Sw21. RNA molecules in DNA∕RNA hybrid intermediates are degraded by RNAse H. The total network has two stable states: either all of the genes Sw21 are ON and all of the Sw12 are OFF, or vice versa.
To demonstrate that this switch design is modular with programmable connectivity, Kim et al. constructed a simple in vitro bistable circuit [Fig. 2B]. In this circuit, two transcriptional switches (Sw12 and Sw21) are connected by mutually inhibitory links. In the “ON state” of a source template, RNA polymerase is able to synthesize a repressor signal (RNA transcript Sw12 or Sw21), which suppresses transcription from the other template (turning it into the “OFF” state). Total transcript concentrations are adjusted by balancing their rate of production and degradation. In the correct parameter region, this feedback circuit has been experimentally shown to exhibit bistable behavior as designed.
Genes for controlling nanodevices
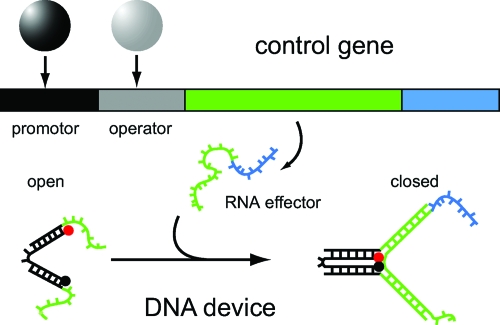
As suggested in Fig. 1, RNA produced from synthetic “genes” may be utilized to drive and control RNA or DNA-based nanodevices. Dittmer et al. (Dittmer et al., 2005) presented the first of such systems in which a fusion of DNA nanotechnology with ideas from synthetic biology was attempted. To this end, a previously introduced DNA nanodevice—so-called DNA tweezers (Yurke et al., 2000)—was operated with RNA effectors rather than DNA “fuel molecules.” The production of the RNA control molecules from artificial gene templates was put under the control of standard regulatory elements taken from the SOS regulon and the lac operon. In Fig. 3, the operation scheme for the gene-controlled closing of DNA tweezers is depicted. In this case, an operator sequence for the repressor protein LacI was put downstream of the promotor sequence, switching OFF transcription of the RNA effector in the presence of LacI. Addition of an inducer molecule (the lactose analogue isopropylthiogalactoside) activated transcription, resulting in a closing of the DNA tweezers by the RNA effectors. This is an example how the response of a molecular machine to environmental changes could be programmed using gene regulatory mechanisms. More generally, genetic mechanisms could be used to coordinate the production and action of nucleic acid nanodevices working in concert, switching them on and off on demand. Obviously, it should be possible to control the behavior of DNA or RNA nanodevices also by purely transcriptional circuits such as that by Kim et al. discussed in the previous paragraph.
Figure 3. An artificial gene with instructions to control a DNA nanodevice (Dittmer et al., 2005).
The gene contains the code for RNA effector strands, which are able to close a DNA tweezers device (Yurke et al., 2000) by hybridization. The promotor itself is under control of an operator, which can be used to make the operation of the nanodevice dependent on an environmental stimulus.
Potentially, the action of DNA or RNA devices could also be coupled to naturally occurring RNA molecules such as mRNA or microRNA. As an example, it is conceivable that these RNA molecules are utilized to trigger the release of a protein from an aptamer-based nanostructure (Dittmer et al., 2004; Beyer and Simmel, 2006).
Artificial development
One of the goals of bionanotechnology is the production of complex structures and materials using biomolecular self-assembly. As mentioned above, “simple” self-assembly based on molecular recognition alone may not be capable of producing all desired structures. For example, it may be necessary to assemble molecular structures in a certain spatial and temporal order.
Similar problems are studied in developmental biology, and it is indeed tempting to think about artificial developmental systems for the assembly of synthetic structures. In biologically motivated work, Isalan et al. recently engineered synthetic spatio-temporal gene networks to emulate Drosophila embryonic pattern formation (Isalan et al., 2005). Embryonic cells were modeled using “gene”-coated paramagnetic beads held in place by magnets in a reaction chamber, forming a spatially extended expression network. In the network, gradients of activators (in this case simply RNA polymerases) were generated from localized sources, switching on bead-immobilized genes for repressor proteins. Diffusion of the repressors led to a spatial modulation of gene expression patterns. Different network connectivities resulted in distinct transitory expression domain patterns, resembling gap formation in the Drosophila embryo. Obviously, the above-mentioned chip-based expression system (Buxboim et al., 2007) should also be of great interest for the study of spatio-temporal effects and the realization of synthetic developmental systems in vitro.
A different – in vivo – approach towards artificial pattern formation was taken by Weiss and co-workers (You et al., 2004). They generated an artificial gene network which responded to the presence of a diffusible inducer (acyl-homoserine lactone, taken from a quorum sensing system) within a certain concentration range—a genetic “band detector.” The band detector was implemented in E. coli and the bacteria were grown as a cell lawn in a Petri dish. Diffusion of the inducer from localized sources selectively turned on expression in bacteria within the “correct” concentration band and thus led to spatial patterning of the biofilm.
