Abstract
The ligand binding domain of the human vitamin D receptor (VDR) was modeled based on the crystal structure of the retinoic acid receptor. The ligand binding pocket of our VDR model is spacious at the helix 11 site and confined at the β-turn site. The ligand 1α,25-dihydroxyvitamin D3 was assumed to be anchored in the ligand binding pocket with its side chain heading to helix 11 (site 2) and the A-ring toward the β-turn (site 1). Three residues forming hydrogen bonds with the functionally important 1α- and 25-hydroxyl groups of 1α,25-dihydroxyvitamin D3 were identified and confirmed by mutational analysis: the 1α-hydroxyl group is forming pincer-type hydrogen bonds with S237 and R274 and the 25-hydroxyl group is interacting with H397. Docking potential for various ligands to the VDR model was examined, and the results are in good agreement with our previous three-dimensional structure-function theory.
The steroid hormone 1α,25-dihydroxyvitamin D3 [1,25-(OH)2D3] is unique not only in its structure but also in its function. Its structure is long in length and flexible unlike any other steroid hormone. In addition to its classical role of regulating calcium metabolism, it is involved in such basic functions as regulation of proliferation and differentiation of cells and the immune response (1). 1,25-(OH)2D3 exerts these effects through a ligand-activated transcription factor, vitamin D receptor (VDR) (2). VDR is a member of the nuclear receptor (NR) superfamily (3), which includes the receptors for the steroid and thyroid hormones and retinoic acids and numerous orphan receptors for which currently no natural ligands are known. All NRs exhibit a common modular structure consisting of six distinct domains with an evolutionary highly conserved DNA binding domain and a moderately conserved ligand binding domain (LBD), which functions as a multifunctional domain. Besides the ligand recognition, it is involved in dimerization and ligand-dependent transactivation. So far, crystallographic structures of six NR-LBDs [retinoid X receptor (4), retinoic acid receptor (RAR) (5, 6), thyroid hormone receptor (7, 8), estrogen receptor (ER) (9–11), progesterone receptor (PR) (12), and peroxisome proliferator-activated receptor (PPAR) (13, 14)] have been solved, including the structure of holo and apo forms, complexes with the natural ligand, synthetic agonists and antagonists, and ternary complexes with the ligand and a coactivator. These structures reveal not only the common fold of NR-LBDs but also the structural role of the ligands in inducing conformational changes in LBD, which makes the recruitment of coactivator possible to initiate the action of the general transcriptional machinery. Crystal structures also afforded structural basis of the mechanism of the action of antagonists.
From structure-function relationship analysis of more than 500 vitamin D analogs, it was shown that variable side-chain structures are accommodated in VDR, whereas only limited structural modifications are tolerated for the A-ring for binding to the VDR (15). On the basis of conformational analysis of vitamin D analogs and through the use of conformationally restricted synthetic analogs, we established a theory on the three-dimensional (3D) conformation-function relationship of vitamin D (16–21). In the theory we proposed three important side-chain arrangements responsible for the action of vitamin D. To develop our structure-function theory of ligands into one that includes the receptor, we need to model the structure of the VDR-LBD and study the interaction between the receptor and various ligands.
The crystal structure of the VDR-LBD or the whole receptor has not been solved. Two models of VDR-LBD have been reported (22, 23), but neither of them was substantiated by mutational analysis. In this paper we report the modeling of hVDR-LBD based on the crystal structure of hRARγ as the template and docking of the hormone and some analogs into the LBD. Our VDR model was additionally corroborated by mutation of polar amino acid residues, which are assumed to interact with the ligand. Evaluation of their ligand binding capacity and transcriptional activity substantiate our model.
Materials and Methods
Sequence Alignment and Molecular Modeling.
The sequence of hVDR-LBD (residues 124–427) was aligned to six NRs (hPPARγ, thyroid hormone receptor α1, hRARγ, human retinoid X receptor α, hERα, and hPR) by software clustalw (version 1.7, default parameters). Parts of the automatically derived alignment then were modified manually. Helix 1 was assigned by comparing with thyroid hormone receptor, RAR, liver X receptor, pregnane X receptor, ecdysone receptor, MB67, and farnesoid X receptor, all of which belong to the same subfamily (group 1) with VDR (24), and the sequence from β-turn to helix 7 (279–320) was aligned in comparison with the x-ray structures of the above-mentioned six NRs.
Molecular modeling and graphical manipulations were performed by using sybyl 6.5 (Tripos Associates, St. Louis). The atomic coordinates of the crystal structure of hRARγ were retrieved from the Brookhaven Protein Data Bank (entry 2LBD).
