Abstract
This paper describes the placement of a crosslinking agent (dibromobimane) between two thiols (Cys-522 and Cys-707) of a fragment, “S1,” of the motor protein, myosin. It turns out that fastening the first anchor of the crosslinker is easy and rapid, but fastening the second anchor (Cys-522) is very temperature dependent, taking 30 min at room temperature but about a week on ice. Moreover, crystallography taken at 4°C would seem to predict that the linkage is impossible, because the span of the crosslinking agent is much less than the interthiol distance. The simplest resolution of this seeming paradox is that structural fluctuations of the protein render the linkage increasingly likely as the temperature increases. Also, measurements of the affinity of MgADP for the protein, as well as the magnetic resonance of the P-atoms of the ADP once emplaced, suggest that binding the first reagent anchor to Cys-707 initiates an influence that travels to the rather distant ADP-binding site, and it is speculated what this “path of influence” might be.
Some years ago, we reported on the possibility of emplacing a new intramolecular crosslink in myosin S1 (1, 2) by using a then newly invented reagent, dibromobimane (3). Extending this work has now led to results of interest beyond the original objective. Our early work, based on identifying N-terminal end groups, indicated that CNBr cutting generated an H-shaped fragment whose “crossbar” is a dibromobimane joining Cys-522 and Cys-707. In the present work, this identification is strengthened by amino acid sequencing from Phe-496 to Glu-509 (interrupted by Trp-510) and from Glu-687 to Leu-711, further establishing the identity of the two cited cysteines. It was noted by Burke (4) that, although he could confirm the 522–707 crosslink at room temperature, the link appeared impossible at 0°C. His observation stimulated us to study the temperature dependence of the crosslinking. As we report here, the reaction with Cys-707 is rapid and depends little on temperature, but the subsequent crosslinking with Cys-522, achievable within 30 min at 25°C, requires a week at 0°C. Crystallography (5, 6) corresponding to near 0°C (as do all current images of S1) indicates an observable interthiol separation of about 23.49 A, compared with the 8-Å length of bimane (the next Cys in the sequence is even farther, at 41.85 Å). This suggests that, after anchoring at Cys-707, the yet unreacted function of dibromobimane and Cys-522 may engage in a slow temperature-dependent search for one another or else are brought together by a structural transition not evident in 4°C images. Although measurements of the ADP-S1 affinity with native S1 conform with the classic Lowey–Luck (7) measurements on intact myosin, it is clear that the affinities are about one-third less with crosslinked S1, or with S1 reacted with monobromobimane (which reacts only Cys-707). Additionally, attaching either bimane alters the nuclear magnetic resonance arising in the P-atoms of the bound β-phosphate. These somewhat disparate observations nevertheless flow together into certain useful conclusions.
1. Establishing the Primary Sequence Environment of the Bimane-Induced Crosslink Between Cys-522 and Cys-707.
As reported (1, 2), the easily induced (at 25°C) dibromobimane crosslink joining the 50-kDa and 20-kDa domains of myosin S1 appeared to be between Cys-522 and Cys-707 of rabbit myosin S1. By using the rabbit amino acid sequence information available at the time, it was expected that after CNBr treatment, the H-shaped fluorescent dibromobimane fragment would be bounded by four ends—Phe-496 through Met-530 from the 50-kDa strand and Glu-687 through Met-778 from the 20-kDa strand. The theoretical average mass to be expected for the isolated fragment is 14,536 g⋅mol−1. The expected N-end groups were actually found (2). At position 517, the sequence gave the option of Met or Glu (presumably resulting from two isoforms). Because of the large size of the fluorescent fragment, the Glu option was strongly preferred. Very recently, K. Maeda has reported (8) a DNA-based rabbit sequence containing Met-517 but points out (personal communication) that at least one other distinct isoform exists. The present work reports sequencing results suggesting that, in our case, 517 is a Glu or a noncleavable Met.
Materials and Methods.
