Abstract
Great advances have been made in electron microscopy (EM) over the past decade, with the result that a number of protein complexes have been solved at near-atomic resolution using EM imaging. However, only a limited number of such complexes are expected to have the high degree of internal order needed to achieve this type of resolution. Many other complexes and polymers will be visualized and reconstructed by EM at an intermediate level of resolution, where the polypeptide chain cannot be directly traced. Crystal and nuclear magnetic resonance structures for components or subunits of these higher-order assemblies are frequently available. One of the greatest strengths of EM continues to be the ability to dock high-resolution structures of components into low or intermediate resolution reconstructions of assemblies to build pseudoatomic models for quaternary structure. This review discusses the strengths and limitations of this approach, with particular emphasis on protein polymers. I discuss how limitations in resolution can lead to ambiguities in building models, and these cannot be always be resolved with available data. The use of homology models for quaternary structure are particularly problematic, given accumulating evidence for the divergence of quaternary structures at the same time that tertiary structure can be conserved.
Most proteins exist within cells and viruses as components of large macromolecular complexes. While some of our earliest insights into protein function came from in vitro biochemical observations of enzyme activity, these assays were typically based upon studying the reactions catalyzed by very dilute solutions of soluble proteins. We now understand that although these assays are extremely useful, isolated molecules acting alone on substrates may not always reflect the densely crowded environments in cells where proteins function in many cases as parts of larger complexes. Highly abundant proteins in the cell, such as actin, tubulin, collagen, and intermediate filaments, form helical filaments, so it is easy to see how most of the protein in a cell can exist in some multimeric or polymeric state. I will focus in this brief article on how very different techniques in structural biology have been successfully combined to give us many new insights into these complexes and polymers.
ADVANCES IN ELECTRON MICROSCOPY
One of the most useful techniques that we have for studying the structure of large macromolecular complexes is electron microscopy (EM). It was shown 40 years ago that two-dimensional electron microscopic images of a protein polymer, the tail of a bacteriophage, could be used to generate a three-dimensional reconstruction of the assembly (DeRosier and Klug, 1968). This application gave rise to the field of three-dimensional electron microscopy, an area that continues to grow. While the original work on bacteriophage tails was done with negatively stained samples, the introduction of electron cryomicroscopy using rapidly frozen, unstained, and fully hydrated specimens (Dubochet et al., 1988) has led to many improvements in resolution. Dramatic advances have been made in EM over the past 5 or 6 years, leading to the structure of an integral membrane protein in its native membrane environment at 1.9 Å resolution (Gonen et al., 2005), and the structures of two viral capsids (Zhang et al., 2008; Yu et al., 2008), the bacterial flagellar filament (Yonekura et al., 2003), and the acetylcholine receptor (Miyazawa et al., 2003), all at better than 4.0 Å resolution. At this resolution the structures are said to be “solved,” since the polypeptide chain can be traced to yield a three-dimensional model. We can clearly expect more such sensational results in the future due to improvements in specimen preparation, imaging, and most importantly, computational image processing. Just as the improvements in the rate at which genomes can be sequenced parallels the advances that have been made in computer processing speed (an exponential given by Moore’s Law for the rate of increase in the number of transistors that can be packaged in an integrated circuit), advances in the field of structure determination by EM also depend heavily upon increased computational capabilities.
MERGING TECHNIQUES
Despite the recent spectacular achievements, very high resolution structures solved by EM may still be exceptional, and in the near future most protein complexes will only be visualized by EM at lower resolutions (perhaps 5–25 Å) where the polypeptide chain cannot be traced and the three-dimensional coordinates of every residue cannot be determined. In some cases, x-ray crystallography can be used to determine at high resolution the structure of a very large complex, such as the large ribosomal subunit (Ban et al., 2000), an entire ribosome (Laurberg et al., 2008) or a RecA-DNA filament (Chen et al., 2008). However, it will more frequently be the case that high resolution structural techniques, such as x-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy, will be combined with low- or medium-resolution EM to yield “pseudoatomic” or “quasiatomic” models of large polymers or complexes. The complementarity of these high- and low-resolution techniques has been exceptional, and I will illustrate this by discussing a number of examples. I will also show how general principles are emerging from some of these studies that may provide new insights into evolutionary mechanisms, insights that would not have been possible without the merger of very different biophysical approaches to understanding structure.
The basic principle is very simple in theory, but as has been said before: in theory, theory and practice are simply related, but in practice that is frequently not the case. If high-resolution structures can be obtained for all of the components of a complex (in the case of a homopolymer, only the structure of a single protomer is needed), and a low- or medium-resolution reconstruction can be obtained for the multimer or polymer, then the high-resolution atomic models can be docked into the lower resolution reconstruction. This docking may be done by “eye,” which can be more elegantly described as a neural net that has been optimized by millions of years of evolution. It can also be done computationally (e.g., Topf et al., 2008), seeking to maximize some function (such as the coefficient of correlation between the reconstructed density and the atomic model being fit). If, and only if, there are no conformational changes between the high-resolution structures of the components and the same molecules within the complexes, and there are no errors in the reconstruction (including noise), then the docking can be done with atomic precision. In practice, it is unlikely that either condition is ever strictly met. Readers of the scientific literature must therefore be able to appreciate the uncertainties, ambiguities, and potential errors that may result from such attempts to build pseudoatomic models. There are many papers that have proposed different approaches to this problem of docking or fitting high-resolution structures into low resolution maps (Rossmann et al., 2005, 2001; Fabiola and Chapman, 2005; Volkmann and Hanein, 2003; Birmanns and Wriggers, 2007, 2003; Trabuco et al., 2008), but I will not attempt to review or summarize them. Rather, I will try to highlight some of the problems and limitations inherent in this type of model building.