Site-Directed Mutagenesis.
The human VDR expression vector pCMX-hVDR was constructed as described (25) and was used as a template for site-directed mutagenesis. Point mutants were created by using a Quick-Change Site-Directed Mutagenesis kit (Stratagene). Five clones of mutated hVDRs (S237A, S275A, S278A, C288A, and H397A) were produced by changing the corresponding amino acid residue into alanine according to the manufacturer's instructions. Escherichia coli DH5α competent cells were transformed with the vectors incorporating the desired mutations. The cDNAs of the clones were purified with a Qiagen Plasmid Midi-Kit (Qiagen, Chatsworth, CA). Each mutant cDNA of clones was sequenced completely to ensure that no other base changes were produced (by cycle sequencing).
Cell-Free Transcription and Translation.
Plasmids (1 μg) containing the cDNA coding for the wild-type or mutant hVDRs were expressed in vitro by using the T7-coupled rabbit reticulocyte lysate system (Promega) according to the manufacturer's instructions. To assess the level and stability of the wild-type and five mutant hVDRs, the reactions also were conducted in the presence of biotinylated lysine-tRNA (Promega) to produce proteins incorporated with biotinylated lysine. The reaction mixture was subjected to SDS/PAGE and electroblotting. Biotinylated proteins were visualized by Streptavidin-horseradish peroxidase followed by chemiluminescent detection (data not shown). The results showed that the level and stability of mutated VDRs were not affected in this in vitro system.
Ligand Binding Assay.
After translation of the wild-type or mutant hVDRs, the lysate was diluted 5-fold with ice-cold TEGWD buffer (20 mM Tris⋅HCl, pH 7.4/1 mM EDTA/1 mM DTT/20 mM sodium tungstate/10% glycerol). The diluted lysate was incubated with increasing concentration of 1,25-(OH)2-[26,27-methyl-3H]D3 (0.056–2.78 nM) for 16 h at 4°C in the presence or absence of a 400-fold molar excess of unlabeled 1,25-(OH)2D3. Bound and unbound ligands were separated by the dextran-charcoal method. Bound ligand was quantitated by using scintillation counting. Data were analyzed by using a Scatchard analysis program, ligand, originally written in basic by Munson (33).
Cell Culture Conditions, Transfection, and Transactivation Assay.
COS-7 cells were cultured in DMEM supplemented with 10% FCS. Cells were seeded in 24-well plates at a density of 2 × 104 per well 24 h before transfection. The next day, the medium was replaced with 250 μl of fresh serum-free medium (Opti-MEM). Then a DNA/Trans IT-LT1 reagent (Mirus, Madison, WI) mixture containing 0.28 μg of a reporter plasmid (SPP×3-TK-Luc), 0.2 μg of wild-type or mutant hVDR expression plasmid (pCMX-hVDR), and 0.02 μg of the internal control plasmid containing sea pansy luciferase expression constructs (pRL-CMV) was prepared according to the manufacturer's procedures and added to each well. The SPP×3-TK-Luc reporter plasmid contains three copies of the mouse osteopontin VDRE. The cells were further incubated for 4 h and the medium was replaced with fresh DMEM containing 5% FCS pretreated with dextran-coated charcoal. The next day, transfected cells were treated with either 10-8 M 1,25-(OH)2D3 or ethanol vehicle and cultured for 16 h. Cells in each well were harvested with a cell lysis buffer, and the luciferase activity was measured with a luciferase assay kit (Toyo Ink, Tokyo) according to the manufacturer's instructions. Transactivation measured by the luciferase activity was normalized with the luciferase activity of the same cells determined by the sea pansy luciferase assay kit (Toyo Ink). All experiments were done in triplicate.
Results
Sequence Alignment.
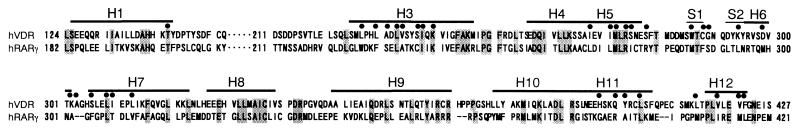
Sequence alignment is most important in homology modeling. The sequence of hVDR-LBD (residues 124–427) was aligned by using clustalw to six NRs (hPPARγ, thyroid hormone receptor α1, hRARγ, human retinoid X receptor α, hERα, and hPR) whose x-ray structures had been reported, and then helix 1 and a sequence between the β-turn to helix 7 (279–320) were modified manually. The resulting alignment of hVDR-LBD, which consists of 11 α-helices and two β-strands forming a β-sheet, is shown in Fig. 1 in comparison with the hRARγ. Our alignment is much the same as that reported by Moras' group (22) but different from the alignment reported by Norman's group (23) at the position of helix 1. That group assigned the residues 147–161 to helix 1, which now must accommodate two consecutive proline residues (P155 and 156), a structural feature not found in α-helices.