Crosslinking of S1 with dibromobimane was performed at 25°C for 2 hr by using 4× excess of reagent (see ref. 3 for details). The linked material, rid of excess reagent, was dialyzed against distilled water and lyophilized. Overnight, at room temperature, such material was cleaved in 70% formic acid, by using 100× excess of CNBr over methionine content. Formic acid was flushed out by N2 gas, and the residue was dissolved in 1% SDS/5% 2-mercaptoethanol, pH 8. Crosslinked fragments were separated on polyacrylamide/0.l% SDS containing 8 M urea. The fluorescent band, of about 15 kDa, derived from the crosslinked part of S1, was cut off from the remaining gel under UV light. The crosslinked fragment was eluted from the gel by electrophoresis, dialyzed against 0.1% SDS, and lyophilized. The dried sample was redissolved in 2% SDS/8 M urea/5% 2-mercaptoethanol/10% ethanol, pH 6.8. Fragments so separated were transferred to polyvinylidenedifluoride membranes. Protein bands were visualized by staining with Coomassie brilliant blue and analyzed with a Perkin–Elmer Applied Biosystems Model 473A protein sequencer, yielding (from the N-end) the sequence FVLEQEEYKKEGIEW for the strand from the 50-kDa domain and the sequence EHELVLHQLRCNGVLEGIRICRKGL for the strand from the 20-kDa domain. The presence of W-510‖ in the former sequence frustrates further sequencing along the strand and thus direct identification of position 517 as a noncleavable E or a cleavable M. However, it follows from the facts (i) dibromobimane crosslinking joins two distinct entities (50 kDa and 20 kDa); joining is signaled in electrophoresis by the appearance of a new band migrating as 105 kDa; (ii) the joined structure contains the two cited sequences; and (iii) of all the thiols of myosin S1, C-707, in the sequence from 20 kDa, is by far the most reactive and thus occupies one function of DBB, that the thiol nearest and most likely to occupy the other function of the dibromobimane crosslinker is Cys-522. Nevertheless, there are additional matters to note. As discussed in Section 2, the 4°C crystallography (8) indicates that two more distant and poorly oriented thiols, Cys-479 and Cys-540, are in the neighborhood. Were the second function of dibromobimane attached to Cys-540, i–iii could still apply, but cleavage at M-530 would have to be ignored, and the resulting expected mass of the fragment would have to be 1,270 g⋅mol−1 greater. Cys-479 had not been found in the original sequence; were it the anchor of the second function, the N-end groups of the resulting would not be those required by ii. Finally, were 517 a slowly cleavable M, it might shed a 22-residue fragment (496–517); we have sought such a fragment but have never found it. Therefore, we proceed on the assumption that dibromobimane crosslinks Cys-522 and Cys-707.
It should be noted that dibromobimane, like N-ethylmaleimide, but not like iodoacetamide, reacts Cys-707 rapidly; its reaction with Cys-522 is very specific, much more so than is that of reagents like iodoacetamide, 1,5-IAEDANS, or monobromobimane.
2. Computer Modeling Associated with Dibromobimane Crosslinking.
The pioneering crystallography of Rayment and associates (5), and more recently the studies of Cohen and associates (6), have enabled exploration of systems and problems such as our present concern. From significant homologies (9), it has become evident that findings obtained in one system can be reasonably transferred (albeit tentatively) to another evolutionarily distant system. Here we transfer S1 information obtained in chicken and Dictyostelium to rabbit, which has never been studied crystallographically. On the other hand, because of experiments to be described next (Section 3), we are circumspect about extrapolating information obtained crystallographically at low temperature, say 4°C, to structural expectations at, say, 25°C, even though for now we can do little else.
Fig. 1 gives the CA-CA distances (in a.u.) between thiols of S1 and supports the conclusions of Sec. 1 regarding location of the dibromobimane crosslink emplaced on myosin S1 in that, among these, Cys-522 is nearest to Cys-707. As shown in Fig. 2, the insertion of the bimane crosslink is accompanied by a significant change in (CA-CA) distance between Cys-522 and Cys-707; before, it is 20.93 A; after, it is 10.94 A. This reduction is accomplished by an unwinding of the helix containing Cys-522 and a modification of the helix between Cys-697 and Cys-707. In Section 4, we report that the affinity of MgADP for myosin S1 is significantly reduced when either bimane reacts with Cys-707, and in Section 5, we report that also NMR signals from bound ADP are similarly altered by either bimane. Because monobromobimane cannot crosslink (for example, to Cys-522), we conclude that the reaction of either bimane with Cys-707 exerts an effect that is in some way transmitted to bound ADP. If the effect results in the partial destabilization of bound MgADP, it may resemble the effect of “activators” of ATPase, reagents that remotely reduce the time taken by the “leaving” reaction (ejection of product). What is the “path of influence” of such an effect? The Cohen work (6) suggests that the binding of ADP causes a partial unwinding of the 697–707 helix; one may speculate that, reciprocally (Fig. 3), perturbation of the helix (or structures coupled to it) may destabilize bound ADP. In this latter direction, the path may be 707–697, to the third to fourth strands, to the P-loop, to the bound ADP. Emplacement of dibromobimane may initiate this path by changing the orientation of Cys-707 or by virtue of its interactions with the adjacent helix and loop (501–525). Alternatively, we may think of these interactions as inhibiting the unwinding of the 697–707 helix that accompanies ADP binding.
Figure 1.