In the context of pattern formation, it would be extremely interesting to study spatio-temporal effects induced by temporally varying chemical “sources.” For instance, genetic oscillators or pulse generators (Basu et al., 2004)—or simpler chemical analogs thereof—could be used to induce spatial patterns via reaction diffusion. In fact, genetic oscillators are discussed as one potential source of segmentation in arthropods (Peel et al., 2005). In more nanotechnology-oriented work along these lines, it has been previously shown that reaction-diffusion systems can be used to produce micro- and even nanoscale patterns (Grzybowski et al., 2005). In the context of DNA nanotechnology it was demonstrated that chemical oscillators can drive DNA conformational changes (Liedl et al., 2006).
Artificial cells
In most of the work on synthetic biosystems mentioned so far, the emphasis was put on single components or modules studied in vitro. For a variety of reasons, it would be extremely interesting to encapsulate these components within artificial cell-like compartments (Szostak et al., 2001; Pohorille and Deamer, 2002; Luisi et al., 2006; Forster and Church, 2006; Murtas et al., 2007). Due to the limited diffusion space, reaction kinetics could be faster and reaction products would not be lost by diffusion—an attractive aspect for nanoscale synthesis. Internal organization of the compartments by supramolecular scaffolds could be used for localized production and the realization of micron-scale assembly lines. If continuous supply of nutrients and disposal of waste products over the compartment boundaries was achieved, the system could be permanently held out of equilibrium. If self-reproduction and some sort of genetic variation could be employed, evolutionary aspects could be studied or even technologically utilized. Finally, small compartments could be used to study stochastic effects on the performance of artificial gene networks, e.g., whether they are robust with respect to fluctuations in enzyme numbers.
Pohorille and Deamer proposed a list of desirable properties for an artificial cell (Pohorille and Deamer, 2002): an information-carrying polymer, such as a nucleic acid, must be synthesized by a template-directed polymerization reaction that occurs in a membrane-bound volume; the monomers of the polymer must be provided externally and transported across the membrane boundary to support the replication process; other small molecules or ions needed for biosynthetic reactions must be delivered from the environment; an external source of chemical energy must be available to drive the biosynthetic reactions. Catalysis, replication and growth must be well regulated so that none of the processes lags behind or gets far ahead of other processes in the cell.
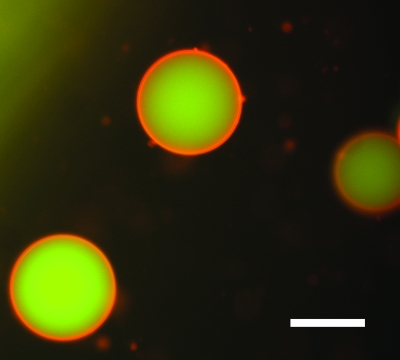
Presumably, the artificial cell “interior” can be constructed similarly to the synthetic biochemical networks mentioned before. As for the encapsulation itself, a very promising approach to mimic a cell-like environment is found in the preparation of lipid bilayer vesicles. First preparations of lipid vesicles date back to the 1960s (Bangham et al., 1965). Thereafter a number of different vesicle types have arisen like small unilamellar vesicles with dimensions from 25 to 100 nm), large unilamellar vesicles with dimensions between 100 nm and 1 μm, and finally giant unilamellar vesicles (GUVs) with dimensions up to 50 μm (Dimitrov and Angelova, 1986; Dimova et al., 2006). An example from our lab for a giant unilamellar vesicle filled with DNA molecules is shown in Fig. 4.
Figure 4. Incorporation of artificial biochemical networks into lipid bilayer vesicles are a promising approach towards realization of artificial cells.
Shown is a fluorescence microscopic image of giant unilamellar vesicles (GUVs, lipids labeled red) filled with fluorescently labeled DNA (green). The GUVs were formed from a lipid mixture containing 90% DPhPC (1,2-diphytanoyl-sn-glycero-3-phosphocholine) and 10% cholesterol by electroswelling using an AC voltage of 1V at 5 Hz for 2 h. The lipids contained 0.1 mol% lipids with a BODIPY label. DNA labeled with Rhodamine Green were incorporated during the electroswelling process. The scale bar is 50 μm.
Gene expression in artificial cells
One step towards assembly of an artificial cell was recently presented by Noireaux et al. (Noireaux and Libchaber, 2004; Noireaux et al., 2005). In this work, a cell-free expression system from E. coli was encapsulated in a lipid vesicle to form a bioreactor. Microdroplets were produced in an oil∕water emulsion and transferred into a feeding solution containing ribonucleotides and amino acids. By doing so, a bilayer was formed and the transcription-translation system together with gene plasmids were isolated in vesicles.
In contrast to in vitro expression experiments in bulk solution, where synthesis of enhanced green fluorescent protein stopped after 2 h, expression time was prolonged in the vesicles for up to 5 h, which is due to the continued supply of the transcription-translation machinery with nutrients through the permeable vesicle membrane.
To solve the problem of limited energy and material resources, Noireaux and Libchaber went even further (Noireaux and Libchaber, 2004). One of the genes encapsulated in the vesicle bioreactor coded for a pore-forming protein (α-hemolysin). After expression inside the vesicle, the protein pores incorporated into the membrane, which increased its selective permeability for nutrients and released osmotic stress. With this “trick,” the vesicle bioreactor sustained expression for up to four days with a maximum protein production of up to 30 μM.