Figure 1.
Sequence alignment of hVDR-LBD with hRARγ-LBD. Shadows indicate identical residues. Circles represent the residues facing LBP. Bars indicate regions of secondary structures.
Modeling of VDR-LBD.
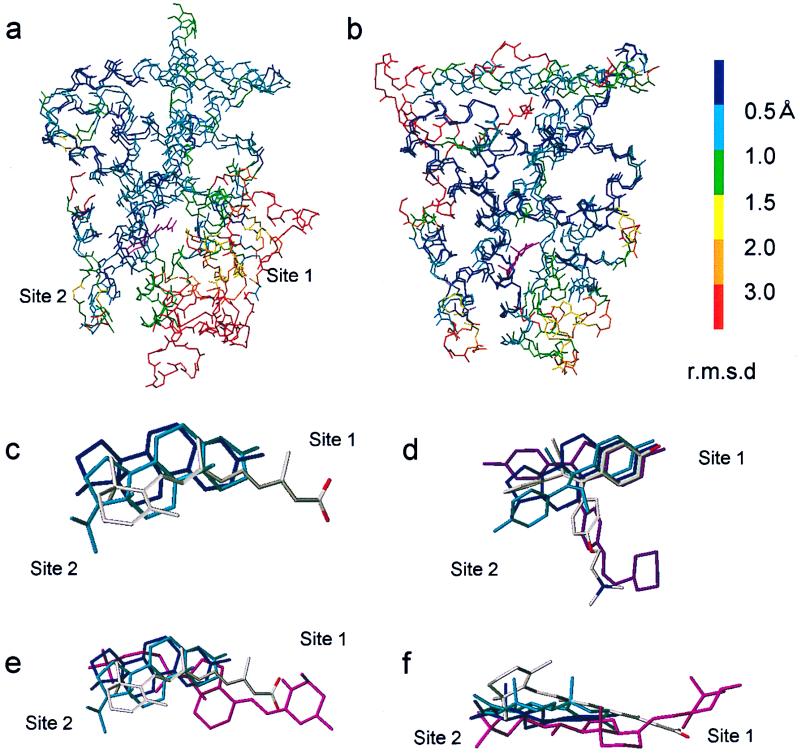
To create a homology model for the hVDR-LBD, we used hRARγ as a template for the following reasons: (i) RAR is a member of the same NR subfamily (group 1) with VDR (24) and has the highest homology with VDR. (ii) The molecular shape of the natural VDR ligand 1,25-(OH)2D3 somewhat resembles that of the RAR ligand retinoic acid. A crucial difference between RAR and VDR is the long insertion between helices 1 and 3. Because this part is impossible to build by homology, we removed loop 1–3 (143–223) for our modeling studies. The elimination of this loop was not considered to have a significant effect on the structures of the ligand binding pocket (LBP) and the transactivation function 2 (AF-2) domain. To confirm this, we overlaid the crystal structures of the NRs in the same group 1 (hRARγ-LBD and hPPARγ-LBD) and their Cα backbones were displayed by colors according to their rms deviation (Fig. 2a). The Cαs of the AF-2 domain and LBP without a β-turn site are in highly conserved regions (blue to green in Fig. 2a). We constructed the Cα framework of VDR by treating all but loops 6–7, 9–10, and 11–12 as structurally conserved regions (SCR) by using a mutation command of sybyl. The structures of loops 6–7, 9–10, and 11–12 were searched computationally from the Protein Databank by using the loop search command and the selected loops were joined to SCR parts. Side chains were attached to the framework, and when they crushed, the χ2 and then χ1, if necessary, were modified to keep the conserved residues in their original conformation. Ligand 1,25-(OH)2D3 was docked (see below), and the VDR/1,25-(OH)2D3 complex was energy-minimized first without a solvent and then in a water-filled box (52.5 × 68.44 × 49.79 Å3) (Tripos force field, Powell method, terminated when the rms force of minimization reached 0.1 kcal/mol Å).
Figure 2.