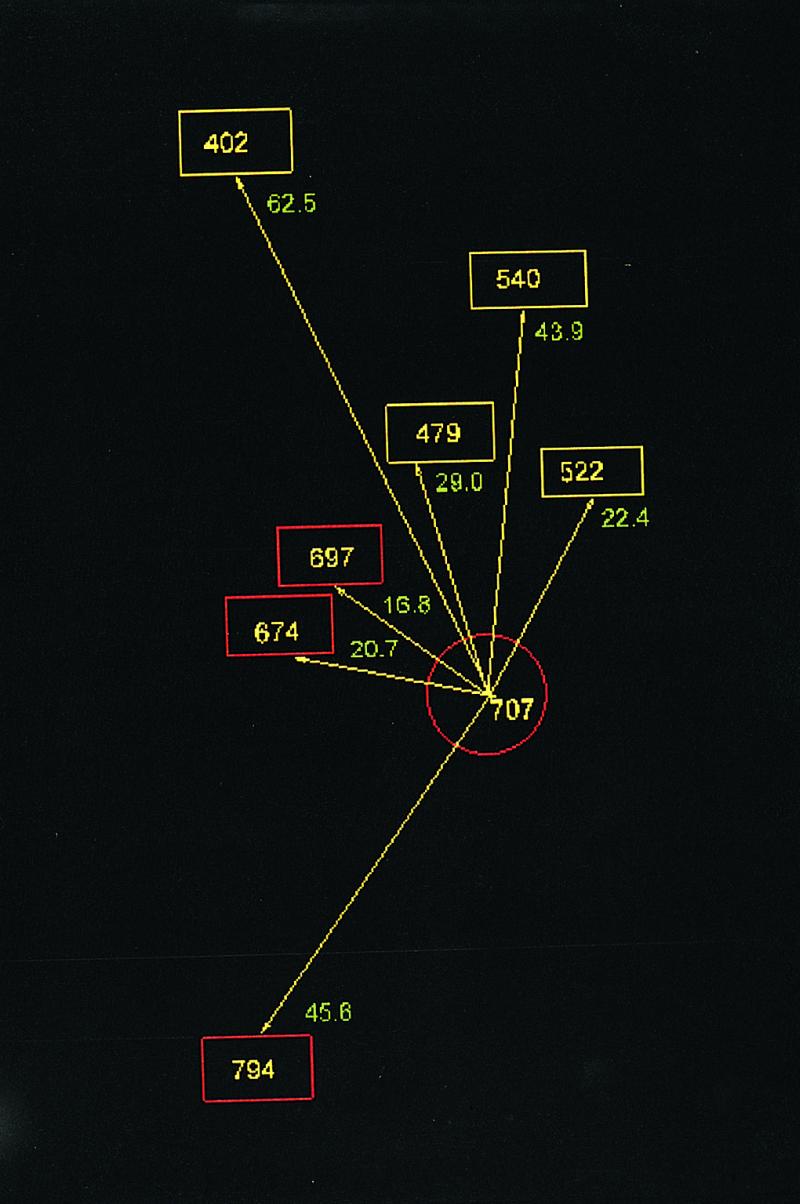
Distances between Cys-707 and other thiols in the 50 kDa domain (cyan) and the 20 kDa domain (red). Residue numeration applies to rabbit muscle (based on ref. 5 and PDP code 2 mys).
Figure 2.
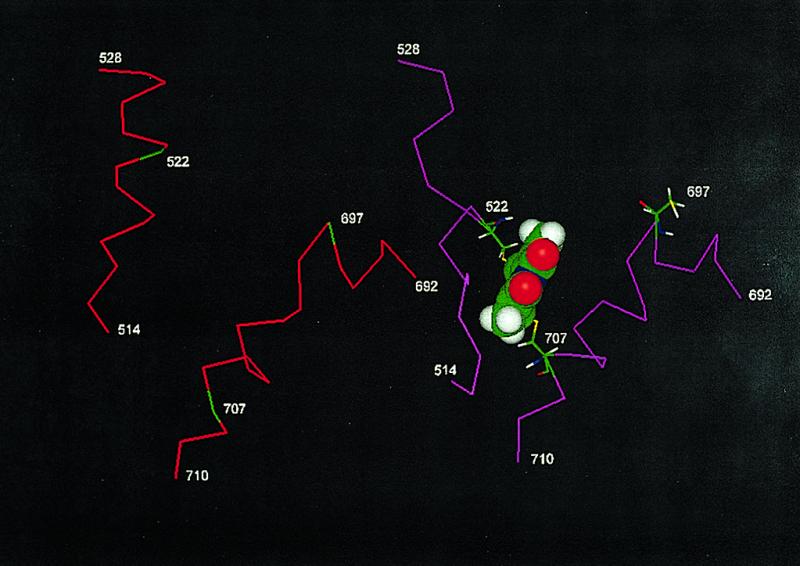
Local conformation (expressed as CA-CA distances) before (red) and after (magenta) a dibromobimane crosslink (in CPK style) is emplaced. This image was constructed from ref. 10, by eliminating the ADP.BeFx ligand of Dictyostellium and replacing Thr-513 and Thr-688 with their homologs in rabbit, viz. Cys-522 and Cys-707.