LIMITATIONS ON ACCURACY
Potential errors in coordinates have been discussed with regards to x-ray crystal structures (Depristo et al., 2004; Wlodawer et al., 2008), with the surprising conclusion that the accuracy of such coordinates has been widely overestimated (Depristo et al., 2004). One of the main sources of errors appears to be the existence of multiple isomers within a crystal, so that the single structure determined may be an artefactual average over several such multiple conformers. These atomic level uncertainties have little, if any, impact on the topic of this review, but they do highlight a problem that arises from structural heterogeneity and polymorphism. Outside of a crystal the potential for multiple states to exist is far larger, and given that these states are not constrained to pack into a single crystal space group, the magnitude of the differences among these states may be far greater than what can be accommodated within a crystal. A growing body of literature is now dealing with using computational image analysis to detect and characterize such conformational heterogeneity in electron micrographs of isolated macromolecular complexes (those not packed into a polymer or crystal) (Elad et al., 2008; Scheres et al., 2007; Fu et al., 2007; Penczek et al., 2006b; Gao et al., 2004; Penczek et al., 2006a).
There is also a growing body of literature dealing with structural heterogeneity within polymers. We have been describing for many years the conformational variability that exists within filaments of actin, called F-actin (for filamentous) (Galkin et al., 2002b, 2001; Orlova and Egelman, 2000; Orlova et al., 1995; Egelman and DeRosier, 1991; Egelman et al., 1982). While F-actin might have been considered exceptional for having such structural plasticity, it now appears that it may be more typical, since other filaments, such as a bacterial type III secretion system polymer (Wang et al., 2006), the bacterial ParM filament (Orlova et al., 2007), the Rad51 recombination filament (Galkin et al., 2006b), an archaeal pilus (Wang et al., 2008) or the flagellar filament from Campylobacter jejuni (Galkin et al., 2008b), display a comparable or greater degree of variability. This heterogeneity and disorder poses great challenges for conventional methods of three-dimensional helical reconstruction (DeRosier and Klug, 1968), where it is necessary to assume that a polymer has a uniform helical symmetry which is imposed over long filaments.
As a result of the problems posed by such variable polymers, we have developed a method of three-dimensional reconstruction of helical filaments that uses a “single particle” type approach (Egelman, 2000, 2007). Due to the very poor signal-to-noise ratio in EM images, particularly those obtained from frozen-hydrated specimens, averaging of many images is needed to produce images that can be reliably interpreted. A two-dimensional crystal provides a means for readily doing this, since every asymmetric unit in the crystal can be simply added together to produce a very high signal-to-noise ratio image. But most proteins or macromolecular complexes do not form two-dimensional crystals. It was realized, however, that images of single particles could be effectively averaged computationally, once the images were aligned (Frank et al., 1981). When these single particles have internal symmetry, such as icosahedral viruses, then a tremendous increase in averaging power occurs, with the result that the three-dimensional reconstructions can now be used to trace the protein backbone in the most favorable cases (Zhang et al., 2008; Yu et al., 2008). Most single particles lack such a high degree of internal symmetry, with the result that reconstructions are at a lower resolution. Nevertheless, the ribosome, which lacks any internal symmetry, can now be reconstructed at a resolution of ∼7 Å (Connell et al., 2007) due to many advances, particularly in computational image analysis. As mentioned earlier, there is a growing realization that individual particles can be in different conformations, and the averaging together of these different states can limit the resolution, or yield artefactual averages. As investigators try to image and reconstruct such single particle complexes at higher resolution, more and more attention is being paid to sorting out conformational heterogeneity.
The single particle approach to reconstruction of helical filaments starts by cutting electron microscopic images of such polymers into many short segments, each of which is treated as a single particle. By this means, many such images can be used in a three-dimensional reconstruction once the orientation of each segment is determined (Fig. 1). A number of such applications have been published by other groups (Sachse et al., 2007, 2008; Jimenez et al., 1999; Li et al., 2002), so our approach is not unique. Since one is not forcing a particular helical symmetry on a long filament, variations in twist and extension do not limit resolution in the same way that they would with traditional approaches. Further, the ability to sort segments into different structural states also eliminates the loss of resolution or artifacts that might result from global averaging.
Figure 1. An electron cryomicrograph of rapidly frozen and unstained Campylobacter flagellar filaments (a) contains three-dimensional information, but this has been projected onto two dimensions.
Segments of these filaments (such as shown by the red box) may be aligned to provide the different views needed to generate a three-dimensional reconstruction (Galkin et al., 2008b), shown as a solid surface in (b). The interpretation of such medium- or low-resolution structures frequently requires high-resolution structures of the component subunits.