Comparison of NR-LBDs and ligands. (a and b) Overlays of Cαs of LBDs of hRARγ and hPPARγ in group 1 (a) and LBDs of hERα and hPR in group 3 (b). rms deviation (rmsd) of each Cα at the same position in the sequence alignment is shown in colors. Ligands are shown in magenta. (c) Overlay of ligands in various NRs: estradiol (E2, navy blue) in ERα, progesterone in PR (cyan), and all-trans-retinoic acid (atom type) in RARγ. (d) Overlay of ER ligands: E2 (navy blue), diethylstilbestrol (cyan), tamoxifen (atom type) and raloxifene (magenta). (e) Overlays of the three ligands shown in c plus 1,25-(OH)2D3 (magenta) in VDR. (f) Different view of e.
The structure of VDR thus modeled was evaluated by using the procheck program. A Ramachandran plot shows that 99% of the residues are either in the most favored or allowed regions and a χ1-χ2 plot displays acceptable values for most of the residues. The backbone of our model is nearly the same as the backbone of the template except for the β-turn site (Fig. 3a).
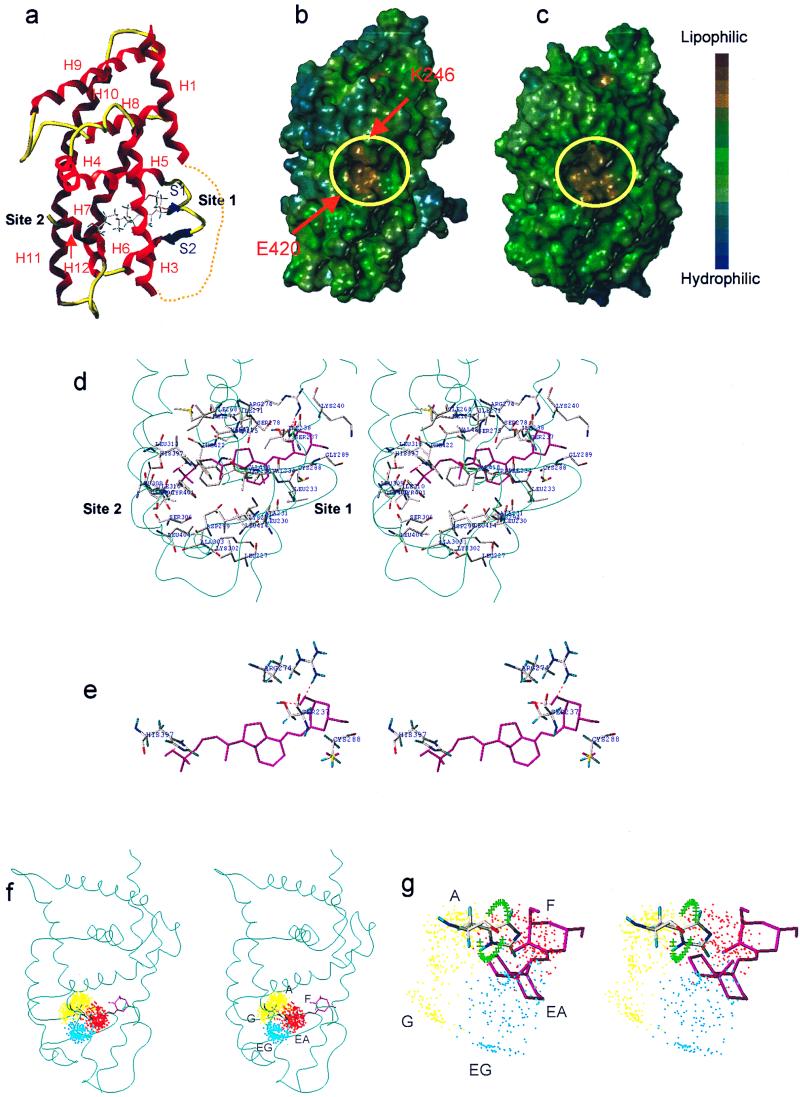
Figure 3.
(a) Ribbon-tube presentation of the VDR-LBD model. Dotted line shows eliminated loop 1–3. (b and c) Lipophilic potential surface of the VDR-LBD (b) and the RARγ-LBD (c). The AF-2 surface is surrounded by yellow circles. Arrows show the charge clump (K246 and E420). (d) Amino acid residues forming the ligand binding cavity and their interaction with 1,25-(OH)2D3 (cyan ribbon represents the backbone, stereo view). (e) The residues interacting with 1,25-(OH)2D3 (stereo view). (f) Side-chain dot map model in VDR (stereo view). (g) Enlargement of f (stereo view). The green crosses show the trace of the histidine NH when the χ2 of H397 was rotated 360° with 10° intervals. The regions about 2 Å away from the crosses are possible hydrogen-bonding areas.