Figure 3.
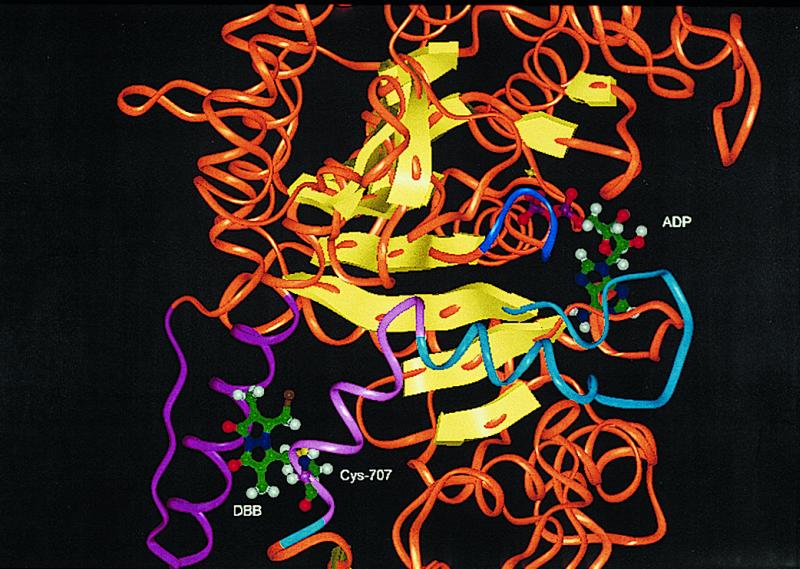
Environment of bound ADP and emplaced dibromobimane. Except for select, otherwise specified, portions, the “trace” is rendered as an orange “oval,” and the β-strands as yellow arrows. The helix that connects Cys-707 and Cys-697 is in light magenta; it connects via a helix and loop (both cyan) to the third β-strand of the β-sheet. The “P-loop” (blue) of the ADP binding site connects to the fourth strand of this β-sheet. The emplaced dibromobimane, shown as attached only to Cys-707, interacts with a helix and loop (both magenta) by steric and hydrophobic contacts. This picture was created from PDB code, 1 mmd, as MMD.ADP.BeFx. ADP placement was obtained by superimposition of the trace of MMD.ADP.BeFx on the trace of MMD.DBB. Sequence numbers are those of rabbit myosin.
3. Kinetics and Thermal Dependence of Dibromobimane Crosslinking.
As already remarked, Burke's observation prompted us to examine why the crosslink of interest seemed impossible at 0°C. From a kinetic study of crosslinking at different temperatures, it proved reasonable to assume as principal chemical transformations (dibromobimane symbolized as Br_Br) to be
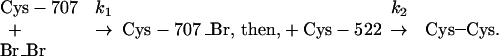 |
From the experimental behavior, we found it justifiable to simplify the system mathematically to
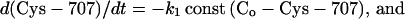 |
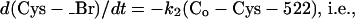 |
we assume that dibromobimane is in excess over either thiol (Co), and the reaction of Cys-707 is essentially complete before the reaction of Cys-522 has begun.
Materials and Methods.
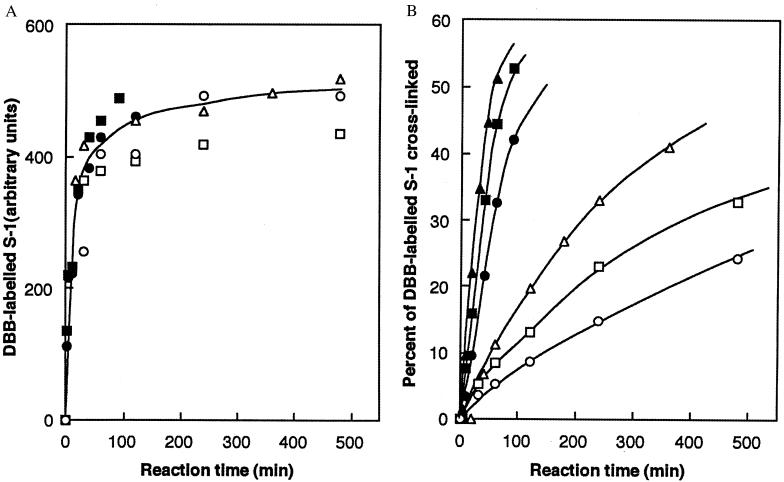
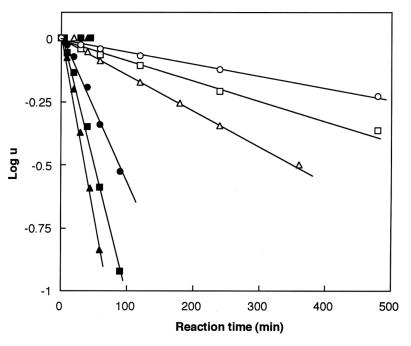
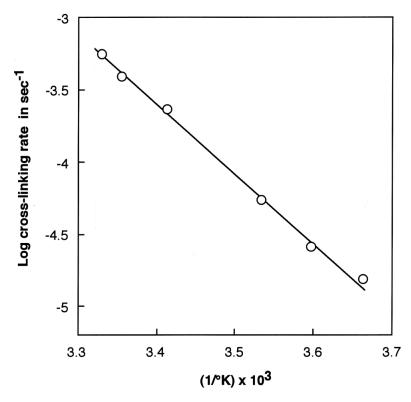
S1 (0.5 mg/ml) was incubated in the dark for various times by using a 4× molar excess of dibromobimane. At desired times, aliquots were removed and quenched with 20 mM 2-mercaptoethanol. Each aliquot was divided into two parts. That for measuring the extent of labeling with dibromobimane was precipitated in trichloroacetate (to a final 5%). After a 15-min centrifugation at 3,500 × g and repeated washing with acetone to remove unreacted dibromobimane, the pellet was redissolved with shaking in 2 ml of 2% sodium dodecylsulfate and 8 M urea buffered by 20 mM Tris, pH 7.5. The intensity of fluorescence of such an aliquot (excitation at 397 nm, emission at 463 nm) measured the dibromobimane bound to S1. That for measuring crosslinking extent was subjected to electrophoresis in 12.5% polyacrylamide gel containing 0.1% sodium dodecylsulfate; the intensity of the band migrating as 105 kDa measured the crosslinking extent. An important empiricism that we have experienced is that the extent of crosslinking of myosin S1 at all temperatures and in all preparations is incomplete and asymptotic to a level of about 60%. Fig. 4 A–B illustrates the kinetic behavior leading us to the simplifications cited above, and Fig. 5 illustrates the first-order behavior of the crosslinking reaction as a function