Abstract
Individuals with partial HSA21 trisomies and mice with partial MMU16 trisomies containing an extra copy of the DYRK1A gene present various alterations in brain morphogenesis. They present also learning impairments modeling those encountered in Down syndrome. Previous MRI and histological analyses of a transgenic mice generated using a human YAC construct that contains five genes including DYRK1A reveal that DYRK1A is involved, during development, in the control of brain volume and cell density of specific brain regions. Gene dosage correction induces a rescue of the brain volume alterations. DYRK1A is also involved in the control of synaptic plasticity and memory consolidation. Increased gene dosage results in brain morphogenesis defects, low BDNF levels and mnemonic deficits in these mice. Epigallocatechin gallate (EGCG) — a member of a natural polyphenols family, found in great amount in green tea leaves — is a specific and safe DYRK1A inhibitor. We maintained control and transgenic mice overexpressing DYRK1A on two different polyphenol-based diets, from gestation to adulthood. The major features of the transgenic phenotype were rescued in these mice.
Introduction
Phenotype mapping, based upon comparisons between genotypes and phenotypes of partial trisomy 21, has revealed that some regions of HSA21 may contain genes involved in specific phenotypes characteristic of Down syndrome (DS) including mental retardation. One such region, DCR-1 [1], [2], contains 19 genes, one of which — DYRK1A [Dual specificity Tyrosine(Y) Regulated Kinase 1A] — is closely associated with Down syndrome phenotypes. A recent study describes a mother and two children presenting a facial phenotype characteristic of DS and with moderate mental retardation. These individuals carry a small duplication of 10 genes including DYRK1A, consistent with a role for DYRK1A as a candidate gene in Down syndrome [3]. DYRK1A is a mammalian ortholog of minibrain in drosophila [4], a gene which is essential for normal postembryonic neurogenesis [5]; as its name implies, the DYRK1A enzyme has dual substrate specificities: autophosphorylation for self activation takes place on the tyrosine-321 residue in the active loop of the catalytic domain [6] and target protein phosphorylation occurs on serine/threonine residues. Many targets have been identified in vitro including FKHR, dynamin1, amphiphysin and tau protein [7], [8], [9]. These findings suggest that DYRK1A is a major player in both cell cycle regulation and synaptic plasticity. DYRK1A levels in the brains of DS subjects with free trisomy were found approximately 1.5-fold higher than those in normal subjects indicating that this protein is overproduced in a gene dosage-dependent manner in Down syndrome [10].
Murine models with partial MMU16 trisomies such as Ts65Dn, Ts1Cje or human HSA21 — all carrying extra copies of several genes, including the DYRK1A gene — have been generated. These models present morphogenesis defects in the cranium and brain [11], [12], together with learning and memory defects [13], detectable in such paradigms as the Morris water maze [14] or object recognition [15] tests. Mice carrying a smaller duplication with 33 genes, encompassing the gene encoding DYRK1A, present brain alterations but do not display abnormal behavior in the Morris water maze. However, deletion of the same region in a model with partial MMU16 trisomy, Ts65Dn, corrects the cognitive deficits seen in the Ts65Dn mice [16]. These results strongly suggest that duplication of genes from this region is necessary to produce the learning impairment seen in the Ts65Dn model of Down syndrome. Transgenic mice have also been developed using a yeast artificial chromosome from this region (YAC 152F7). cDNA mapping experiments [17] and human genome sequencing [18] showed that YAC152F7 contains five genes: PIGP, TTC3, DSCR9, DSCR3 and DYRK1A. This murine model presents both brain abnormalities and learning impairments [19], [20], [21]. On the contrary, transgenic mice for the YAC 141G6 bearing extra copies of all genes included in YAC 152F7 except for DYRK1A did not display any brain or behavioural alterations. Similar phenotypic alterations have been obtained in mice transgenic for a human BAC [22] carrying only human DYRK1A and with a murine BAC clone carrying only murine Dyrk1a (data not shown). In a previous study, using regional MRI, we found that morphological alterations throughout the brain in the YAC tg152F7 were not uniform: the total brain volume was 14% greater in transgenic mice than in wild-type mice, with an effect 2,5 greater (25%/10%) in the ventral region (including the thalamic-hypothalamic region) than in the cortex (10% greater volume) [23]. Unbiased stereological cell counts of NeuN-positive neurons revealed a greater cell density in the thalamic-hypothalamic area and a lower cell density in the somatosensory cortex in transgenic mice than in wild-type mice. These results indicate that the phenotypic effects of overexpressing the DYRK1A gene in the brain differ in a region-specific manner. These morphological alterations are inverse with those described in a model with heterozygous deletion of Dyrk1a [24] (including a smaller brain volume than in wild-type mice, particularly in the ventral region). The invalidated heterozygous model also displays learning impairments and an altered pyramidal cell phenotype [25].