The LBP is in the same position as in RAR. Its cavity is wide at site 2 and narrow at site 1 (Fig. 3d). Generally, there are two ligand-anchoring sites at two extremities of NR-LBP, one facing a β-turn called site 1 and the other facing helix 11 called site 2 (notation by Moras and coworkers, ref. 26). The amino acid residues forming the ligand binding cavity are shown in Figs. 1 and 3d. The cavity is mostly lined by hydrophobic residues but hydrophilic residues are exposed at the two extremities (S237, K240, R274, S275, S278, and C288 at site 1; H397 and Q400 at site 2) (Fig. 3d). These hydrophilic residues are potential interaction sites for the 1α-, 3β-, and 25-hydroxyl groups of 1,25-(OH)2D3.
The surface structure of the VDR model mapped with the lipophilic potential is shown in Fig. 3b compared with the surface of RAR (Fig. 3c). The circled highly lipophilic part is the AF-2 surface to which a coactivator binds. In our model the AF-2 surface is successfully created in a position similar to that of RAR. At the opposite terminals of the AF-2 surface, positively (K) and negatively (E) charged residues, so-called charge clump (13), are exposed. These two residues are completely conserved among NRs that we aligned (see above). The charge clump allows a coactivator to place its LXXLL motif in the right orientation and right position. In our VDR model, the residues forming the charge clump (E420 at helix 12 and K246 at helix 3) are perfectly positioned at the right places of the AF-2 surface (Fig. 3b).
Docking of Vitamin D.
Before docking 1,25-(OH)2D3 into VDR-LBP, we analyzed the docking mode of all known 3D structures of NRs. All NR-ligand complexes were overlaid by coinciding their Cα at their signature region and the proteins were deleted (Fig. 2c). Fig. 2c demonstrates that all of the ligands are harbored in the same region in all NRs and are aligned at site 2. However, site 1 appears to be specific to each receptor because the position is variable depending on the length of the cognate ligand. When positions of various ligands in a particular NR are compared, the structures acceptable at site 2 appear to be variable but those satisfying the site 1 requirements are stringent as the example of various ER ligands shows (Fig. 2d).
In docking the ligand into the LBP, it is important to determine the orientation and the conformation of the ligand. From structure-function studies of vitamin D, we obtained the following conclusions about the conformation of vitamin D: (i) a wide variety of side-chain structures is acceptable in the LBP (19–21); (ii) the A-ring adopts the α-conformation with the 1α-hydroxyl in the axial and 3β-hydroxyl in the equatorial orientation (M. Shimizu, personal communication; see also ref. 27); and (iii) the seco B-ring part adopts the extended 6,7-s-trans conformation (28). There are four possibilities for the orientation in the LBP. Concerning the orientation with respect to the long molecular axis, we assumed that the side chain of the ligand is accommodated in the wide cavity at site 2 and the A-ring in the narrow cavity at site 1. This satisfies the known structure-function relationship of vitamin D. With respect to the short axis, either the β-face up (the same orientation as in all of the other steroid hormone/receptor complexes) or the β-face down orientation can be permissible in the LBP. We finally chose the former orientation for the following reasons: (i) the above-mentioned evidence (Fig. 2c) and (ii) positioning of the potent hydrogen bonding residues facing the LBP (see below). Having determined the orientation, we docked 1,25-(OH)2D3 as follows: (i) The ligand was overlaid with other NR ligands (Fig. 2 e and f) aligning at site 2. (ii) The ligand position was manually adjusted so as to minimize the van der Waals bump between the ligand and the amino acid residues facing the LBP. (iii) Hydrogen bonding residues were searched computationally. The resulting structure of VDR/1,25-(OH) 2D3 complex is shown in Fig. 3d. The 1α-hydroxyl group forms pincer-type hydrogen bonds with S237 (hydrogen acceptor, 1.84 Å) and R274 (hydrogen donor, 1.80 Å) (Fig. 3e). Pincer-type hydrogen bonds are commonly observed at site 1 in the crystal structures of NR/natural ligand complexes and appear to play an important role to rigidly anchor ligands in their LBP. The 25-hydroxyl group forms a hydrogen bond with the imidazole ring of H397 (helix 11), which acts as the hydrogen donor (1.98 Å). A similar bond is observed between H524 in ER and the 17-hydroxyl group of estradiol (E2) in the ER/E2 complex (9). We were not able to specify an amino acid residue interacting with the 3β-hydroxyl group of the ligand. One candidate is K240 at helix 3; however, the mutant VDR (K240A) has been known to cause only a small effect (1.3-fold decrease) on the affinity for the ligand as reported by Vaisanen et al. (29). It has been known that removal of either the 1α- or 25-hydroxyl group significantly decreases (1/500–1,000) the affinity for VDR but the removal of the 3β-hydroxyl group has a much smaller effect (1/17) (15). As described below, the C288A mutant significantly reduced both the affinity for 1,25-(OH)2D3 and transcriptional activity. This finding suggests that C288 may experience some interaction with the ligand. It has been demonstrated by Swamy et al.‖ that affinity labeling using 1,25-(OH)2D3 3β-bromoacetate took place exclusively at C288. In our model, the distance between the sulfur atom of C288 and 3-oxygen atom of 1,25-(OH)2D3 is 6.5 Å. Altogether we assume that C288 has hydrophobic contact with the ligand. These data also support the β-face up orientation of the ligand in our model.