of time. Fig. 6, log k2 vs. 1/T, yields the “heat of activation” (as typically cited from the theory of absolute reaction rates) viz. ca. 8.4 kcal; no “break” in the vicinity of 15°C was found in this plot. Two significant findings emerge from these results: (i) the reaction of Cys-707 with dibromobimane has virtually no temperature dependence, and (ii) the 707–522 crosslinking reaction is extremely slow; the same (limited) extent of crosslinking is achieved at all reactions tested, however.
Figure 4.
(A) Fluorescence of S1 (in arbitrary units) acquired by reaction with dibromobimane, depicted as a function of time. The different symbols represent measurements made at various temperatures: open circles, 0°C; open squares, 5°C; open triangles, 10°C; closed circles, 20°C; closed squares, 25°C; closed triangles, 30°C. (B) Percent of dibromobimane-labeled S1 crosslinked (measured by the amount of 105-kDa band that forms on union of the 20-kDa and 50-kDa domains), depicted as a function of time, to show the first-order nature of the reaction. Symbols are as in A.
Figure 5.
Logarithm of the as-yet-uncrosslinked fraction of S1, u, as a function of time. Experimental measurements indicated that the maximum fraction of S1 that could be crosslinked was approximately 0.6. Symbols are as in Fig. 4A; rates are estimated from Fig. 4B.
Figure 6.
Rate of crosslinking S1 as a function of reciprocal absolute temperature. These data yield a heat of activation of ca. 8.5 kcal.
4. Equilibrium Dialysis of the MgADP-S1, MgADP-S1.Dibromobimane, and MgADP.Monobromobimane Systems.
Originally for the purpose of setting conditions for NMR experiments, subsequently also for strategic reasons, we undertook to measure the equilibrium constant for binding of MgADP to various systems at 0°C under the conditions we use in NMR studies (the only temperature at which it is feasible to apply the method to S1). The method that we used for these measurements is a variant of the original method (7) and assumes the ADP-S1 1:1 stoichiometry now well established by Lowey and others. As a membrane permeable to MgADP but not to S1 separates compartments ′ and ", it is possible to assert conservations, Ao = v′[A] + v′[AM] + v"[A]" and Mo = v"[M] + v"[AM], and the Mass Law, [AM]/K[A][M], where A stands for MgADP, M for S1, K is in liter·mol−1, subscripted symbols are in mol·liter−1, v, v", and V = v′ + v" are in liters. From the foregoing equations, it follows that after the binding equilibrium is established,
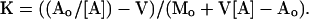 |
All variables on the right-hand side are directly measurable, so K is determined. The binding constant measurements for native S1 were obtained by essentially the same technique used by Lowey and Luck with myosin but were not intended to reexamine their measurements (for example, our solvent conditions necessarily differed from theirs); however, it is evident that there is a quantitative similarity between the two results as regards the unmodified (“native”) samples.