Stochastic effects in artificial cells
One of the motivations for the construction of synthetic biosystems with a reduced number of components is the prospect of a quantitative description of their behavior. All of the systems’ parameters are supposed to be known, and some of them can even be set or varied deliberately. A deterministic description of chemical reaction systems is based on the assumption that concentrations can be treated as continuous variables, whose time evolution is governed by a set of coupled ordinary differential equations, the reaction rate equations. Studying synthetic reaction networks in small compartments like vesicles or micelles, however, will inevitably lead to the occurrence of stochastic effects, as some of the reactants will only be present at low copy numbers (Gillespie, 1977). One then has to consider discrete molecule numbers, and the chemical reaction network has to be treated as a stochastic process. Number fluctuations are expected to be significant in small systems and may strongly influence the overall behavior of the networks.
As a simple example for stochastic effects in a synthetic gene network, the influence of low copy numbers on the behavior of the in vitro bistable switch introduced above (Kim et al., 2006) will be discussed here briefly. Typical reactant concentrations for the bistable switch are in the 1–100 nM range. Assuming a reaction volume of 10 fl (for a vesicle with a diameter of 2.5 μm), typical reactant numbers are between 5 and 500.
Based on the rate equations given in Kim et al., 2006, we simulated the behavior of the bistable switch both deterministically and stochastically (Fig. 5). The deterministic simulations shown in Fig. 5A were performed using the built in ODE solver of the pathway simulation package COPASI (Hoops et al., 2006). Stochastic simulations [Figs. 5B, 5C] of the bistable switch were programmed in MATLAB based on the Gillespie algorithm (Gillespie, 1977; Gibson and Bruck, 2000). Parameters, concentrations and rate constants for the deterministic simulation were chosen in accordance with Kim et al., 2006, the corresponding parameters for the stochastic solver were adjusted for a reaction volume of 10 fl. Figure 5A shows the evolution of concentrations of switches Sw12 and Sw21 in the “ON” state [cf. Fig. 2B] for initial conditions for which the stable configuration is (Sw12 OFF∕Sw21 ON). Obviously, the behavior of both deterministic and stochastic solutions is qualitatively similar in this case and stochasticity mainly results in “noise” added to the traces [cf. insets of Figs. 5A, 5B]. However, when a number of trajectories is calculated using the stochastic model, a wide variation of the “switching times” is found, i.e., the time at which the system locks into its stable configuration. Variation of switching times over 3000 s would be clearly noticeable, if Kim et al.’s in vitro bistable switch was operated in artificial cell compartments.
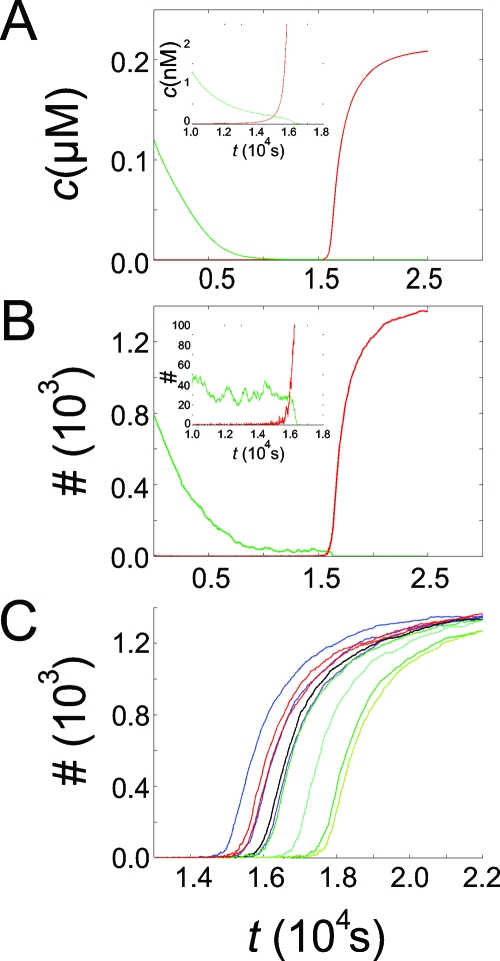
Figure 5. Simulation of an in vitro bistable circuit ( Kim et al. , 2006 ).
A: deterministic simulation based on rate equations, B: stochastic simulation based on the Gillespie algorithm. Shown is the temporal evolution of concentrations∕numbers of genes Sw12 (green) and Sw21 (red) in the “ON” state. The initial conditions for the simulation are chosen in the region of attraction of the state (Sw12 OFF, Sw21 ON). At roughly t=15 000 s in the simulations, all of the genes Sw21 turn ON and genes Sw12 turn OFF. The insets show a zoom of the same simulation runs close to the switching time. In the stochastic case, there is considerable noise in the number of genes in the different states, which is more noticeable in the insets. In part C of the figure, the result of ten consecutive stochastic simulations of the switching event is shown. The switching times vary considerably over a range of 3000 s.
Effects of biochemical noise and stochasticity have been previously studied in natural gene networks (Blake et al., 2003; Pedraza and van Oudenaarden, 2005; Mettetal and van Oudenaarden, 2007), and also in synthetic circuits implemented in bacteria (Elowitz and Leibler, 2000;Hooshangi et al., 2005; Tian and Burrage, 2006). Apart from molecular number fluctuations, biochemical noise originates from fluctuations in reaction rates and also from fluctuations in environmental conditions, which influence gene expression levels (Pedraza and van Oudenaarden, 2005). In biology, robustness with respect to fluctuations is achieved by an intricate combination of feedback mechanisms, redundancy, modularity, and decoupling of organizational levels (Kitano, 2004). Stochastic effects are also expected to play an important role for molecular devices and networks, when implemented in the context of small reaction compartments or artificial cells. Accordingly, robustness and noise tolerance represent great challenges for the design of future artificial biochemical networks.