We investigated the possibility of correcting these phenotypes through modulation of DYRK1A activity. Bain and colleagues described the properties of 30 inhibitors, tested with 25 different kinases: one of the most specific inhibitors of DYRK1A was epigallocatechin gallate, with an apparent IC50 of 0.33 µM [26]. DYRK1A inhibition by EGCG was also demonstrated in NIH3T3 cells and a mutation analysis indicated a mechanism involving a non competitive inhibition [27]. EGCG can cross the blood-brain barrier [28] and the placental barrier [29]. EGCG is the major catechin in green tea leaves (40 to 50% of the total catechins amount). Indeed, feeding a green tea drink to mice (usually drinking 3–5 ml/day) is equivalent to administering 0.6 mg/day pure EGCG, which is effective in the treatment of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced Parkinson's disease (MPTP), through its effects on dopaminergic neurons [30].
We maintained mice on two polyphenol-based diets from gestation to adulthood. Using the phenotypic tools that we have previously designed we compared the effects of these diets on batches of control and transgenic animals.
Results
Link between the Dyrk1a gene copy number and the brain phenotypes
We generated double transgenic mice by crossing YACtg152F7 with Dyrk1a (+/−) mice. Resultant double transgenic mice had normal levels of DYRK1A mRNA in the brain (Fig 1A) and a corrected body weight (data not shown); MRI analyses revealed a corrected brain volume and a corrected regional morphology (fig 1B). Figure 1C shows a linear regression analysis of the relationship between brain volume and Dyrk1a gene copy number.
Figure 1. Phenotypic correction by genotype correction of DYRK1A copy number.
Wild type (WT), YACtg152F7 transgenic (TG), dyrk1a (+/−) (HT) and double transgenics (TGxHT) generated by three different crossings. A: brain DYRK1A mRNA levels determined by quantitative PCR; B: in vivo MRI assessment of total brain volume (mm3); ** for p<0.01. (Mann-Whitney-Wilcoxon test; statistical significance considered to be p<0.05); C: linear regression analysis of brain volume and DYRK1A gene dosage. (R2 = 0.977).
GTP (Green tea polyphenols) diet and brain morphogenesis
Thus brain volume is regulated by DYRK1A gene copy number. Indeed inhibiting the activity of DYRK1A may counteract the phenotypic effects of its overexpression.
We used a green tea infusion, which was prepared daily. Mice were maintained on this diet from gestation (started during initial mating period) and through adulthood, until they were killed for examination. The study included four groups of mice: wild-type (wt) and transgenic (tg), fed with water or with green tea. The active compounds in green tea infusions are thought to be predominantly polyphenols and caffeine, which is less abundant in green tea than in black tea [31].
The amount of the protein dyrk1a in samples prepared from dissected hypothalamus-thalamus region was assessed by western blot analysis for the four groups of animals: this amount is not modified by the diet.(fig 2A and B)
Figure 2. Average DYRK1A protein levels in thalamus-hypothalamus for each genotype-treatment group.
A: western blot assessment of dyrk1a and actin levels; B: average Dyrk1a protein levels for wild type (WT, n = 6), YACtg152F7 transgenic (TG, n = 6) water-fed (H2O) and in wild type (WT, n = 3), YACtg152F7 transgenic (TG, n = 7) green tea-fed (GTP). ** for p<0.01.
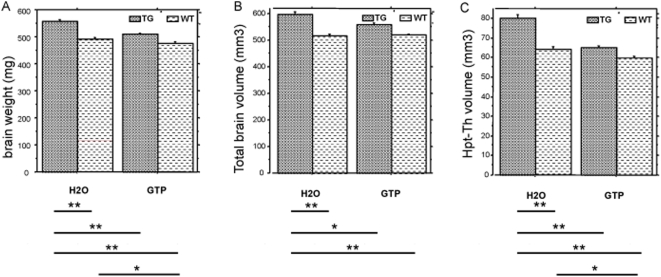
We assessed total brain volume using in vivo MRI and determined brain weight after dissection (fig 3). Brains from transgenic mice fed with green tea were 7.1% heavier, and had 7.5% greater volume, than wild-type mice green tea fed; the transgenic fed water had 13.2% heavier brains, with 15% greater volume than wt water fed. The regional volume of the thalamic-hypothalamic region was 24.4% greater in water-fed YACtg152F7 mice as compared to WT, and only 9% greater in green tea-fed YACtg152F7 mice as compared to WT. There are small but significant differences in brain weight and volume of the thalamus-hypothalamus region between water-fed and green tea-fed wild-type mice. Two-way ANOVA revealed significant effects of genotype and treatment and an interaction between treatment and genotype effect (supplementary table 1 in Data S1).
Figure 3. Effect of GTP treatment on DYRK1A-induced brain alterations.
A: weight of total brain (mg) in wild type (WT, n = 26), YACtg152F7 (TG, n = 13) water-fed (H2O) and in wild type (WT, n = 13), YACtg152F7 transgenic (TG, n = 18) green tea-fed GTP; B: in vivo MRI assessment of total brain volume (mm3) in wild type (n = 10) and YACtg152F7 transgenic (n = 10) water-fed (H2O) and in wild type (n = 9) and YACtg152F7 transgenic (n = 11) green tea-fed GTP; C: in vivo MRI assessment of hypothalamus-thalamus volume (mm3) in wild type (n = 6) and YACtg152F7 transgenic (n = 6) water-fed (H2O) and in wild type (n = 5) and YACtg152F7 transgenic (n = 7) green tea-fed (GTP). (Details of the MRI experiments in supp. data). ** for p<0.01; * for p<0.05.