ACTIN AS AN EXAMPLE
Since the crystal structure of monomeric actin, called G-actin (for globular) was first solved many years ago (Kabsch et al., 1990), actin provides a perfect example of the potential complexity in fitting a high-resolution structure of a subunit into a low-resolution EM reconstruction of a multimer, polymer, or macromolecular complex. Just as a ribosome can exist in different conformational states (Scheres et al., 2007), an actin filament cannot be reduced to a single structural model. It is clear that there can be large variability in both the twist and the tilt of subunits within the same filament (Galkin et al., 2002b; Schmid et al., 2004). It is also unknown to what extent there are conformational changes between the various crystal structures of G-actin and the protomer within the F-actin polymer. An initial attempt to build a model for F-actin based upon the rigid-body docking of a crystal structure of G-actin into a filament model, constrained by x-ray fiber diffraction data (Holmes et al., 1990), suggested that the possible conformational changes might be small. All subsequent EM studies have been in agreement with this conclusion, with the exception of observations that the smallest subdomain of actin, subdomain 2 (containing ∼40 of the 375 residues in the protein), can be highly variable in conformation within the filament (Orlova and Egelman, 1993).
When actin is complexed with actin-binding proteins (and hundreds of such proteins exist!) a new degree of complexity emerges. Not only does one have the problem of fitting the actin subunit into the reconstruction, but the actin-binding protein may have even greater degrees of freedom. For instance, one common motif present in many actin-binding proteins is the Calponin Homology (CH) domain, named after the protein calponin where one such motif exists (Bramham et al., 2002; Goldsmith et al., 1997; Matsudaira, 1991). In some actin-binding proteins, such as spectrin, dystrophin, utrophin, and α-actinin, tandem CH-domains exist within the regions of these proteins shown to bind F-actin. In one protein, fimbrin, four CH-domains are found (Klein et al., 2004), and two different actin-binding interfaces are responsible for fimbrin crosslinking two actin filaments. When the resolution is limited in reconstructions of F-actin decorated with either these proteins, or fragments of these proteins, great ambiguities can exist in the interpretation. That has generated a certain amount of controversy in the literature about how these domains interact with F-actin, and whether there is a single conserved mode of interaction between a CH-domain and actin (Lehman et al., 2004; Sutherland-Smith et al., 2003; Hanein et al., 1998; Galkin et al., 2003, 2002a). The demonstration that when calponin binds F-actin the CH-domain within calponin is never attached to actin shows that there cannot be a conserved mode of interaction between CH-domains and actin (Galkin et al., 2006a). It also shows that the fitting of atomic models into low-resolution reconstructions, as was done for the calponin CH-domain (Bramham et al., 2002), is potentially problematic when the resolution is insufficient to generate unambiguous fits. This type of ambiguity that may be present at low resolution led us to generate an incorrect model for the Rad51-DNA filament (Yang et al., 2001). The ambiguities arise for several reasons. One is that when the resolution is limited, multiple solutions may exist that cannot be distinguished. While one solution may score in some way higher than others, the confidence in such scoring must be limited given the caveats about noise and error in the reconstructions and conformational changes from the high resolution model being docked. Given these examples, it would be reassuring if the confidence in docking atomic models into low-resolution reconstructions could be reduced to a simple formula. Unfortunately, there are many factors that are involved, including the asymmetry of the molecule being docked, the quality of the reconstruction, and the extent of conformational change between the subunit whose structure is known and the subunit in the polymer or complex.
AMBIGUITIES DISAPPEAR AT HIGHER RESOLUTION
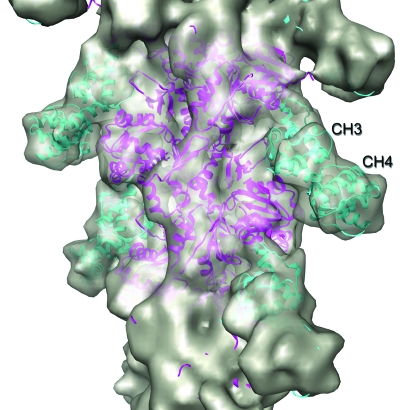
As resolution improves, not only do ambiguities in docking or fitting disappear, but new insights can emerge into structure and function. A reconstruction of tobacco mosaic virus at better than 5 Å resolution (Sachse et al., 2007) showed significant differences at low radius with a model determined by x-ray fiber diffraction (Namba et al., 1989; Namba and Stubbs, 1986), and these differences may be important to how the protein coat binds RNA and switches between assembly and disassembly. A recent result obtained at lower resolution (Fig. 2) is informative when one looks at the general question of docking atomic structures into EM reconstructions. A fragment of fimbrin containing two CH-domains (CH3 and CH4) was used to decorate F-actin, and this has been solved at ∼12 Å resolution using cryo-EM and a single particle approach to helical reconstruction (Galkin et al., 2008a). The single particle method was essential, since the heavily decorated actin filaments still had great variability in twist, conformation, degree of binding, etc. The resolution that was achieved meant that the individual CH-domains could be docked into the reconstruction separately, and the asymmetry between the two CH-domains at this resolution was great enough so that there was no ambiguity between which “blob” was CH3 and which was CH4 (Fig. 2). This resolution was also high enough to see that the two CH-domains needed to be rotated with respect to each other from the conformation in which they were seen in a crystal structure (Klein et al., 2004). At lower resolution, one might have had a complete ambiguity about which domain was CH3 and which was CH4, as well as being forced to fit the tandem CH-domains as a rigid body as seen in the crystal. Clearly the interaction between CH3 and actin is very different than between CH4 and actin, showing that the structural conservation of the CH-domain cannot define its interaction with actin. The specific mode of interaction between the two CH-domains and F-actin is different than that either observed or proposed for other tandem CH-domains (Hanein et al., 1998; Galkin et al., 2002a). Since the CH-domain in the eponymous protein calponin never even interacts with actin (Galkin et al., 2006a; Gimona and Mital, 1998), the presence of a CH-domain tells us much more about the evolutionary history of a protein than it does about the current function or interactions of that protein. This conclusion may be depressing for those who would like to reduce biology to a set of conserved interactions among a set of structurally conserved domains!