TOWARDS IN VIVO IMPLEMENTATION OF DNA NANODEVICES
In vivo implementation of artificial nanodevices differs from the pure “bottom up” approach to biological nanotechnology as it utilizes already existing, living biological machinery—with their incompletely known inner workings. Nevertheless, in vivo operation of synthetic molecular devices may represent a faster road to successful applications than the cumbersome and extremely challenging construction of artificial cell-like systems.
In vivo implementation of molecular devices based on DNA or RNA is conceivable in two very different ways: in vitro preparation and chemical modification of nanodevices, followed by packaging and delivery; or transfection of cells with artificial genes containing the blueprint and instructions for the nanodevices (cf. Fig. 1). Whereas several nanomechnical devices based on DNA have been operated using RNA effectors in vitro (Dittmer and Simmel, 2004; Dittmer et al., 2005; Zhong and Seeman, 2006), delivery or in vivo assembly of such structures has not yet been attempted. For in vivo assembly, nanodevices based on intramolecularly folded single strands are favorable. An example for that are so-called “intramers” (Famulok et al., 2001; Famulok and Mayer, 2006). These are intracellular RNA aptamers, which are produced from an engineered expression system, which is transfected into live cells. Intramers have already been demonstrated to be useful in targeting disease-related proteins in vivo. A similar approach is conceivable for other RNA-based nanodevices. As for in vivo production of RNA-based nanostructures, the tRNA-technique mentioned above may be of great use (Ponchon and Dardel, 2007). In general, however, it may not be straightforward to transfer DNA-based in vitro technology to RNA-based in vivo systems. Currently, computational tools are being developed to support RNA-based nanoconstruction (Yingling and Shapiro, 2007). An overview of potential applications of DNA-based nanodevices in vivo and challenges for their implementation has been recently given in Simmel, 2007.
In the remaining paragraphs, we will briefly discuss two recent examples, where in vivo synthetic biosystems have been engineered very much in the spirit of DNA nanotechnology: artificial riboregulators, and in vivo computers based on RNA interference.
Artificial riboregulators
A few years ago, “riboswitches” have been found to play an important role in gene regulation in bacteria (Mandal and Breaker, 2004). Riboswitches are natural RNA aptamer structures incorporated into mRNA transcripts. Depending on the presence or absence of the aptamer target molecules, the transcripts may adopt one of several alternative conformations. Depending on the conformation, transcription may be terminated, or translation of mRNA into protein may be inhibited. In this way, riboswitches exert genetic control on the transcriptional or translational level. It has been shown that cells perform complex tasks such as logical computations using riboswitch circuits (Sudarsan et al., 2006). As has been discussed in the previous paragraphs, switchable structures made from DNA and RNA are a central topic in DNA nanotechnology, and coupling these switches to gene regulation has been attempted in a variety of different ways. In this sense, the construction of artificial riboswitches very well represents a fusion of ideas from DNA nanotechnology and synthetic biology. In fact, artificial riboregulators have been recently devised and implemented in prokaryotic (Isaacs et al., 2004) and also eukaryotic (Bayer and Smolke, 2005) cells. For example, Isaacs et al. constructed partly self-complementary mRNA molecules, which folded back onto themselves in such a way that the ribosome binding site was blocked, and hence translation was inhibited. This strategy for post-transcriptional regulation had been found before in natural riboswitches.
In vivo computing
Another form of regulatory RNA are microRNAs, which are components of the natural RNA interference (RNAi) machinery (Hannon, 2002). In RNAi, a gene silencing process is triggered by the presence of double-stranded RNA molecules, which are cleaved by the enzyme Dicer into short RNA duplexes—so-called short interfering RNAs (siRNA). One of the RNA strands contained in the siRNA duplex is bound by the so-called RNA-induced silencing complex, which induces site-specific degradation of mRNAs containing a complementary sequence. Excitingly, this natural RNAi process can be used to knock down genes using synthetic siRNA molecules (Elbashir et al., 2001). Shortly after the discovery of RNAi, it was found that also endogenous regulatory RNA molecules exist—so-called microRNAs (He and Hannon, 2004). Recently, Benenson and colleagues utilized the RNAi process to perform logical computations in vivo (Rinaudo et al., 2007). To this end, the computation was implemented into genes, whose expression was regulated by natural microRNAs. A reporter gene (coding for a fluorescent protein) was only expressed, when a certain logical combination of several endogenous molecular inputs was present in vivo. Hence, the fluorescence of the modified organisms represented their current logical “molecular state.” Apart from computing, microRNAs quite generally seem to be prime candidates for coupling artificial DNA or RNA-based nanodevices to life processes in vivo.
CONCLUSION
Bionanotechnology, and DNA nanotechnology in particular, aims at the construction of artificial molecular structures and machines from biomolecules, utilizing self-assembly and self-organization phenomena. In fact, DNA and RNA molecules have been successfully harnessed to realize a variety of supramolecular structures, nanomechanical devices and molecular computers.