Properties of Mutant hVDRs.
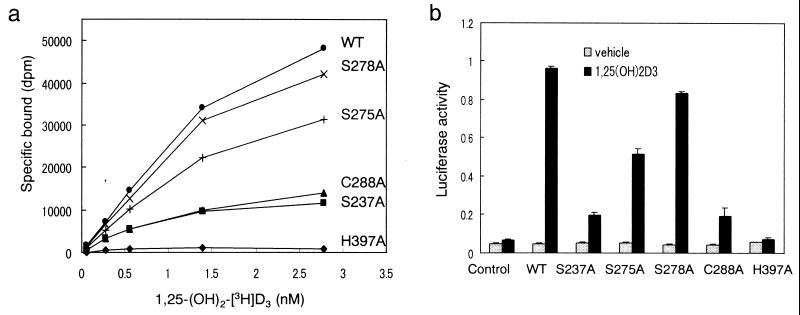
To substantiate our model, we prepared VDR mutants in which the following residues were individually replaced by alanine: S237, S275, S278, C288, and H397. All of these residues are conserved in the VDRs from various species and supposed to interact with the ligand in our model. The binding ability of the wild-type and mutant hVDRs for 1,25-(OH)2[3H]D3 was evaluated and the results are shown in Fig. 4a. In H397A, the ability to bind to the natural ligand was completely abolished. The affinity for the ligand was significantly reduced in S237A and C288A whereas the reduction in S275A and S278A was to a lesser extent. These results indicate that for ligand binding H397 is essential and S237 and C288 play an important role.
Figure 4.
Binding capability and transcriptional activity of the wild-type (WT) and mutant VDRs. (a) Specific binding to 1,25-(OH)2D3. The WT and the mutant hVDRs were synthesized in vitro in a rabbit reticulocyte lysate. The lysate was incubated with increasing concentrations of 1,25-(OH)2D3 for 16 h at 4°C. Bound and unbound ligands were separated by dextran-coated charcoal. (b) Transcriptional activity. COS-7 cells were cotransfected with WT or mutant hVDR expression vectors, SPPx3-TK-Luc as a reporter plasmid and pRL-CMV vector as an internal control. Before harvesting, cells were treated for 16 h with 10−8 M 1,25-(OH)2D3. Transactivation was determined by luciferase activity and normalized to the internal control.
Transcriptional activity was evaluated by using a dual luciferase assay system (Toyo Ink). The results are shown in Fig. 4b. The transcriptional activity of the five mutant VDRs was in parallel with their binding affinity for 1,25-(OH)2D3. The activity was completely abolished in H397A, significantly reduced in S237A and C288A, and weakly reduced in S275A and S278A. Immunoblotting of each VDR was conducted to assess the levels and stability of these proteins and confirmed that the mutations do not affect the expression or stability of the hVDR protein in COS-7 cells (data not shown).
We did not prepare a mutant of R274 but a mutant R274L has been known in human type II rickets (30). This mutant lacks activity in both ligand binding and transactivation. Thus, all evidence indicates that both H397 and R274 play a crucial role in ligand binding and firmly supports our model.
Docking of Other Ligands.