Materials and Methods.
Because equilibrium dialysis experiments are long lasting (about 36 hr), we pretreated our proteins to eliminate trace contaminants (adenylate kinase, 5′-adenylic acid deaminase) by successive DEAE chromatography, passage through cellulose phosphate, eluted into 75 mM imidazole + 250 mM KCl, pH 7.0, and precipitation by 60% saturation ammonium sulfate (see ref. 9). After ammonium sulfate treatment, proteins were desalted by using 2.6 cm × 40 cm Sephadex G-25 columns equilibrated in the solvent (71 mM Pipes/143 mM KCl/7 mM MgCl2/0.14 mM EDTA/2 mM NaN3, pH 7.0); subsequently, D2O was added, because it was also in NMR experiments (see below). In preliminary tests, proteins thus purified did not show significant conversion of ADP to AMP and IMP after 3-day incubations. The concentrations of proteins (as measured by the Bradford method) routinely used were 30–50 μM, and the corresponding MgADP was in ca. 1.5 molar excess. Scienceware/Bel-Art 1-ml cells were used. Spectra/Por (Spectrum Laboratories, Laguna Hills, CA) membrane discs with a cutoff above 12–14 kDa were pretreated in 10 mM EDTA for 30 min at 60°C before use. A membrane was placed between each pair of half cells to separate compartments ′ and ", which were then filled initially with “protein-only” and “ADP-only” solutions, now containing also D2O to a final concentration of 30%; stirring magnetic minibars were included in each half cell before the assembly was sealed. Of the variables constituting K, [A] was measured spectrophotometrically; Mo, Ao, v′, and v" were measured gravimetrically on a microbalance after ascertaining the appropriate densities of the fluids micropipetted into the system. We believe the aggregate accuracy of these manipulations to be within 3% of the cited values. Multiple cells were incubated at 0°C for 36 hr with gentle stirring. No visible aggregation of proteins was observed. After dialysis, the cells were disassembled in a cold room, and samples from v" were drawn for measurement in a spectrophotometer at 259 nm, assuming a molar extinction coefficient of 15.4 × 103 cm−1. Absorbances at 248.5 and 280 nm were also remeasured to check for protein leakages or conversions from ADP to AMP and IMP.
Table 1 summarizes our results, which have been used to set some of the conditions in Section 5. Additionally, however, comparison of the K values obtained with dibromobimane- and monobromobimane-modified proteins suggests that they are about the same. Because monobromobimane cannot crosslink, this finding implies that the agency of change of binding constant, and perhaps of NMR signal (see below), is not crosslinking per se, but is probably a structural change induced in the Cys-707 region and transmitted to the binding site of MgADP in S1.
Table 1.
Measurements of the affinity of MgADP for S1 at 0°C, in liter/mol × 105, for native S1 (N), S1 modified with dibromobimane (D), and monobromobimane (MBB)
| Measurement | Mean ± SD |
|---|---|
| N (n = 11): 1.65, 0.41, 0.21, 1.36, 0.69, 0.20, 0.61, 0.64, 0.19, 0.93, 0.26 | 0.64 ± 0.47 |
| D (n = 7): 0.51, 0.16, 0.09, 0.25, 0.15, 0.18, 0.16 | 0.22 ± 0.13 |
| M (n = 4): 0.11, 0.26, 0.28, 0.11 | 0.19 ± 0.07 |
The solvent is 50 mM Pipes/100 mM KCl/5 mM MgCl2/0.1 mM EDTA/0.1 mM EGTA/2 mM NaN3, in 30% D2O, pH 7.09 (at 0°C before adding D2O).
5. On the Phosphorus Resonance of β-Phosphate of S1-Bound MgADP.
Shriver and Sykes (11, 12) introduced the phosphorus resonance of S1-bound MgADP as a probe of the ADP-S1 relation, and the relation was reexamined more recently by Tanokura and Ebashi (13). In both reports, the temperature dependence of the signal was studied. To our present application, however, the relevant temperature is 0°C. Unfortunately, the previous measurements at this temperature differ from one another and from what we have found. We believe, however, that our results yield a consistent view the ADP-S1 relation in different chemical derivatizations of S1.
Materials and Methods.