Transcending the potential of “simple” molecular self-assembly, however, many other properties of biological systems would be desirable for technological systems, for example, environmental responsiveness, robustness, fault tolerance and self-healing, self-reproduction, evolvability, growth and differentiation. Some of these properties can probably only be realized in compartmentalized nonequilibrium systems containing complex molecular interaction networks.
As demonstrated in this perspective, there have recently been many efforts to integrate components developed in DNA nanotechnology into larger networks. The genetic nature of its “building materials,” DNA and RNA, makes DNA nanotechnology compatible with genetic processes. Genes may be used to produce RNA structures, gene regulation may be used to control nanodevices or to feed molecular computers, and in turn DNA or RNA switches may be used for unconventional gene regulatory processes.
Integration of DNA or RNA-based systems is possible both in vitro and in vivo. In vitro, there have been efforts to incorporate synthetic gene networks within artificial cell-like compartments, but there are also efforts to directly operate nucleic acid devices in live cells. Here utilization of riboregulators and RNAi processes seem particularly promising. In either case, stochastic effects are expected to play an important role in systems behavior and will have to be considered for stable operation of artificial biosystems in small compartments.
The research trends surveyed in this perspective indicate that part of the current efforts in DNA nanotechnology and DNA computing will be absorbed in a future synthetic biology. But the relationship between nanotechnology and synthetic biology is mutual (Ball, 2005). Current DNA nanotechnology offers components and concepts for synthetic biology. On the other hand, synthetic biology could be the ultimate “bionanotechnology.”
ACKNOWLEDGMENTS
We thank Erik Winfree and Jongmin Kim for many useful discussions. Financial support by the Human Frontier Science Program (young investigator Grant No. RGY74) and the Nanosystems Initiative Munich is gratefully acknowledged.
References
- Adleman, L M (1994). “Molecular computation of solutions to combinatorial problems.” Science 10.1126/science.7973651 266, 1021–1024. [DOI] [PubMed] [Google Scholar]
- Ball, P (2005). “Synthetic biology for nanotechnology.” Nanotechnology 10.1088/0957-4484/16/1/R01 16, R1–R8. [DOI] [Google Scholar]
- Bangham, A D, et al. (1965). “Diffusion of univalent ions across lamellae of swollen phospholipids.” J. Mol. Biol. 13, 238. [DOI] [PubMed] [Google Scholar]
- Basu, S, et al. (2004). “Spatiotemporal control of gene expression with pulse-generating networks.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0307571101 101, 6355–6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bath, J, et al. (2005). “A free-running DNA motor powered by a nicking enzyme.” Angew. Chem., Int. Ed. 10.1002/anie.200501262 44, 4358–4361. [DOI] [PubMed] [Google Scholar]
- Bath, J, and Turberfield, A J (2007). “DNA nanomachines.” Nat. Nanotechnol. 10.1038/nnano.2007.104 2, 275. [DOI] [PubMed] [Google Scholar]
- Bayer, T S, and Smolke, C D (2005). “Programmable ligand-controlled riboregulators of eukaryotic gene expression.” Nat. Biotechnol. 10.1038/nbt1069 23, 337. [DOI] [PubMed] [Google Scholar]
- Benenson, Y, et al. (2001). “Programmable and autonomous computing machine made of biomolecules.” Nature (London) 10.1038/35106533 414, 430–434. [DOI] [PubMed] [Google Scholar]
- Benenson, Y, et al. (2004). “An autonomous molecular computer for logical control of gene expression.” Nature (London) 10.1038/nature02551 429, 423–429. [DOI] [PubMed] [Google Scholar]
- Beyer, S, and Simmel, F C (2006). “A modular DNA signal translator for the controlled release of a protein by an aptamer.” Nucleic Acids Res. 10.1093/nar/gkl075 34, 1581–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake, W J, et al. (2003). “Noise in eukaryotic gene expression.” Nature (London) 10.1038/nature01546 422, 633–637. [DOI] [PubMed] [Google Scholar]
- Buxboim, A, et al. (2007). “A single-step photolithographic interface for cell-free gene expression and active biochips.” Small 10.1002/smll.200600489 3, 500–510. [DOI] [PubMed] [Google Scholar]
- Chen, J H, and Seeman, N C (1991). “Synthesis from DNA of a molecule with the connectivity of a cube.” Nature (London) 10.1038/350631a0 350, 631–633. [DOI] [PubMed] [Google Scholar]
- Chin, J W (2006). “Modular approaches to expanding the functions of living matter.” Nat. Chem. Biol. 10.1038/nchembio789 2, 304–311. [DOI] [PubMed] [Google Scholar]
- Chworos, A, et al. (2004). “Building programmable jigsaw puzzles with RNA.” Science 10.1126/science.1104686 306, 2068–2072. [DOI] [PubMed] [Google Scholar]