On the basis of a series of conformation-function relationship studies on vitamin D and analogs, we have reported that the regions occupied by the vitamin D side chain are classified into five areas, A, G, EA, EG, and F. Of these, the A, EA, and F regions are important for VDR binding and cell differentiation, and all three regions would be harbored in VDR-LBP. In Fig. 3 f and g, we draw the five spatial regions with yellow (upper A and lower G in Fig. 3f), red (F), and cyan (upper EA and lower EG in Fig. 3f) dots in the LBP of our VDR model. Thus, the side chain of 20-epi-1,25-(OH)2D3 would be harbored in a region visualized by the upper cyan dots (EA) and 22-oxa-1,25-(OH)2D3 in the red dot region (F). The green crosses in Fig. 3g show the trace of the histidine NH when the χ2 of H397 was rotated 360° with 10° intervals. Thus H397 can form a hydrogen bond at a space about 2 Å away from these crosses, showing a possibility that the same H397 is used to form a hydrogen bond with the 25-hydroxyl group of analogs in EA and F regions. In ER, H524 (helix 11) forms hydrogen bond with different ligands by rotating the χ2 of the histidine. In our model the imidazole ring of H397 cannot be accessed by side-chain conformations located in regions G and EG, which may be the reason vitamin D analogs show low potency when their side chains occupy these regions.
Discussion
It generally is accepted for homology modeling of proteins that the sequence identity of the objective and the template must be higher than 30%. Identity of the sequence between the LBDs of VDR and RAR is only 25% (similarity 45%) even excluding the loop 1–3. However, sequence identity and conservation of 3D structures are not going in parallel. Among the NR-LBDs, identity of signature regions, which constitute the AF-2 surface, is nearly 60% (31). Conversely, identity is low in the sequences constituting the LBP. However, this seems to be important to endow the specificity to each NR. As Fig. 2 a and b show, the framework structures at the LBP and AF2 regions are highly conserved among the NRs in the same group. So, as far as the 3D structures of the AF-2 and LBP are concerned, NR models created by homology modeling are thought to be considerably reliable.
We assigned the key residues forming hydrogen bonds with the two important hydroxyl groups of 1,25-(OH)2D3, namely S237 and R274 with the 1α-hydroxyl group and H397 with the 25-hydroxyl group. The importance of these residues for ligand binding and transactivation was confirmed by mutational analysis. Mutation studies reported previously are compatible with this model (29, 32). In our model the ligand does not directly interact with helix 12 unlike other NRs. However, analogs with epi-oriented and extended side chains, such as 24,26,27-trihomo-22-oxa-20-epivitamin D3 (KH1060), can form hydrophobic interaction between their terminal alkyl groups and the hydrophobic residues at helix 12 facing the ligand binding cavity, which explains the highest potency of KH1060 among vitamin D analogs.
Our model is similar to two other models (22, 23) in the structure of Cα framework and in the residues lining LBP but is quite different from them in the docking mode. In Moras and coworkers' model (22), 1,25-(OH)2D3 is docked with its A-ring heading helix 11, and in Norman et al.'s model (23), the ligand adopts a 6,7-s-cis conformation, though orientation of the long axis of the ligand is the same as ours.
Examination of the activities of mutants toward EA- and F-oriented analogs is important to further support our model structure and structure-function theory.
Acknowledgments
Technical support by M. Yanagisawa for the mutational analysis is gratefully acknowledged.
Abbreviations
- VDR
vitamin D receptor
- RAR
retinoic acid receptor
- 1,25-(OH)2D3
1α,25-dihydroxyvitamin D3
- LBP
ligand binding pocket
- LBD
ligand binding domain
- NR
nuclear receptor
- ER
estrogen receptor
- PR
progesterone receptor
- PPAR
peroxisome proliferator-activated receptor
- 3D
three-dimensional
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.020522697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.020522697
Swamy, N., Ray, R., et al. First International Conference on Chemistry and Biology of Vitamin D Analogs, September 26–28, 1999, Providence, RI.