Figure 2. A three-dimensional reconstruction of the complex between F-actin and a fragment of fimbrin, obtained from electron cryomicrographs, is shown as a gray surface (Galkin et al., 2008a).
The resolution of this reconstruction (∼12 Å) allows for the unambiguous docking of atomic structures of actin (magenta ribbons) and the fimbrin ABD2 (Actin Binding Domain 2, cyan ribbons) into the reconstruction. The resolution is also sufficient to see that the two major domains in actin must be more closed in this complex than they are in either the crystal structure of G-actin (Schutt et al., 1993) or in reconstructions from naked F-actin filaments (Galkin et al., 2008a), and the ribbon model shown for actin (magenta) has been modified in this way. Similarly, the fimbrin ABD2, containing CH3 and CH4, has been perturbed from the crystal structure (Klein et al., 2004) in this docking by a rotation of CH4 with respect to CH3.
DIVERGENCE OF POLYMER STRUCTURE
The problem that we are discussing, how to fit high-resolution structures into low resolution maps, is also compounded by the fact that polymer structures can diverge rather unexpectedly over the course of evolution. Several examples have recently been published that show how protein subunits that have a relatively conserved tertiary structure can assemble into polymers that have very different quaternary structures. We have known for a number of years that bacterial homologs of actin exist, such as ParM, which is involved in plasmid segregation (Moller-Jensen et al., 2002). The crystal structure of ParM (van den Ent et al., 2002) confirmed that it has largely the same fold as actin, hexokinase, and HSP70 (Bork et al., 1992), supporting the notion that all of these proteins diverged from a common ancestral protein. Yet filaments of ParM have been shown to have the opposite helical hand as F-actin (Orlova et al., 2007; Popp et al., 2008), establishing that they cannot have the same subunit-subunit contacts that exist in F-actin. One will thus need high resolution structures of the ParM filament to begin modeling the details of the subunit-subunit interfaces that exist.
One of the triumphs of electron microscopy has been the generation of a complete atomic model of the Salmonella flagellar filament using electron cryomicrographs (Yonekura et al., 2003). Bacterial flagellin is fairly conserved in the coiled-coil domains responsible for the assembly of the flagellar filament (Beatson et al., 2006), so the expectation was that all flagellar filaments would therefore have this same architecture. The finding that Campylobacter flagellar filaments are assembled from seven protofilaments (Galkin et al., 2008b), rather than 11 as in Salmonella, means that the subunit-subunit contacts in Campylobacter must be quite different than in Salmonella, even though the tertiary structures of the protein subunits must be very similar. Due to the rather featureless nature of the coiled-coils at low resolution, the ∼15 Å resolution obtained in the EM reconstruction (Galkin et al., 2008b) is actually insufficient to generate a unique model of how these subunits pack together in the Campylobacter flagellar filaments. In contrast, much lower resolution reconstructions of F-actin (at 20–25 Å) can be used to uniquely orient a subunit (Egelman et al., 1997) into a filament model. In the case of the globular heads of myosin, the structural asymmetry is large enough that ∼25 Å resolution was sufficient to provide an unambiguous fit of crystal structures into a cryo-EM map (Woodhead et al., 2005).
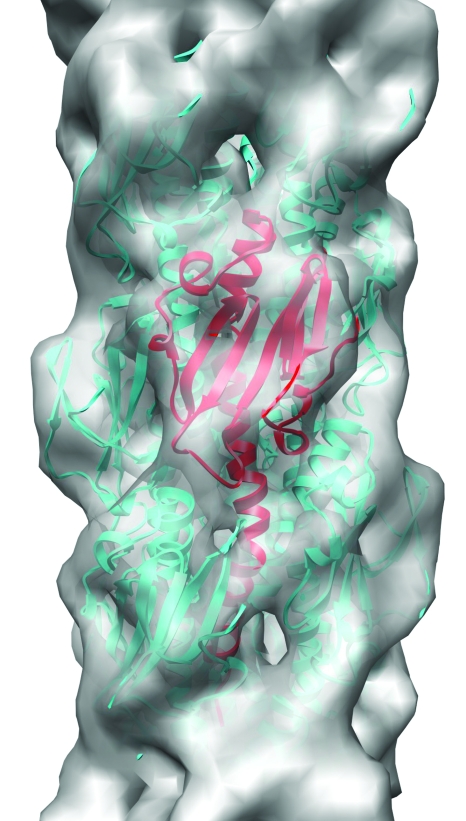
The last example regards bacterial type IV pili, which are involved in functions ranging from motility to adhesion to natural transformation. A cryo-EM reconstruction of the type IV pilus from Neisseria gonorrhoeae at ∼12 Å resolution (Fig. 3) proved sufficient to be able to generate a model for the pilus built from a crystal structure of the subunit (Craig et al., 2006). A putative type IV pilin exists in archaea (Szabo et al., 2007), and it might be expected that it would assemble in the same manner as the bacterial homolog, with hydrophobic N-terminal α-helices forming a core. Surprisingly, the archaeal pili have a hollow lumen with a very different packing symmetry than observed in Neisseria gonorrhoeae (Wang et al., 2008). The resolution of the reconstruction (∼15 Å) is simply too low to begin building any reasonable homology model for both the subunit and the packing in the pilus.