- Dimitrov, D S, and Angelova, M I (1986). “Swelling and electroswelling of lipids—theory and experiment.” Stud. Biophys. 113, 15–20. [Google Scholar]
- Dimova, R, et al. (2006). “A practical guide to giant vesicles. Probing the membrane nanoregime via optical microscopy.” J. Phys.: Condens. Matter 10.1088/0953-8984/18/28/S04 18, S1151–S1176. [DOI] [PubMed] [Google Scholar]
- Dittmer, W U, et al. (2004). “A DNA-based machine that can cyclically bind and release thrombin.” Angew. Chem., Int. Ed. 10.1002/anie.200353537 43, 3550–3553. [DOI] [PubMed] [Google Scholar]
- Dittmer, W U, et al. (2005). “Using gene regulation to program DNA-based molecular devices.” Small 10.1002/smll.200500074 1, 709–712. [DOI] [PubMed] [Google Scholar]
- Dittmer, W U, and Simmel, F C (2004). “Transcriptional control of DNA-based nanomachines.” Nano Lett. 10.1021/nl049784v 4, 689–691. [DOI] [Google Scholar]
- Douglas, S M, et al. (2007). “DNA-nanotube-induced alignment of membrane proteins for NMR structure determination.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0700930104 104, 6644–6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S M, et al. (2001). “Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells.” Nature (London) 10.1038/35078107 411, 494. [DOI] [PubMed] [Google Scholar]
- Elowitz, M B, and Leibler, S (2000). “A synthetic oscillatory network of transcriptional regulators.” Nature (London) 10.1038/35002125 403, 335–338. [DOI] [PubMed] [Google Scholar]
- Ezziane, Z (2006). “DNA computing: applications and challenges.” Nanotechnology 10.1088/0957-4484/17/2/R01 17, R27. [DOI] [Google Scholar]
- Famulok, M, et al. (2001). “Intramers as promising new tools in functional proteomics.” Chem. Biol. 10.1016/S1074-5521(01)00070-9 8, 931. [DOI] [PubMed] [Google Scholar]
- Forster, A C, and Church, G M (2006). “Towards synthesis of a minimal cell.” Molecular Syst. Biol. 2, 45. [DOI] [PMC free article] [PubMed]
- Gardner, T S, et al. (2000). “Construction of a genetic toggle switch in Escherichia coli.” Nature (London) 10.1038/35002131 403, 339–342. [DOI] [PubMed] [Google Scholar]
- Garibotti, A V, et al. (2007). “A simple DNA-based translation system.” Nano Lett. 10.1021/nl0628605 7, 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, M A, and Bruck, J (2000). “Efficient exact stochastic simulation of chemical systems with many species and many channels.” J. Phys. Chem. A 10.1021/jp993732q 104, 1876–1889. [DOI] [Google Scholar]
- Gillespie, D T (1977). “Exact stochastic simulation of coupled chemical reactions.” J. Phys. Chem. 10.1021/j100540a008 81, 2340–2361. [DOI] [Google Scholar]
- Goodman, R P, et al. (2005). “Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication.” Science 10.1126/science.1120367 310, 1661–1665. [DOI] [PubMed] [Google Scholar]
- Grzybowski, B A, et al. (2005). “Micro- and nanotechnology via reaction diffusion.” Soft Mater. 10.1039/b501769f 1, 114–128. [DOI] [Google Scholar]
- Guo, P X, et al. (1998). “Inter-RNA interaction of phage phi 29 pRNA to form a hexameric complex for viral DNA transportation.” Mol. Cell 10.1016/S1097-2765(00)80124-0 2, 149. [DOI] [PubMed] [Google Scholar]
- Halpin, D R, and Harbury, P B (2004). “DNA display II. Genetic manipulation of combinatorial chemistry libraries for small-molecule evolution.” PLoS Biol. 2, 1022–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon, G J (2002). “RNA interference.” Nature (London) 10.1038/418244a 418, 244. [DOI] [PubMed] [Google Scholar]
- Hartwell, L H, et al. (1999). “From molecular to modular cell biology.” Nature (London) 10.1038/35011540 402, C47–C52. [DOI] [PubMed] [Google Scholar]
- He, L, and Hannon, G J (2004). “Micrornas: small RNAs with a big role in gene regulation.” Nat. Rev. Genet. 10.1038/nrg1379 5, 522. [DOI] [PubMed] [Google Scholar]
- Hoops, S, et al. (2006). “Copasi—a complex pathway simulator.” Bioinformatics 10.1093/bioinformatics/btl485 22, 3067–3074. [DOI] [PubMed] [Google Scholar]
- Hooshangi, S, et al. (2005). “Ultrasensitivity and noise propagation in a synthetic transcriptional cascade.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0408507102 102, 3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs, F J, et al. (2004). “Engineered riboregulators enable post-transcriptional control of gene expression.” Nat. Biotechnol. 10.1038/nbt986 22, 841–847. [DOI] [PubMed] [Google Scholar]
- Isalan, M, et al. (2005). “Engineering gene networks to emulate Drosophila embryonic pattern formation.” PLoS Biol. 10.1371/journal.pbio.0030064 3, 488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger, L, and Chworos, A (2006). “The architectonics of programmable RNA and DNA nanostructures.” Curr. Opin. Struct. Biol. 10.1016/j.sbi.2006.07.001 16, 531. [DOI] [PubMed] [Google Scholar]