GTP diet and learning deficit
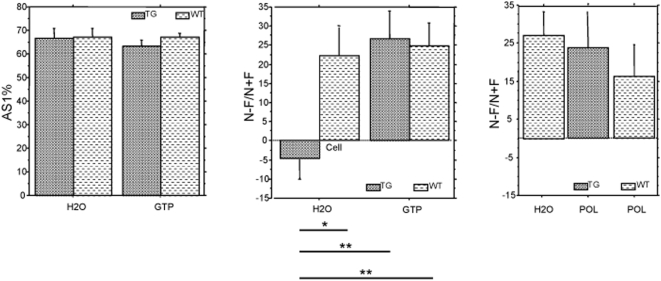
A learning deficit was also observed in murine models overexpressing the DYRK1A gene, using the Morris water maze with direct and reverse platform paradigms. These defects may indicate a long-term memory deficit. We used two paradigms, spontaneous alternation in the Y–maze and object recognition, to assess the effects on short-term and long-term memory, respectively [32], [33]. The Y-maze consists of three arms, each of which the animal is free to visit. The rate of spontaneous alternation (visiting each arm in turn) was greater than 50% for each of the four groups. Similar findings were obtained for both of two test sessions (number of alternations/total number of possible alternations ×100 (AS1), fig 4A), indicating all mouse groups had similar levels of working memory. Additionally, the number of entries did not differ between the first and the second halves of the first session, between the first and second sessions and between groups of mice. This demonstrated both short-term habituation and long-term habituation of the mice, neither of which were affected by treatment or genotype. In the object recognition test, each mouse is placed individually in an open-field setting, in which one object is placed at the left-hand side and a second object at the right-hand side. The test comprises five daily sessions. After the training sessions one object is substituted for a novel object. Exploration time was lower with the transgenic mice than with wild-type mice, and lower with GTP-treated mice than with water-fed mice during both the acquisition and retention sessions. Wild-type animals showed a clear long-term memory acquisition during the retention session. By contrast, the memory index 100×(N−F/N+F) was close to zero for water-fed transgenic animals: these animals were unable to recognize object novelty in the object recognition task. The GTP-treated transgenic mice showed discrimination indices equivalent to that of wild type mice (fig 4B). The main constituents of green tea infusion are catechins and caffeine. We used purified polyphenols from green tea (polyphenon 60; see composition in sup Table III) to assess the effect of a purified compound. This second experiment incorporated three groups of mice: water-fed controls, polyphenon-treated controls and polyphenon-treated transgenic mice. Each of the three groups had similar exploration times in the novel object recognition paradigm and recognized the novel object, demonstrating similar object novelty discrimination behavior (fig 4C).
Figure 4. Effect of GTP treatment on short- and long-term memory.
In wild type (WT, n = 10) and YACtg152F7 transgenic (TG, n = 10) water-fed (H2O) and in wild type (WT, n = 10) and YACtg152F7 transgenic (TG, n = 10) green tea-fed (GTP, n = 10). A: Spontaneous alternation test with two sessions of ten minutes each separated by 24 h: number of alternations/total number of possible alternations ×100 (AS1). B: Object recognition test: difference in exploration time between the new and familiar objects, in percentage of total time spent exploring the two objects; 100×(N−F/N+F). ** for p<0.01, * for p<0.05. according to Wilcoxon test results (two-way ANOVA in supplementary data). C: Object recognition test on WT H2O-fed (n = 10), WT polyphenon-fed (n = 10) and TG polyphenon-fed (n = 6) (no significant differences between the three groups).
Effect of GTP diet on markers of synaptic plasticity
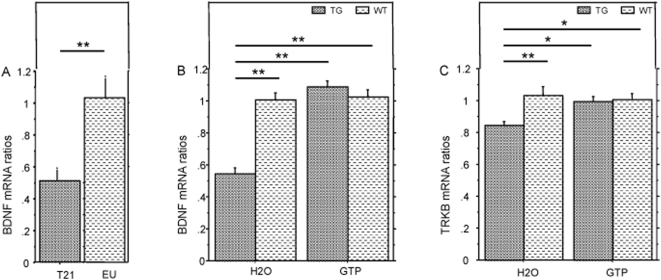
The ability of animals to learn and remember may be encoded at the synaptic level: synaptic plasticity has been linked to various factors including levels of the neurotrophic factor, BDNF [34], [35]. We used quantitative RT-PCR to measure BDNF mRNA levels in hippocampi of human fetal brains from individuals with trisomy 21 and from age-matched diploid individuals (19–21 weeks) to determine potential BDNF deficiency in trisomy 21 patients. BDNF mRNA levels were 50% lower in these patients than in age-matched diploid individuals (Fig 5A). BDNF transcription is regulated by CREB [36]. We have previously found low levels pCREB in the brains of YACtg152F7 mice [20]. In this study, we compared BDNF mRNA levels in hippocampi from transgenic mice with those from water-fed wild-type mice and BDNF mRNA levels of GTP-fed tg with wt mice. Tg mice had a lower level of BDNF in the hippocampus than wt mice; this defect was corrected by GTP treatment (Fig 5B). We also found that mRNA levels for the BDNF plasma membrane receptor, TRKB, were significantly lower in YACtg152F7 mice than in wt mice. This defect was also corrected by GTP treatment (Fig 5C).