Figure 3. The type IV pilus has been reconstructed at ∼12 Å resolution from electron cryomicrographs ( Craig et al., 2006 ) and is shown as a gray surface.
This resolution provides a unique fit for the crystal structure of the component pilin (one subunit shown in red ribbons, while the surrounding subunits are shown in cyan).
IN SUMMARY
The existence of high resolution structures of protein subunits, determined by x-ray crystallography or NMR spectroscopy, combined with low resolution EM reconstructions of complexes or polymers, has given us a huge opportunity to merge techniques and develop “pseudoatomic models” of the higher-order structures. Our own work in this area has revealed an unexpected degree of polymorphism and variability in the way that polymers are assembled. A general conclusion that emerges from these observations is that conservation of tertiary structure in proteins is much stronger than any conservation of quaternary structure. Large changes in sequence can take place with relatively small changes in tertiary structure, exemplified by actin-like proteins where a common fold can exist with unrecognizable sequence identity among many members of this superfamily (Bork et al., 1992). On the other hand, single amino acid changes can lead to large changes in quaternary structure, such as in the sickle-cell hemoglobin pathology (Eaton and Hofrichter, 1990). Polymers formed by proteins with similar folds, actin and ParM, can be very different (Orlova et al., 2007; Popp et al., 2008), so that an atomic model of one such polymer does not necessarily lead to an atomic model for the other. Electron microscopic studies have revealed this divergence of quaternary structure, but the ultimate interpretation of these results may likely require high resolution structures of subunit-subunit interfaces (Chen et al., 2008), biochemical probes, mutational analysis, and spectroscopy. Thus, the challenge of correctly fitting atomic structures of subunits into the lower resolution EM reconstructions of the higher order complexes or polymers that they form is exacerbated by the dynamics and conformational heterogeneity that can exist within these complexes.
ACKNOWLEDGMENTS
This work was supported by NIH Grant Nos. EB001567 and GM081303. The author is indebted to people in his laboratory, as well as many collaborators, for his own work that has been discussed.
References
- Ban, N, Nissen, P, Hansen, J, Moore, P B, and Steitz, T A (2000). “The complete atomic structure of the large ribosomal subunit at 2.4 A resolution.” Science 10.1126/science.289.5481.905 289, 905–920. [DOI] [PubMed] [Google Scholar]
- Beatson, S A, Minamino, T, and Pallen, M J (2006). “Variation in bacterial flagellins: from sequence to structure.” Trends Microbiol. 14, 151–155. [DOI] [PubMed] [Google Scholar]
- Birmanns, S, and Wriggers, W (2003). “Interactive fitting augmented by force-feedback and virtual reality.” J. Struct. Biol. 144, 123–131. [DOI] [PubMed] [Google Scholar]
- Birmanns, S, and Wriggers, W (2007). “Multi-resolution anchor-point registration of biomolecular assemblies and their components.” J. Struct. Biol. 157, 271–280. [DOI] [PubMed] [Google Scholar]
- Bork, P, Sander, C, and Valencia, A (1992). “An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins.” Proc. Natl. Acad. Sci. U.S.A. 89, 7290–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramham, J, Hodgkinson, J L, Smith, B O, Uhrin, D, Barlow, P N, and Winder, S J (2002). “Solution structure of the calponin CH domain and fitting to the 3D-helical reconstruction of F-actin: calponin.” Structure (London) 10, 249–258. [DOI] [PubMed] [Google Scholar]
- Chen, Z, Yang, H, and Pavletich, N P (2008). “Mechanism of homologous recombination from the RecA-ssDNA∕dsDNA structures.” Nature (London) 453, 489–494. [DOI] [PubMed] [Google Scholar]
- Connell, S R, et al. (2007). “Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors.” Mol. Cell 25, 751–764. [DOI] [PubMed] [Google Scholar]
- Craig, L, Volkmann, N, Arvai, A S, Pique, M E, Yeager, M, Egelman, E H, and Tainer, J A (2006). “Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions.” Mol. Cell 23, 651–662. [DOI] [PubMed] [Google Scholar]
- Depristo, M A, de Bakker, P I, and Blundell, T L (2004). “Heterogeneity and inaccuracy in protein structures solved by x-ray crystallography.” Structure (London) 10.1016/j.str.2004.02.031 12, 831–838. [DOI] [PubMed] [Google Scholar]
- DeRosier, D J, and Klug, A (1968). “Reconstruction of three-dimensional structures from electron micrographs.” Nature (London) 10.1038/217130a0 217, 130–134. [DOI] [PubMed] [Google Scholar]