- Jermutus, L, et al. (1998). “Recent advances in producing and selecting functional proteins by using cell-free translation.” Curr. Opin. Biotechnol. 10.1016/S0958-1669(98)80042-6 9, 534–548. [DOI] [PubMed] [Google Scholar]
- Joyce, G F (2007). “Forty years of in vitro evolution.” Angew. Chem., Int. Ed. [DOI] [PubMed]
- Kauffman, S, and Ellington, A D (1999). “Thinking combinatorially.” Curr. Opin. Chem. Biol. 10.1016/S1367-5931(99)80040-4 3, 256–259. [DOI] [PubMed] [Google Scholar]
- Kim, J, et al. (2004). “Neural network computation by in vitro transcriptional circuits.” Adv. Neural Inf. Process. Syst. 17, 681–688. [Google Scholar]
- Kim, J, et al. (2006). “Construction of an in vitro bistable circuit from synthetic transcriptional switches.” Mol. Syst. Biol. art no. 68 2006. [DOI] [PMC free article] [PubMed]
- Kitano, H (2004). “Biological robustness.” Nat. Rev. Genet. 10.1038/nrg1471 5, 826–837. [DOI] [PubMed] [Google Scholar]
- Koltermann, A, and Kettling, U (1997). “Principles and methods of evolutionary biotechnology.” Biophys. Chem. [DOI] [PubMed]
- LaBean, T H, and Li, H Y (2007). “Constructing novel materials with DNA.” Nano Today 2, 26. [Google Scholar]
- Li, X, and Liu, D R (2004). “DNA-templated organic synthesis: nature’s strategy for controlling chemical reactivity applied to synthetic molecules.” Angew. Chem., Int. Ed. 10.1002/anie.200400656 43, 4848–4870. [DOI] [PubMed] [Google Scholar]
- Liedl, T, et al. (2006). “A surface-bound DNA switch driven by a chemical oscillator.” Angew. Chem., Int. Ed. 10.1002/anie.200600353 45, 5007. [DOI] [PubMed] [Google Scholar]
- Lu, Y, and Liu, J W (2006). “Functional DNA nanotechnology: emerging applications of DNAzymes and aptamers.” Curr. Opin. Biotechnol. 10.1016/j.copbio.2006.10.004 17, 580. [DOI] [PubMed] [Google Scholar]
- Luisi, P L, et al. (2006). “From never born proteins to minimal living cells: two projects in synthetic biology.” Origins Life Evol. Biosphere 10.1007/s11084-006-9033-6 36, 605–616. [DOI] [PubMed] [Google Scholar]
- Mandal, M, and Breaker, R R (2004). “Gene regulation by riboswitches.” Nat. Rev. Mol. Cell Biol. 5, 451–463. [DOI] [PubMed] [Google Scholar]
- Mao, C D, et al. (1999). “A nanomechanical device based on the B-Z transition of DNA.” Nature (London) 10.1038/16437 397, 144–146. [DOI] [PubMed] [Google Scholar]
- Mettetal, J T, and van Oudenaarden, A (2007). “Necessary noise.” Science 10.1126/science.1146747 317, 463–464. [DOI] [PubMed] [Google Scholar]
- Murtas, G, et al. (2007). “Protein synthesis in liposomes with a minimal set of enzymes.” Biochem. Biophys. Res. Commun. 363, 12–17. [DOI] [PubMed] [Google Scholar]
- Noireaux, V, et al. (2003). “Principles of cell-free genetic circuit assembly.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.2135496100 100, 12672–12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noireaux, V, et al. (2005). “Toward an artificial cell based on gene expression in vesicles.” Phys. Biol. 10.1088/1478-3975/2/3/P01 2, P1–P8. [DOI] [PubMed] [Google Scholar]
- Noireaux, V, and Libchaber, A (2004). “A vesicle bioreactor as a step toward an artificial cell assembly.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0408236101 101, 17669–17674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedraza, J M, and van Oudenaarden, A (2005). “Noise propagation in gene networks.” Science 10.1126/science.1109090 307, 1965–1969. [DOI] [PubMed] [Google Scholar]
- Peel, A D, et al. (2005). “Arthropod segmentation: beyond the Drosophila paradigm.” Nat. Rev. Genet. 10.1038/nrm1824 6, 905–916. [DOI] [PubMed] [Google Scholar]
- Penchovsky, R, and Breaker, R R (2005). “Computational design and experimental validation of oligonucleotide-sensing allosteric ribozymes.” Nat. Biotechnol. 10.1038/nbt1155 23, 1424. [DOI] [PubMed] [Google Scholar]
- Pohorille, A, and Deamer, D (2002). “Artificial cells: prospects for biotechnology.” Trends Biotechnol. 10.1016/S0167-7799(02)01909-1 20, 123. [DOI] [PubMed] [Google Scholar]
- Ponchon, L, and Dardel, F (2007). “Recombinant RNA technology: the tRNA scaffold.” Nat. Methods 4, 571. [DOI] [PubMed] [Google Scholar]
- Qian, H (2005). “Cycle kinetics, steady state thermodynamics and motors, —a paradigm for living matter physics.” J. Phys.: Condens. Matter [DOI] [PubMed]
- Rackham, O, and Chin, J W (2005). “A network of orthogonal ribosome center dot mRNA pairs.” Nat. Chem. Biol. 10.1038/nchembio719 1, 159–166. [DOI] [PubMed] [Google Scholar]
- Rinaudo, K, et al. (2007). “A universal RNAi-based logic evaluator that operates in mammalian cells.” Nat. Biotechnol. 10.1038/nbt1307 25, 795. [DOI] [PubMed] [Google Scholar]