Figure 5. Effect of GTP treatment on BDNF and TRKB mRNA levels.
mRNA levels determined by quantitative PCR. A: BDNF in fetal human hippocampus (EU: normal karyotype (n = 4) and T21: trisomy 21 (n = 5); B: BDNF in adult hippocampus from wild type (WT; n = 6), YACtg152F7 transgenic (TG; n = 10), water-fed (H2O) WT (n = 8), TG (n = 3), green tea-fed (GTP); C: TRKB in adult hippocampus from wild type (WT; n = 4), YACtg152F7 transgenic (TG; n = 18), water-fed (H2O) WT (n = 4), TG (n = 4) green tea-fed (GTP). ** for p<0.01; * for p<0.05.
Discussion
In this study we have shown that modulation of the DYRK1A gene copy number can correct brain morphogenesis alterations. It is possible to compensate the deficit in murine Dyrk1a heterozygote by the addition of one copy of human DYRK1A; the correction operates also on the phenotypes seen in the YACtg152F7 model and induced by the overexpression. These results established Dyrk1a as a key player in control of brain development and brain morphogenesis: the brain volume, as shown by MRI experiments is strongly related to Dyrk1a gene copy number (Fig 1C). We also demonstrated that chronic administration of polyphenols from green tea can have a similar, although less efficient than normalizing the gene copy number, corrective effect on brain alterations indicating that the diet is bringing the level of active DYRK1A to a value between the values produced in the transgenic and the wild type situations. The effect of the polyphenols is also visible when comparing water fed and green tea fed wild type animals: the diet induces a significant reduction of brain weight and thalamus-hypothalamus volume suggesting that the diet induced reduction of active Dyrk1a is equivalent to a genic content below 2 copies. Polyphenol treatment had no effect on the results to the spontaneous alternation paradigm: transgenic animals do not show any impairment for this task and behave similarly to the control animals. Using a novel object recognition paradigm to assess long term memory, transgenic mice with three copies of DYRK1A were clearly impaired: polyphenol treatment ameliorates cognitive deficits in tgYAC152F7 mice. Other groups have shown an effect of polyphenols on brain functions: in a study designed to determine whether cognition could be influenced by a flavanol-rich diet, Van Praag et al [37] found that memory, hippocampal vascularisation and neuronal spine density were enhanced in mice fed a (-)epicatechin-containing diet compared with controls. The polyphenol treatment does not modify the amount of DYRK1A protein. Our results suggest either a direct effect of EGCG on the activity of DYRK1A or an indirect effect, acting via a downstream target in the DYRK1A pathway. DYRK1A phosphorylation of endocytic proteins has been observed in cell cultures: this phosphorylation modifies the interactions of dynamin 1, synaptojanin 1 and amphiphysin 1 with other members of the endocytic accessory proteins and potentially regulates the assembly of protein complexes. Additionally, EGCG treatment in HEK 293 cells results in decreased phosphorylation of amphiphysin, a DYRK1A target [8]. Modulation of these trafficking proteins is generally thought to influence synaptic plasticity. LTP and its opposing process, long-term depression, are widely considered the major cellular mechanisms that underlie learning and memory. LTP was reduced and LTD was augmented in comparison to diploid controls in the isolated hippocampus of Ts65Dn and Ts1Cje, two DS murine models carrying a partial trisomy 16 encompassing the DYRK1A gene [38], [39], [40]. LTP levels can be rescued in hippocampal slices from Ts65Dn mice by EGCG treatment [41]. This is consistent with our observations of long-term memory impairment rescue. These effects may be due to EGCG action on DYRK1A resulting in modulation of the endocytic apparatus and/or modulation of BDNF-related pathways. BDNF is one of the neurotrophins involved in the regulation of neuron survival and differentiation; it has also acute effects on synaptic transmission. It has been proposed that it acts as a modulator of GABAergic inhibition [42]. In hippocampal slices synaptic inhibition was enhanced by reduced BDNF expression in bdnf+/− animals [43]. Further experimentation will be needed to elucidate effects of timing of treatment on the various phenotypic outcomes. Although correction of morphogenesis defects may require an early treatment, treatment during adulthood may result in improvement of cognition performance; indeed, this was recently demonstrated using GABAa receptor antagonists [15]. However it should be also mentioned the EGCG is a potent antioxidant and a free radical scavenger and has protective effects in ischemia [44] and in atherosclerosis [45]. It can decrease the amyloidosis in the brain by acting on APP processing [46]. Therefore other EGCG targets may explain the observed rescue of phenotypes that are primarily caused by the overexpression of Dyrk1a. Nevertheless our results suggest a central role for Dyrk1a in CNS functioning and highlight a potential clinical benefit of DYRK1A inhibitors, particularly of natural polyphenol extracts; these extracts are already used as dietary supplements for the treatment of other disorders and have been shown to be well tolerated at doses similar to those used in the present study. Such treatment may potentially be used to improve the cognitive performances of Down syndrome patients.