- Dubochet, J, Adrian, M, Chang, J J, Homo, J C, Lepault, J, McDowall, A W, and Schultz, P (1988). “Cryo-electron microscopy of vitrified specimens.” Q. Rev. Biophys. 21, 129–228. [DOI] [PubMed] [Google Scholar]
- Eaton, W A, and Hofrichter, J (1990). “Sickle cell hemoglobin polymerization.” Adv. Protein Chem. 40, 63–279. [DOI] [PubMed] [Google Scholar]
- Egelman, E H (2000). “A robust algorithm for the reconstruction of helical filaments using single-particle methods.” Ultramicroscopy 10.1016/S0304-3991(00)00062-0 85, 225–234. [DOI] [PubMed] [Google Scholar]
- Egelman, E H (2007). “The iterative helical real space reconstruction method: surmounting the problems posed by real polymers.” J. Struct. Biol. 10.1016/j.jsb.2006.05.015 157, 83–94. [DOI] [PubMed] [Google Scholar]
- Egelman, E H, and DeRosier, D J (1991). “Angular disorder in actin: is it consistent with general principles of protein structure?” J. Mol. Biol. 217, 405–408. [DOI] [PubMed] [Google Scholar]
- Egelman, E H, Francis, N, and DeRosier, D J (1982). “F-actin is a helix with a random variable twist.” Nature (London) 10.1038/298131a0 298, 131–135. [DOI] [PubMed] [Google Scholar]
- Egelman, E H, Orlova, A, and McGough, A (1997). “Only one F-actin model.” Nat. Struct. Biol. 4, 683–684. [DOI] [PubMed] [Google Scholar]
- Elad, N, Clare, D K, Saibil, H R, and Orlova, E V (2008). “Detection and separation of heterogeneity in molecular complexes by statistical analysis of their two-dimensional projections.” J. Struct. Biol. 162, 108–120. [DOI] [PubMed] [Google Scholar]
- Fabiola, F, and Chapman, M S (2005). “Fitting of high-resolution structures into electron microscopy reconstruction images.” Structure (London) 13, 389–400. [DOI] [PubMed] [Google Scholar]
- Frank, J, Verschoor, A, and Boublik, M (1981). “Computer averaging of electron micrographs of 40S ribosomal subunits.” Science 214, 1353–1355. [DOI] [PubMed] [Google Scholar]
- Fu, J, Gao, H, and Frank, J (2007). “Unsupervised classification of single particles by cluster tracking in multi-dimensional space.” J. Struct. Biol. 157, 226–239. [DOI] [PubMed] [Google Scholar]
- Galkin, V E, Orlova, A, Cherepanova, O, Lebart, M C, and Egelman, E H (2008a). “High-resolution cryo-EM structure of the F-actin-fimbrin∕plastin ABD2 complex.” Proc. Natl. Acad. Sci. U.S.A. 105, 1494–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin, V E, Orlova, A, Fattoum, A, Walsh, M P, and Egelman, E H (2006a). “The CH-domain of calponin does not determine the modes of calponin binding to F-actin.” J. Mol. Biol. 359, 478–485. [DOI] [PubMed] [Google Scholar]
- Galkin, V E, Orlova, A, Lukoyanova, N, Wriggers, W, and Egelman, E H (2001). “Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-Actin subunits.” J. Mol. Biol. 153, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin, V E, Orlova, A, VanLoock, M S, and Egelman, E H (2003).“Do the utrophin tandem calponin homology domains bind F-actin in a compact or extended conformation?” J. Mol. Biol. 331, 967–972. [DOI] [PubMed] [Google Scholar]
- Galkin, V E, Orlova, A, VanLoock, M S, Rybakova, I N, Ervasti, J M, and Egelman, E H (2002a). “The utrophin actin-binding domain binds F-actin in two different modes: implications for the spectrin superfamily of proteins.” J. Cell Biol. 157, 243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin, V E, VanLoock, M S, Orlova, A, and Egelman, E H (2002b). “A new internal mode in F-actin helps explain the remarkable evolutionary conservation of actin’s sequence and structure.” Curr. Biol. 12, 570–575. [DOI] [PubMed] [Google Scholar]
- Galkin, V E, Wu, Y, Zhang, X P, Qian, X, He, Y, Yu, X, Heyer, W D, Luo, Y, and Egelman, E H (2006b). “The Rad51∕RadA N-terminal domain activates nucleoprotein filament ATPase activity.” Structure (London) 14, 983–992. [DOI] [PubMed] [Google Scholar]
- Galkin, V E, Yu, X, Bielnicki, J, Heuser, J, Ewing, C P, Guerry, P, and Egelman, E H (2008b). “Divergence of quaternary structures among bacterial flagellar filaments.” Science 320, 382–385. [DOI] [PubMed] [Google Scholar]
- Gao, H, Valle, M, Ehrenberg, M, and Frank, J (2004). “Dynamics of EF-G interaction with the ribosome explored by classification of a heterogeneous cryo-EM dataset.” J. Struct. Biol. 147, 283–290. [DOI] [PubMed] [Google Scholar]
- Gimona, M, and Mital, R (1998). “The single CH domain of calponin is neither sufficient nor necessary for F-actin binding.” J. Cell. Sci. 111, 1813–1821. [DOI] [PubMed] [Google Scholar]