31P measurements were made at the NMR Center of the Indiana University—Purdue University at Indianapolis (IUPUI) Department of Physics, at 121.4 MHz, by using a Varian UNITY-300 NMR spectrometer equipped with a high-stability variable temperature controller and a 10-mm broadband probe. Typical sample conditions (intended to resemble refs. 1–3) were myosin S1, 0.7 mM/ADP, 1 mM/Ap5A, 0.2–0.3 mM/K-Pipes, 50 mM/KCl, 100 mM/MgCl2, 5 mM/EDTA, 0.1 mM/EGTA, 0.1 mM/NaN3, 1.4 mM/D2O, 30%, pH 7.0 (before D2O). A 1.6-mm (i.d.) capillary containing phenylphosphonate, 4 mM, and MnCl2, 0.4 mM, in D2O, pH 7.0, served as external reference. The parameters for each spectrum were: pulse width, 700; number of data points, 8,192; sweep width, 7,500 Hz; recycle delay, 1.6 s; number of scans, 28,000–32,000; line broadening, 20 Hz. In this report, the temperature was 0°C.
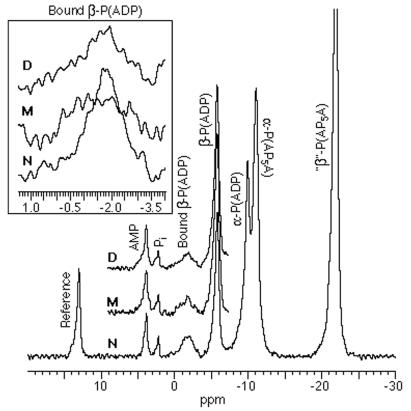
Fig. 7 presents the NMR observations pertinent to this paper. The central figure shows the 31P-spectra of ADP interacting with myosin S1 (N, D, and M) in the presence of a reference substance, an adenylate kinase inhibitor, and Mg2+ ions at 0°C, pH 7. In each myosin S1, we are especially concerned with a peak at about −2 ppm generated by the β-phosphate atoms of bound ADP and a peak at about −5 ppm generated by the same atoms of unbound ADP. Inset for each myosin S1 isolates and enlarges the −2-ppm peak. In N, we never found the broad but clear −2 ppm peak (cf. ref. 13) to have a doublet structure (cf. refs. 11, 12) but rather found it to have the symmetrical shape shown in the Inset. On the other hand, the shapes and areas of the peak are clearly different in D and M from what they are in N, suggesting that these differences mirror differences in S1-ADP affinities (Section 4). When the areas of the peaks are obtained by integration, we find that the “bound” area of N is about 20% greater than the “bound” area of either D or M; correspondingly, the “unbound” area of N is about 20% less than that of the “unbound” area of either D or M; moreover, the areas of D and M are negligibly different from one another. These results imply that we are properly accounting for all ADP. However, the agreement between the ratios of areas measured by NMR and the same ratios calculated from the equilibrium dialysis experiments (Section 4) is only semiquantitative (20% vs. 10%), probably because of the difference in the concentrations (one to two orders of magnitude higher in the NMR work) used in the two techniques.** Nevertheless, the NMR data, like the equilibrium dialysis data, indicate that the affinity of ADP for S1 is equally reduced when S1 is treated with either MBB (not crosslinked) or with DBB (crosslinked).
Figure 7.
The 31P chemical shift spectra of MgADP interacting with myosin S1 (in various states of derivatization) in the presence of a reference substance and an adenylate kinase inhibitor at 0°C, pH 7; N, native; M, derivatized with monobromobimane; D, derivatized with dibromobimane. (Inset) The −2-ppm peaks have been isolated and enlarged to show changes resulting from derivatization. Note that N shows no doublet structure but clearly generates a peak (see refs. 11–13).
Conclusions and Discussion
Together with earlier work by us (1, 2) and by Morris Burke (4), the results of Section 1 can be taken to strengthen the conclusion that when the fluorescent crosslinker dibromobimane (3) reacts with myosin S1, it quickly reacts with Cys-707 and then relatively slowly with Cys-522 to form the crosslink. Emplacing such an easy-to-find crosslink may be useful in future applications, but two incidental results may have deeper meaning for understanding myosin. One is that the beautiful results provided by crystallography at one temperature (e.g., 4°C) must be combined with knowledge about thermal fluctuations to obtain an effective or “average” structure at, say, physiological temperature. As discussed in Section 2, connecting points 23 A apart by using a span of only 8 A seems impossible. But the experiments of Section 3 show that it is possible and that the probability of doing so goes up with temperature in a very understandable way with a characteristic (8.4 kcal) that is very common. The second result to note is “crosstalk” between sites. It is well known that modification of Cys-707 affects ATPase.‡‡ Section 4 reports changes in affinity for ADP when a bimane attaches to Cys-707 and Section 5 reports how a probe, the resonance of β-phosphate atoms on ADP directly, “feels” the attachment. In Section 3, we speculate about the likely “path of influence” whereby Cys-707 communicates with the environment of bound ADP. This second result is an effort to get at the nature of “communication” or “allostery” in myosin, that is, to understand such effects in quasimechanical terms. Neither of our central results is totally new; for example, the significant role of fluctuations in generating the “effective structure” of proteins has been outlined by Cooper (14), and we and others have imagined “paths of influence” (15) in theorizing about chemomechanical transduction, but we hope that specific results, as in the present work, help to “flesh out” the more general ideas.