Materials and Methods
Animal housing, genotyping and treatment
Transgenic mice were generated on an FVB inbred background [19]. Line 12 contains one copy of YAC152F7. The FVB background that was used for micro-injection of the transgenic fragments carries a recessive mutation (rd) inducing retinal degeneration. To avoid an effect of rd on visual cues, we used F1 male offspring from C57BL/6J females and transgenic FVB males for the behavioral experiments and corresponding mRNA experiments. Mice with heterozygous deletion of Dyrk1a [24] were bred on a Swiss background and transgenic FVB males (YACtg152F7) were crossed with Dyrk1a (+/−) females to generate offspring with the four different genotypes. All animals were bred under SPF conditions and were treated in compliance with animal welfare policies from the Ministry of Agriculture (law 87848). JMD has the habilitation 75–369 for experiences on vertebrates and these experiments have been approved by the CREEA N°4. Mice were genotyped as previously described [20], [23]. Treatment was prepared daily. In the first set of experiments, mice were fed green tea ad libitum (1 g in 100 ml water for 5 min at 100°C), corresponding to 0.6–1 mg EGCG per day [30]; in the second set of experiments, mice were given a solution of polyphenon 60 (0.8 g/l, Sigma), equivalent to 1.2 mg per day.
Quantitative PCR experiments
mRNA was isolated (Microfastrack, Invitrogen) and reverse transcribed (Ambion), and qRT-PCR (Light cycler, Roche) was performed with primer sequences described in supplementary table 2 in Data S1.
Protein extraction and western blotting
Adult mice (3 months old) were sacrificed by decapitation. Brains were removed and dissected on ice to separate the cortex, hippocampus and the thalamus/hypothalamus before being frozen in liquid nitrogen and stored at −80°C. Individual thalamus/hypothalamus were homogenized in NP-40 modified lysis buffer [20 mM Tris-HCl (pH 7.4); 140 mM NaCl; 10% glycerol; 1% NP-40; 2 mM EDTA] containing proteases inhibitors (1 mM Pefa-Bloc; 2 µg/ml Aprotinin, leupeptin and pepstatine; 5 µg/ml E64) and phophatase inhibitors (2 mM NaF; 1 mM Na3VO4 and PhosSTOP phosphatase inhibitors cocktail). Protein concentration was determined using the Bio Rad Bradford protein assay following the manufacturer's instructions. For western blotting, equal amounts of total proteins (30–50 µg) were subjected to SDS-PAGE electrophoresis and transferred into nitrocellulose membranes. The latter were blocked with TBS (Tris buffered saline pH 7.5)-Tween 0,1% containing 5% of non-fat dried milk and incubated with the Dyrk1A antibody (Abnova, diluted 1∶1000 in the blocking buffer) at 4°C overnight. Excess of primary antibody was removed by rinsing with TBS-T, followed by 1 h incubation with the species-appropriate horseradish peroxidase-conjugated secondary antibody (Anti-mouse Ig diluted 1∶40000 in the blocking solution) at room temperature. Immunoblots were subsequently probed with anti-ß actin antibody (Sigma, diluted 1 ∶4000) as loading control. Signals were revealed by enhanced chemiluminescence with the LAS300 Image Reader and results analyzed with science Lab 2005 –MultiGauge software (Life science Fujufilm).
MRI experiments
We performed in vivo MRI studies on animals from three litters, using a horizontal 7T magnet connected to an INOVA console (Varian, CA) and equipped with 120 mT/m gradient coils and home-built 1H coils. Experimental conditions are given in the supplementary methods section Data S1. Brain structure volumes were estimated, using a visual guide visible on each image set. MRI analysis protocols were established and carried out independently by two investigators. The same analysis was performed twice, once with each of two software packages — WINMRI (Bruker) and AMIRA, (TGS Inc., San Diego, CA) — and by each investigator, to evaluate the reproducibility of volume measurements. Each 2D coronal data set consisted of 41 images. As brain volume depends on age and transgenic state, the number of slices taken was not constant and was therefore not used to delimit the brain. In scans taken between the appearance of the cerebellum and the disappearance of the olfactory bulbs, brain volumes were extracted by manually outlining regions of interest and multiplying by slice thickness (0.5 mm).
Y alternation
Working memory, exploratory activity and short-term habituation were assessed using a spontaneous alternation task: the natural tendency of a mouse when placed in the Y-maze is to move from one arm of the maze to another. During the test, mice were placed at the centre of the maze and the sequence of entries into the three arms was noted over a period of 10 min. (see supp methods and supp Data S1).
Novel Object recognition
A novel object recognition test was used to assess the declarative memory (see supp methods in Data S1). The performance is measured on the whole retention session with: the time spent exploring the two objects (N+F), the difference of exploration time between the new object and the familiar object (N−F), the difference of exploration time between the new object and the familiar object in percent of the time spent exploring the two objects (%(N−F)/(N+F) = 100×(N−F)/(N+F)).
Memory of the object is considered to be present for a group on a given period of time if animals spend more time in exploring the new object than the familiar one, i.e. if (N−F) and %(N−F)/(N+F) are higher than zero.