- Rothemund, P WK (2006). “Folding DNA to create nanoscale shapes and patterns.” Nature (London) 10.1038/nature04586 440, 297–302. [DOI] [PubMed] [Google Scholar]
- Rothemund, P WK, et al. (2004). “Algorithmic self-assembly of DNA Sierpinski triangles.” PLoS Biol. 10.1371/journal.pbio.0020424 2, 2041–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann, J, et al. (2006). “DNA-encoded chemical libraries.” J. Biotechnol. 126, 568–581. [DOI] [PubMed] [Google Scholar]
- Schuster, P (1997). “Genotypes with phenotypes: adventures in an RNA toy world.” Biophys. Chem. [DOI] [PubMed]
- Seeman, N C (1982). “Nucleic acid junctions and lattices.” J. Theor. Biol. 10.1016/0022-5193(82)90002-9 99, 237–240. [DOI] [PubMed] [Google Scholar]
- Seeman, N C (2004). “Nanotechnology and the double helix.” Sci. Am. 290, 64. [DOI] [PubMed] [Google Scholar]
- Seeman, N C (2005). “From genes to machines: DNA nanomechanical devices.” Trends Biochem. Sci. 10.1016/j.tibs.2005.01.007 30, 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman, N C, and Lukeman, P S (2005). “Nucleic acid nanostructures: bottom-up control of geometry on the nanoscale.” Rep. Prog. Phys. 10.1088/0034-4885/68/1/R05 68, 237–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, W B, and Seeman, N C (2004). “A precisely controlled DNA biped walking device.” Nano Lett. 10.1021/nl049527q 4, 1203–1207. [DOI] [Google Scholar]
- Shih, W M, et al. (2004). “A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron.” Nature (London) 10.1038/nature02307 427, 618–621. [DOI] [PubMed] [Google Scholar]
- Shin, J S, and Pierce, N A (2004). “A synthetic DNA walker for molecular transport.” J. Am. Chem. Soc. 10.1021/ja047543j 126, 10834–10835. [DOI] [PubMed] [Google Scholar]
- Shu, D, et al. (2004). “Bottom-up assembly of RNA arrays and superstructures as potential parts in nanotechnology.” Nano Lett. 10.1021/nl0494497 4, 1717–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmel, F C (2007). “Towards biomedical applications for nucleic acid nanodevices.” Nanomedicine 2, 817–830. [DOI] [PubMed] [Google Scholar]
- Spirin, A S, et al. (1988). “A continuous cell-free translation system capable of producing polypeptides in high yield.” Science 10.1126/science.3055301 242, 1162–1164. [DOI] [PubMed] [Google Scholar]
- Stojanovic, M N, and Stefanovic, D (2003). “A deoxyribozyme-based molecular automaton.” Nat. Biotechnol. 10.1038/nbt862 21, 1069–1074. [DOI] [PubMed] [Google Scholar]
- Sudarsan, N, et al. (2006). “Tandem riboswitch architectures exhibit complex gene control functions.” Science 10.1126/science.1130716 314, 300–304. [DOI] [PubMed] [Google Scholar]
- Szostak, J W, et al. (2001). “Synthesizing life.” Nature (London) 10.1038/35053176 409, 387–390. [DOI] [PubMed] [Google Scholar]
- Tian, T H, and Burrage, K (2006). “Stochastic models for regulatory networks of the genetic toggle switch.” Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0507818103 103, 8372–8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman, S, et al. (2007). “An autonomous polymerization motor powered by DNA hybridization.” Nat. Nanotechnol. 10.1038/nnano.2007.225 2, 490. [DOI] [PubMed] [Google Scholar]
- Wilson, D S, and Szostak, J W (1999). “In vitro selection of functional nucleic acids.” Annu. Rev. Biochem. 10.1146/annurev.biochem.68.1.611 68, 611–647. [DOI] [PubMed] [Google Scholar]
- Winfree, E, et al. (1998). “Design and self-assembly of two-dimensional DNA crystals.” Nature (London) 10.1038/28998 394, 539–544. [DOI] [PubMed] [Google Scholar]
- Yan, H, et al. (2002). “A robust DNA mechanical device controlled by hybridization topology.” Nature (London) 10.1038/415062a 415, 62–65. [DOI] [PubMed] [Google Scholar]
- Yan, H, et al. (2003a). “Parallel molecular computations of pairwise exclusive or (XOR) using DNA “String tile” self-assembly.” J. Am. Chem. Soc. 10.1021/ja036676m 125, 14246–14247. [DOI] [PubMed] [Google Scholar]
- Yan, H, et al. (2003b). “DNA-templated self-assembly of protein arrays and highly conductive nanowires.” Science 10.1126/science.1089389 301, 1882–1884. [DOI] [PubMed] [Google Scholar]
- Yingling, Y G, and Shapiro, B A (2007). “Computational design of an RNA hexagonal nanoring and an RNA nanotube.” Nano Lett. 10.1021/nl070984r 7, 2328–2334. [DOI] [PubMed] [Google Scholar]
- You, L C, et al. (2004). “Programmed population control by cell-cell communication and regulated killing.” Nature (London) 10.1038/nature02491 428, 868–871. [DOI] [PubMed] [Google Scholar]
- Yurke, B, et al. (2000). “A DNA-fueled molecular machine made of DNA.” Nature (London) 10.1038/35020524 406, 605–608. [DOI] [PubMed] [Google Scholar]
- Zhong, H, and Seeman, N C (2006). “RNA used to control a DNA rotary nanomachine.” Nano Lett. 10.1021/nl062183e 6, 2899. [DOI] [PMC free article] [PubMed] [Google Scholar]