Acknowledgments
At various times in this work the authors benefited from the scientific counsel of Prof. Thomas James of the University of California San Francisco (UCSF), Prof. Nageswara Rao of Indiana University–Purdue University at Indianapolis, Prof. Morris Burke of Case-Western Reserve University, and Prof. John Shriver of the Southern Illinois University. The authors are grateful for National Science Foundation Grant MCB-9603670 to M.F.M., H.M.H., K.U., and M.K. M.F.M. is much indebted as well for a grant from the UCSF Academic Senate, and for the continued support and encouragement of the University of the Pacific Dental School, and of Prof. Regis Kelly of UCSF.
Footnotes
Tryptophan fluorescence (excitation at 293 nm; emission at 340 nm) at the appropriate cycle was confirmed.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.030523997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.030523997
The concentration difference may generate discrepancies in two ways. There is a difficulty in the measurement of the NMR peak areas, and also there is a difficulty in attempting to account for hydrolytic losses of ADP because of inadvertent catalytic contaminants existing at relatively high concentration under NMR conditions.
The specific effects, however, are diverse. For example, the classical Cys-707 reagents, iodoacetamide, 1,5-IAEDANS, etc., “activate” ATPase, presumably by destabilizing the bound ADP product. Reagents of small bulk such as methyl methane sulfonate or reagents that are “improperly oriented,” such as other isomers of IAEDANS, have little effect. Certain reagents, such as emplaced bimane, presumably interfere with early steps, e.g., with the conversion of M·ATP to M·ADP·Pi and therefore “inhibit” ATPase.
References
- 1.Mornet D, Ue K, Morales M F. Proc Natl Acad Sci USA. 1985;81:1658–1662. doi: 10.1073/pnas.82.6.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ue K. Biochemistry. 1987;26:1889–1804. doi: 10.1021/bi00381a016. [DOI] [PubMed] [Google Scholar]
- 3.Kosower E M, Kosower N S. Methods Enzymol. 1995;12:133–166. doi: 10.1016/0076-6879(95)51117-2. [DOI] [PubMed] [Google Scholar]
- 4.Agarwal R, Burke M. Arch Biochem Biophys. 1991;290:1–6. doi: 10.1016/0003-9861(91)90583-5. [DOI] [PubMed] [Google Scholar]
- 5.Rayment I, Rypnewski W R, Schmidt-Base K, Smith R, Tomchik D R, Benning M, Winkelmann D, Wesenberg G, Holden H M. Science. 1993;261:50–59. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 6.Houdusse A, Kalabokis V N, Himmel D, Szent-Gyorgyi A G, Cohen C. Cell. 1999;97:459. doi: 10.1016/s0092-8674(00)80756-4. [DOI] [PubMed] [Google Scholar]
- 7.Lowey S, Luck S M. Biochemistry. 1969;8:3195–3199. doi: 10.1021/bi00836a010. [DOI] [PubMed] [Google Scholar]
- 8.Maeda, K., Hostinova, E., Roesch-Kleinhauf, A., Schuster, R., Gasperik, J. & Wittinghofer, A. (1995) National Center for Biotechnology Information, Accession No. U32574.
- 9.Warrick H M, Spudich J A. Annu Rev Cell Biol. 1987;3:379–421. doi: 10.1146/annurev.cb.03.110187.002115. [DOI] [PubMed] [Google Scholar]
- 10.Fisher A J, Smith C A, Thoden J B, Smith R, Sutoh K, Holden H M, Rayment I. Biochemistry. 1995;34:8960–8972. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- 11.Shriver J W, Sykes B D. Biochemistry. 1981;20:2004–2012. doi: 10.1021/bi00510a041. [DOI] [PubMed] [Google Scholar]
- 12.Shriver J W, Sykes B D. Biochemistry. 1981;20:6357–6362. doi: 10.1021/bi00525a011. [DOI] [PubMed] [Google Scholar]
- 13.Tanokura M, Ebashi S. J Biochemistry. 1993;113:19–21. doi: 10.1093/oxfordjournals.jbchem.a123996. [DOI] [PubMed] [Google Scholar]
- 14.Cooper A. Prog Biophys Mol Biol. 1984;44:181–214. doi: 10.1016/0079-6107(84)90008-7. [DOI] [PubMed] [Google Scholar]
- 15.Botts J, Takashi R, Torgerson P, Hozumi T, Muhlrad A, Mornet D, Morales M F. Proc Natl Acad Sci USA. 1984;81:2060–2064. doi: 10.1073/pnas.81.7.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]