Data analysis
Data were analyzed using a Mann-Whitney-Wilcoxon test and statistical significance was recorded for p<0.05. A second analysis was used to assess the effects of treatment and genotype (two-way ANOVA results are presented in supplementary Data S1).
Supporting Information
(0.15 MB DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by EU grants: T21 targets(FP5) and AnEUploidy (FP6). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Delabar JM, Theophile D, Rahmani Z, Chettouh Z, Blouin JL, et al. Molecular mapping of twenty-four features of Down syndrome on chromosome 21. Eur J Hum Genet. 1993;1(2):114–24. doi: 10.1159/000472398. [DOI] [PubMed] [Google Scholar]
- 2.Korenberg JR, Aaltonen J, Brahe C, Cabin D, Creau N, et al. Report and abstracts of the Sixth International Workshop on Human Chromosome 21 Mapping. Cold Spring Harbor, New York, USA. May 6–8,1996. Cytogenet Cell Genet. 1997;79(1–2):21–52. doi: 10.1159/000134681. [DOI] [PubMed] [Google Scholar]
- 3.Ronan A, Fagan K, Christie L, Conroy J, Nowak NJ, et al. Familial 4.3 Mb duplication of 21q22 sheds new light on the Down syndrome critical region. J Med Genet. 2007;Jul;44(7):448–51. doi: 10.1136/jmg.2006.047373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guimera J, Casas C, Pucharcos C, Solans A, Domenech A, et al. A human homologue of Drosophila minibrain (MNB) is expressed in the neuronal regions affected in Down syndrome and maps to the critical region. Hum Mol Genet. 1996;Sep;5(9):1305–10. doi: 10.1093/hmg/5.9.1305. [DOI] [PubMed] [Google Scholar]
- 5.Tejedor F, Zhu XR, Kaltenbach E, Ackermann A, Baumann A, et al. Minibrain: a new protein kinase family involved in postembryonic neurogenesis in Drosophila. Neuron. 1995;Feb;14(2):287–301. doi: 10.1016/0896-6273(95)90286-4. [DOI] [PubMed] [Google Scholar]
- 6.Lochhead PA, Sibbet G, Morrice N, Cleghon V. Activation-loop autophosphorylation is mediated by a novel transitional intermediate form of DYRKs. Cell. 2005;Jun 17;121(6):925–36. doi: 10.1016/j.cell.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 7.Woods YL, Rena G, Morrice N, Barthel A, Becker W, et al. The kinase DYRK1A phosphorylates the transcription factor FKHR at Ser329 in vitro, a novel in vivo phosphorylation site. Biochem J. 2001;May 1;355(Pt 3):597–607. doi: 10.1042/bj3550597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murakami N, Xie W, Lu RC, Chen-Hwang MC, Wieraszko A, et al. Phosphorylation of amphiphysin I by minibrain kinase/dual-specificity tyrosine phosphorylation-regulated kinase, a kinase implicated in Down syndrome. J Biol Chem. 2006;Aug 18;281(33):23712–24. doi: 10.1074/jbc.M513497200. Epub 2006 May 29. [DOI] [PubMed] [Google Scholar]
- 9.Woods YL, Cohen P, Becker W, Jakes R, Goedert M, et al. The kinase DYRK phosphorylates protein-synthesis initiation factor eIF2Bepsilon at Ser539 and the microtubule-associated protein tau at Thr212: potential role for DYRK as a glycogen synthase kinase 3-priming kinase. Biochem J. 2001;May 1;355(Pt 3):609–15. doi: 10.1042/bj3550609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dowjat WK, Adayev T, Kuchna I, Nowicki K, Palminiello S, et al. Trisomy-driven overexpression of DYRK1A kinase in the brain of subjects with Down syndrome. Neurosci Lett. 2007;Feb 8;413(1):77–81. doi: 10.1016/j.neulet.2006.11.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baxter LL, Moran TH, Richtsmeier JT, Troncoso J, Reeves RH. Discovery and genetic localization of Down syndrome cerebellar phenotypes using the Ts65Dn mouse. Hum Mol Genet. 2000;Jan 22;9(2):195–202. doi: 10.1093/hmg/9.2.195. [DOI] [PubMed] [Google Scholar]
- 12.Aldridge K, Reeves RH, Olson LE, Richtsmeier JT. Differential effects of trisomy on brain shape and volume in related aneuploid mouse models. Am J Med Genet A. 2007;May 15;143A(10):1060–70. doi: 10.1002/ajmg.a.31721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reeves RH, Irving NG, Moran TH, Wohn A, Kitt C, et al. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat Genet. 1995;1995 Oct;11(2):177–84. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- 14.Escorihuela RM, Vallina IF, Martínez-Cué C, Baamonde C, Dierssen M, et al. Impaired short- and long-term memory in Ts65Dn mice, a model for Down syndrome. Neurosci Lett. 1998;May 15;247(2–3):171–4. doi: 10.1016/s0304-3940(98)00317-6. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez F, Morishita W, Zuniga E, Nguyen J, Blank M, et al. Pharmacotherapy for cognitive impairment in a mouse model of Down syndrome. Nat Neurosci. 2007;Apr;10(4):411–3. doi: 10.1038/nn1860. [DOI] [PubMed] [Google Scholar]