- Goldsmith, S C, Pokala, N, Shen, W, Fedorov, A A, Matsudaira, P, and Almo, S C (1997). “The structure of an actin-crosslinking domain from human fimbrin.” Nat. Struct. Biol. 4, 708–712. [DOI] [PubMed] [Google Scholar]
- Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S C, and Walz, T (2005). “Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.” Nature (London) 10.1038/nature04321 438, 633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanein, D, Volkmann, N, Goldsmith, S, Michon, A M, Lehman, W, Craig, R, DeRosier, D, Almo, S, and Matsudaira, P (1998). “An atomic model of fimbrin binding to F-actin and its implications for filament crosslinking and regulation.” Nat. Struct. Biol. 5, 787–792. [DOI] [PubMed] [Google Scholar]
- Holmes, K C, Popp, D, Gebhard, W, and Kabsch, W (1990). “Atomic model of the actin filament.” Nature (London) 10.1038/347044a0 347, 44–49. [DOI] [PubMed] [Google Scholar]
- Jimenez, J L, Guijarro, J I, Orlova, E, Zurdo, J, Dobson, C M, Sunde, M, and Saibil, H R (1999). “Cryo-electron microscopy structure of an SH3 amyloid fibril and model of the molecular packing.” EMBO J. 10.1093/emboj/18.4.815 18, 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch, W, Mannherz, H G, Suck, D, Pai, E F, and Holmes, K C (1990). “Atomic structure of the actin: DNase I complex.” Nature (London) 10.1038/347037a0 347, 37–44. [DOI] [PubMed] [Google Scholar]
- Klein, M G, Shi, W, Ramagopal, U, Tseng, Y, Wirtz, D, Kovar, D R, Staiger, C J, and Almo, S C (2004). “Structure of the actin crosslinking core of fimbrin.” Structure (London) 12, 999–1013. [DOI] [PubMed] [Google Scholar]
- Laurberg, M, Asahara, H, Korostelev, A, Zhu, J, Trakhanov, S, and Noller, H F (2008). “Structural basis for translation termination on the 70S ribosome.” Nature (London) 454, 852–857. [DOI] [PubMed] [Google Scholar]
- Lehman, W, Craig, R, Kendrick-Jones, J, and Sutherland-Smith, A J (2004). “An open or closed case for the conformation of calponin homology domains on F-actin?” J. Muscle Res. Cell Motil. 25, 351–358. [DOI] [PubMed] [Google Scholar]
- Li, H, DeRosier, D J, Nicholson, W V, Nogales, E, and Downing, K H (2002). “Microtubule structure at 8 A resolution.” Structure (London) 10.1016/S0969-2126(02)00827-4 10, 1317–1328. [DOI] [PubMed] [Google Scholar]
- Matsudaira, P (1991). “Modular organization of actin crosslinking proteins.” TIBS 16, 87–92. [DOI] [PubMed] [Google Scholar]
- Miyazawa, A, Fujiyoshi, Y, and Unwin, N (2003). “Structure and gating mechanism of the acetylcholine receptor pore.” Nature (London) 10.1038/nature01748 424, 949–955. [DOI] [PubMed] [Google Scholar]
- Moller-Jensen, J, Jensen, R B, Lowe, J, and Gerdes, K (2002). “Prokaryotic DNA segregation by an actin-like filament.” EMBO J. 21, 3119–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba, K, Pattanayek, R, and Stubbs, G (1989). “Visualization of protein-nucleic acid interactions in a virus. Refined structure of intact tobacco mosaic virus at 2.9 A resolution by x-ray fiber diffraction.” J. Mol. Biol. 10.1016/0022-2836(89)90391-4 208, 307–325. [DOI] [PubMed] [Google Scholar]
- Namba, K, and Stubbs, G (1986). “Structure of tobacco mosaic virus at 3.6 A resolution: implications for assembly.” Science 10.1126/science.3952490 231, 1401–1406. [DOI] [PubMed] [Google Scholar]
- Orlova, A, and Egelman, E H (1993). “A conformational change in the actin subunit can change the flexibility of the actin filament.” J. Mol. Biol. 10.1006/jmbi.1993.1393 232, 334–341. [DOI] [PubMed] [Google Scholar]
- Orlova, A, and Egelman, E H (2000). “F-actin retains a memory of angular order.” Biophys. J. 78, 2180–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova, A, Garner, E C, Galkin, V E, Heuser, J, Mullins, R D, and Egelman, E H (2007). “The structure of bacterial ParM filaments.” Nat. Struct. Mol. Biol. 14, 921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova, A, Prochniewicz, E, and Egelman, E H (1995). “Structural dynamics of F-actin. II. Co-operativity in structural transitions.” J. Mol. Biol. 245, 598–607. [DOI] [PubMed] [Google Scholar]
- Penczek, P A, Frank, J, and Spahn, C M (2006a). “A method of focused classification, based on the bootstrap 3D variance analysis, and its application to EF-G-dependent translocation.” J. Struct. Biol. 154, 184–194. [DOI] [PubMed] [Google Scholar]
- Penczek, P A, Yang, C, Frank, J, and Spahn, C M (2006b). “Estimation of variance in single-particle reconstruction using the bootstrap technique.” J. Struct. Biol. 154, 168–183. [DOI] [PubMed] [Google Scholar]