References
- 1.Feldman D, Glorieux F H, Pike J W, editors. Vitamin D. New York: Academic; 1997. [Google Scholar]
- 2.DeLuca H F. J Bone Miner Metab. 1990;8:1–9. [Google Scholar]
- 3.Evans R M. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Nature (London) 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 5.Renaud J P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Nature (London) 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 6.Klaholz B P, Renaud J P, Mitschler A, Zusi C, Chambon P, Gronemeyer H, Moras D. Nat Struct Biol. 1998;5:199–202. doi: 10.1038/nsb0398-199. [DOI] [PubMed] [Google Scholar]
- 7.Wagner R L, Apriletti J W, McGrath M E, West B L, Baxter J D, Fletterick R. Nature (London) 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 8.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brzozowski A M, Pike A C, Dauter Z, Hubbard R E, Bonn T, Engstrom O, Ohman L, Greene G L, Gustafsson J A, Carlquist M. Nature (London) 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 10.Tanenbaum D M, Wang Y, Williams S P, Sigler P B. Proc Natl Acad Sci USA. 1998;95:5998–6003. doi: 10.1073/pnas.95.11.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 12.Williams S P, Sigler P B. Nature (London) 1998;393:392–396. doi: 10.1038/30775. [DOI] [PubMed] [Google Scholar]
- 13.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Nature (London) 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 14.Uppenberg J, Svensson C, Jaki M, Bertilsson G, Jendeberg L, Berkenstam A. J Biol Chem. 1998;273:31108–31112. doi: 10.1074/jbc.273.47.31108. [DOI] [PubMed] [Google Scholar]
- 15.Bouillon R, Okamura W H, Norman A W. Endocr Rev. 1995;16:200–257. doi: 10.1210/edrv-16-2-200. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto K, Takahashi J, Hamano K, Yamada S, Yamaguchi K, DeLuca H F. J Org Chem. 1993;58:2530–2537. [Google Scholar]
- 17.Yamamoto K, Ohta M, DeLuca H F, Yamada S. Bioorg Med Chem Lett. 1995;5:979–984. [Google Scholar]
- 18.Ishida H, Shimizu M, Yamamoto K, Iwasaki Y, Yamada S, Yamaguchi K. J Org Chem. 1995;60:1828–1833. [Google Scholar]
- 19.Yamamoto K, Sun W-Y, Ohta M, Hamada K, DeLuca H F, Yamada S. J Med Chem. 1996;39:2727–2737. doi: 10.1021/jm9600048. [DOI] [PubMed] [Google Scholar]
- 20.Yamada S, Yamamoto K, Masuno H, Ohta M. J Med Chem. 1998;41:1467–1475. doi: 10.1021/jm970761l. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto K, Ooizumi H, Umesono K, Verstuyf A, Bouillon R, DeLuca H F, Shinki T, Suda T, Yamada S. Bioorg Med Chem Lett. 1999;9:1041–1046. doi: 10.1016/s0960-894x(99)00129-8. [DOI] [PubMed] [Google Scholar]
- 22.Wurtz J-M, Guillot B, Moras D. In: Vitamin D: Chemistry, Biology, and Clinical Applications of the Steroid Hormone. Norman A W, Bouillon R, Thomasset M, editors. Riverside: Univ. of California-Riverside Printing and Reprographics; 1997. pp. 165–172. [Google Scholar]
- 23.Norman A W, Adams D, Collins E D, Okamura W H, Fletterick R J. J Cell Biochem. 1999;74:323–333. [PubMed] [Google Scholar]
- 24.Laudet V, Hanni C, Coll J, Catzeflis F, Stehelin D. EMBO J. 1992;11:1003–1013. doi: 10.1002/j.1460-2075.1992.tb05139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Umesono K, Murakami K K, Thompson C C, Evanas R M. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fagart J, Wurtz J M, Souque A, Hellal-Levy C, Moras D, Rafestin-Oblin M E. EMBO J. 1998;17:3317–3325. doi: 10.1093/emboj/17.12.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sicinski R R, Prahl J M, Smith C M, DeLuca H F. J Med Chem. 1998;41:4662–4674. doi: 10.1021/jm9802618. [DOI] [PubMed] [Google Scholar]
- 28.Bouillon R, Sarandese L A, Allewaert K, Zhao J, Mascarenas J L, Mourino A, Vrielynck S, de Clercq P, Vandewalle M J. J Bone Miner Res. 1993;8:1009–1015. doi: 10.1002/jbmr.5650080815. [DOI] [PubMed] [Google Scholar]
- 29.Vaisanen S, Rouvinen J, Maenpaa P H. FEBS Lett. 1998;440:203–207. doi: 10.1016/s0014-5793(98)01436-7. [DOI] [PubMed] [Google Scholar]
- 30.Kristjansson K, Rut A R, Hewison M, O'Riordan J L H, Hughes M R. J Clin Invest. 1993;92:12–16. doi: 10.1172/JCI116539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wurtz J M, Bourguet W, Renaud J P, Vivat V, Chambon P, Moras D, Gronemeyer H. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- 32.Nakajima S, Hsieh J-C, MacDonald P N, Galligan M A, Haussler C A, Whitfield G K, Haussler M R. Mol Endocrinol. 1994;8:159–172. doi: 10.1210/mend.8.2.8170472. [DOI] [PubMed] [Google Scholar]
- 33.Munson P J, Rodbard D. Anal Biochem. 1980;107:220–239. doi: 10.1016/0003-2697(80)90515-1. [DOI] [PubMed] [Google Scholar]