- 16.Olson LE, Roper RJ, Sengstaken CL, Peterson EA, Aquino V, et al. Trisomy for the Down syndrome ‘critical region’ is necessary but not sufficient for brain phenotypes of trisomic mice. Hum Mol Genet. 2007;Apr 1;16(7):774–82. doi: 10.1093/hmg/ddm022. [DOI] [PubMed] [Google Scholar]
- 17.Dahmane N, Ghezala GA, Gosset P, Chamoun Z, Dufresne-Zacharia MC, et al. Transcriptional map of the 2.5-Mb CBR-ERG region of chromosome 21 involved in Down syndrome. Genomics. 1998;Feb 15;48(1):12–23. doi: 10.1006/geno.1997.5146. [DOI] [PubMed] [Google Scholar]
- 18.Hattori M, Fujiyama A, Taylor TD, Watanabe H, Yada T. The DNA sequence of human chromosome 21. Nature. 2000;May 18;405(6784):311–9. doi: 10.1038/35012518. et al.; Chromosome 21 mapping and sequencing consortium. (2000) Erratum in: Nature 2000 Sep 7;407(6800):110. [DOI] [PubMed] [Google Scholar]
- 19.Smith DJ, Stevens ME, Sudanagunta SP, Bronson RT, Makhinson M, et al. Functional screening of 2 Mb of human chromosome 21q22.2 in transgenic mice implicates minibrain in learning defects associated with Down syndrome. Nat Genet. 1997;May;16(1):28–36. doi: 10.1038/ng0597-28. [DOI] [PubMed] [Google Scholar]
- 20.Branchi I, Bichler Z, Minghetti L, Delabar JM, Malchiodi-Albedi F, et al. Transgenic mouse in vivo library of human Down syndrome critical region 1: association between DYRK1A overexpression, brain development abnormalities, and cell cycle protein alteration. J Neuropathol Exp Neurol. 2004;May;63(5):429–40. doi: 10.1093/jnen/63.5.429. [DOI] [PubMed] [Google Scholar]
- 21.Chabert C, Jamon M, Cherfouh A, Duquenne V, Smith DJ, et al. Functional analysis of genes implicated in Down syndrome: 1. Cognitive abilities in mice transpolygenic for Down Syndrome Chromosomal Region-1 (DCR-1). Behav Genet. 2004;Nov;34(6):559–69. doi: 10.1007/s10519-004-5584-3. [DOI] [PubMed] [Google Scholar]
- 22.Ahn KJ, Jeong HK, Choi HS, Ryoo SR, Kim YJ, et al. DYRK1A BAC transgenic mice show altered synaptic plasticity with learning and memory defects. Neurobiol Dis. 2006;Jun;22(3):463–72. doi: 10.1016/j.nbd.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Sebrié C, Chabert C, Ledru A, Guedj F, Po C, et al. Increased dosage of DYRK1A and brain volumetric alterations in a YAC model of partial trisomy 21. Anat Records (Hoboken) 2008;Mar;291(3):254–62. doi: 10.1002/ar.20640. [DOI] [PubMed] [Google Scholar]
- 24.Fotaki V, Dierssen M, Alcantara S, Martinez S, Marti E, et al. Dyrk1A haploinsufficiency affects viability and causes developmental delay and abnormal brain morphology in mice. Mol Cell Biol. 2002;Sep;22(18):6636–47. doi: 10.1128/MCB.22.18.6636-6647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bain J, McLauchlan H, Elliott M, Cohen P. The specificities of protein kinase inhibitors: an update. Biochem J. 2003;2003 Apr 1;371(Pt 1):199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adayev T, Chen-Hwang MC, Murakami N, Wegiel J, Hwang YW. Kinetic properties of a MNB/DYRK1A mutant suitable for the elucidation of biochemical pathways. Biochemistry. 2006;Oct 3;45(39):12011–9. doi: 10.1021/bi060632j. [DOI] [PubMed] [Google Scholar]
- 28.Lin LC, Wang MN, Tseng TY, Sung JS, Tsai TH. Pharmacokinetics of (-)-epigallocatechin-3-gallate in conscious and freely moving rats and its brain regional distribution. J Agric Food Chem. 2007;Feb 21;55(4):1517–24. doi: 10.1021/jf062816a. Epub 2007 Jan 27. [DOI] [PubMed] [Google Scholar]
- 29.Chu KO, Wang CC, Chu CY, Choy KW, Pang CP, et al. Uptake and distribution of catechins in fetal organs following in utero exposure in rats. Hum Reprod. 2007;Jan;22(1):280–7. doi: 10.1093/humrep/del353. Epub 2006 Sep. [DOI] [PubMed] [Google Scholar]
- 30.Choi JY, Park CS, Kim DJ, Cho MH, Jin BK, et al. Prevention of nitric oxide-mediated 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced Parkinson's disease in mice by tea phenolic epigallocatechin 3-gallate. Neurotoxicology. 2002;Sep;23(3):367–74. doi: 10.1016/s0161-813x(02)00079-7. [DOI] [PubMed] [Google Scholar]