- Popp, D, et al. (2008). “Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability.” EMBO J. 27, 570–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann, M G, Bernal, R, and Pletnev, S V (2001). “Combining electron microscopic with x-ray crystallographic structures.” J. Struct. Biol. 136, 190–200. [DOI] [PubMed] [Google Scholar]
- Rossmann, M G, Morais, M C, Leiman, P G, and Zhang, W (2005). “Combining x-ray crystallography and electron microscopy.” Structure (London) 13, 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse, C, Chen, J Z, Coureux, P D, Stroupe, M E, Fandrich, M, and Grigorieff, N (2007). “High-resolution electron microscopy of helical specimens: a fresh look at tobacco mosaic virus.” J. Mol. Biol. 371, 812–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse, C, Fandrich, M, and Grigorieff, N (2008). “Paired beta-sheet structure of an Abeta(1-40) amyloid fibril revealed by electron microscopy.” Proc. Natl. Acad. Sci. U.S.A. 105, 7462–7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres, S H, Gao, H, Valle, M, Herman, G T, Eggermont, P P, Frank, J, and Carazo, J M (2007). “Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization.” Nat. Methods 4, 27–29. [DOI] [PubMed] [Google Scholar]
- Schmid, M F, Sherman, M B, Matsudaira, P, and Chiu, W (2004). “Structure of the acrosomal bundle.” Nature (London) 10.1038/nature02881 431, 104–107. [DOI] [PubMed] [Google Scholar]
- Schutt, C E, Myslik, J C, Rozycki, M D, Goonesekere, N C W, and Lindberg, U (1993). “The structure of crystalline profilin: β-actin.” Nature (London) 10.1038/365810a0 365, 810–816. [DOI] [PubMed] [Google Scholar]
- Sutherland-Smith, A J, Moores, C A, Norwood, F L, Hatch, V, Craig, R, Kendrick-Jones, J, and Lehman, W (2003). “An atomic model for actin binding by the CH domains and spectrin-repeat modules of utrophin and dystrophin.” J. Mol. Biol. 329, 15–33. [DOI] [PubMed] [Google Scholar]
- Szabo, Z, Stahl, A O, Albers, S V, Kissinger, J C, Driessen, A J, and Pohlschroder, M (2007). “Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases.” J. Bacteriol. 189, 772–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topf, M, Lasker, K, Webb, B, Wolfson, H, Chiu, W, and Sali, A (2008). “Protein structure fitting and refinement guided by cryo-EM density.” Structure (London) 16, 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco, L G, Villa, E, Mitra, K, Frank, J, and Schulten, K (2008). “Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics.” Structure (London) 16, 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent, F, Moller-Jensen, J, Amos, L A, Gerdes, K, and Lowe, J (2002). “F-actin-like filaments formed by plasmid segregation protein ParM.” EMBO J. 21, 6935–6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann, N, and Hanein, D (2003). “Docking of atomic models into reconstructions from electron microscopy.” Methods Enzymol. 374, 204–225. [DOI] [PubMed] [Google Scholar]
- Wang, Y A, Yu, X, Ng, S YM, Jarrell, K F, and Egelman, E H (2008). “The structure of an archaeal pilus.” J. Mol. Biol. 381, 456–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y A, Yu, X, Yip, C, Strynadka, N C, and Egelman, E H (2006). “Structural polymorphism in bacterial EspA filaments revealed by cryo-EM and an improved approach to helical reconstruction.” Structure (London) 14, 1189–1196. [DOI] [PubMed] [Google Scholar]
- Wlodawer, A, Minor, W, Dauter, Z, and Jaskolski, M (2008). “Protein crystallography for non-crystallographers, or how to get the best (but not more) from published macromolecular structures.” FEBS J. 275, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhead, J L, Zhao, F Q, Craig, R, Egelman, E H, Alamo, L, and Padron, R (2005). “Atomic model of a myosin filament in the relaxed state.” Nature (London) 436, 1195–1199. [DOI] [PubMed] [Google Scholar]
- Yang, S, VanLoock, M S, Yu, X, and Egelman, E H (2001). “Comparison of Bacteriophage T4 UvsX and Human Rad51 filaments suggests that RecA-like polymers may have evolved independently.” J. Mol. Biol. 312, 999–1009. [DOI] [PubMed] [Google Scholar]
- Yonekura, K, Maki-Yonekura, S, and Namba, K (2003). “Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy.” Nature (London) 10.1038/nature01830 424, 643–650. [DOI] [PubMed] [Google Scholar]
- Yu, X, Jun, L, and Zhou, Z H (2008). “3.88 Å structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy.” Nature (London) 453, 415–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X, Settembre, E, Xu, C, Dormitzer, P R, Bellamy, R, Harrison, S C, and Grigorieff, N (2008). “Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction.” Proc. Natl. Acad. Sci. U.S.A. 105, 1867–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]