- 31.Yang DJ, Hwang LS, Lin JT. Effects of different steeping methods and storage on caffeine, catechins and gallic acid in bag tea infusions. J Chromatogr A. 2007;Jul 13;1156(1–2):312–20. doi: 10.1016/j.chroma.2006.11.088. Epub 2006 Dec 11. [DOI] [PubMed] [Google Scholar]
- 32.O'Doherty A, Ruf S, Mulligan C, Hildreth V, Errington ML, et al. An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science. 2005;Sep 23;309(5743):2033–7. doi: 10.1126/science.1114535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bozon B, Davis S, Laroche S. A requirement for the immediate early gene zif268 in reconsolidation of recognition memory after retrieval. Neuron. 2003;Nov 13;40(4):695–701. doi: 10.1016/s0896-6273(03)00674-3. [DOI] [PubMed] [Google Scholar]
- 34.Hall J, Thomas KL, Everitt BJ. Rapid and selective induction of BDNF expression in the hippocampus during contextual learning. Nat Neurosci. 2000;Jun;3(6):533–5. doi: 10.1038/75698. [DOI] [PubMed] [Google Scholar]
- 35.Heldt SA, Stanek L, Chhatwal JP, Ressler KJ. Hippocampus-specific deletion of BDNF in adult mice impairs spatial memory and extinction of aversive memories. Mol Psychiatry. 2007;Jul;12(7):656–70. doi: 10.1038/sj.mp.4001957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barco A, Patterson S, Alarcon JM, Gromova P, Mata-Roig M, et al. Gene expression profiling of facilitated L-LTP in VP16-CREB mice reveals that BDNF is critical for the maintenance of LTP and its synaptic capture. Neuron. 2005;Oct 6;48(1):123–37. doi: 10.1016/j.neuron.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 37.Van Praag H, Lucero MJ, Yeo GW, Stecker K, Heivand N, et al. Plant-derived flavanol (-)epicatechin enhances angiogenesis and retention of spatial memory in mice. J Neurosci. 2007;May 30;27(22):5869–78. doi: 10.1523/JNEUROSCI.0914-07.2007. Erratum in: J Neurosci. 2007 Aug 1;27(31):ii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siarey RJ, Carlson EJ, Epstein CJ, Balbo A, Rapoport SI, et al. Increased synaptic depression in the Ts65Dn mouse, a model for mental retardation in Down syndrome. Neuropharmacology. 1999;Dec;38(12):1917–20. doi: 10.1016/s0028-3908(99)00083-0. [DOI] [PubMed] [Google Scholar]
- 39.Siarey RJ, Villar AJ, Epstein CJ, Galdzicki Z. Abnormal synaptic plasticity in the Ts1Cje segmental trisomy 16 mouse model of Down syndrome. Neuropharmacology. 2005;Jul;49(1):122–8. doi: 10.1016/j.neuropharm.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 40.Kleschevnikov AM, Belichenko PV, Villar AJ, Epstein CJ, Malenka RC, et al. Hippocampal long-term potentiation suppressed by increased inhibition in the Ts65Dn mouse, a genetic model of Down syndrome. J Neurosci. 2004;2004 Sep 15;24(37):8153–60. doi: 10.1523/JNEUROSCI.1766-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie W, Ramakrishna N, Wieraszko A, Hwang YW. Promotion of Neuronal Plasticity by (–)-Epigallocatechin-5 3-Gallate. Neurochem Res. 2007 doi: 10.1007/s11064-007-9494-7. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka T, Saito H, Matsuki N. Inhibition of GABAA synaptic responses by brain-derived neurotrophic factor (BDNF) in rat hippocampus. J Neurosci. 1997;May 1;17(9):2959–66. doi: 10.1523/JNEUROSCI.17-09-02959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olofsdotter K, Lindvall O, Asztély F. Increased synaptic inhibition in dentate gyrus of mice with reduced levels of endogenous brain-derived neurotrophic factor. Neuroscience. 2000;101(3):531–9. doi: 10.1016/s0306-4522(00)00428-0. [DOI] [PubMed] [Google Scholar]
- 44.Lee H, Bae JH, Lee SR. Protective effect of green tea polyphenol EGCG against neuronal damage and brain edema after unilateral cerebral ischemia in gerbils. J Neurosci Res. 2004;Sep 15;77(6):892–900. doi: 10.1002/jnr.20193. [DOI] [PubMed] [Google Scholar]
- 45.Chyu KY, Babbidge SM, Zhao X, Dandillaya R, Rietveld AG, et al. Differential effects of green tea-derived catechin on developing versus established atherosclerosis in apolipoprotein E-null mice. Circulation. 2004;May 25;109(20):2448–53. doi: 10.1161/01.CIR.0000128034.70732.C2. Epub 2004 May 10. Links. [DOI] [PubMed] [Google Scholar]
- 46.Rezai-Zadeh K, Shytle D, Sun N, Mori T, Hou H, et al. Green tea epigallocatechin-3-gallate (EGCG) modulates amyloid precursor protein cleavage and reduces cerebral amyloidosis in Alzheimer transgenic mice. J Neurosci. 2005;Sep 21;25(38):8807–14. doi: 10.1523/JNEUROSCI.1521-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(0.15 MB DOC)