Summary
In the mitochondria of trypanosomatids, the majority of mRNAs undergo massive U-insertion/deletion editing. Throughout the processes of pre-mRNA polyadenylation, guide RNA (gRNA) uridylylation and annealing to mRNA, and editing reactions, several multi-protein complexes must engage in transient interactions to produce a template for protein synthesis. Here we report the identification of a protein complex essential for gRNA stability. The gRNA binding complex (GRBC) interacts with gRNA processing, editing and polyadenylation machineries and with the mitochondrial edited mRNA stability (MERS1) factor. RNAi knockdown of the core subunits, GRBC1 and 2, led to the elimination of gRNAs thus inhibiting mRNA editing. Inhibition of MERS1 expression selectively abrogated edited mRNAs. Homologous proteins unique to the order of Kinetoplastida, GRBC1 and 2, form a stable 200 kDa particle which directly binds gRNAs. Systematic analysis of RNA-mediated and RNA-independent interactions involving GRBC and MERS1 suggests a unified model for RNA processing in the kinetoplast mitochondria.
Introduction
The mitochondrial DNA of kinetoplastid protozoans is a dense catenated structure comprised of several maxicircles and thousands of minicircles. Maxicircle DNA represents a typical mitochondrial genome encoding ribosomal RNAs and proteins. Polycistronic transcripts of a maxicircle’s conserved region are processed by nucleolytic cleavage into rRNAs and pre-mRNAs, which are then 3′ uridylylated (Adler et al., 1991) and 3′ adenylated (Bhat et al., 1992), respectively. The uracil insertion/deletion RNA editing is required for the expression of ~2/3 of mitochondrial genes (reviewed in (Aphasizhev and Aphasizheva, 2008; Simpson et al., 2004; Stuart et al., 2005)). The cascade of editing reactions is directed by trans-acting gRNAs (Blum et al., 1990). These molecules are thought to be independently transcribed, primarily from the minicircle DNA, and 3′ uridylylated prior to hybridization with pre-edited mRNAs. Thus, maxicircle and minicircle genomes interact at the RNA level in a protein-mediated process to produce functional mRNAs.
Multi-protein complexes that carry out a cascade of editing reactions were studied in Trypanosoma brucei (Aphasizhev et al., 2003c; Carnes et al., 2008; Law et al., 2005; Law et al., 2007; Panigrahi et al., 2003; Panigrahi et al., 2006) and Leishmania tarentolae (Aphasizhev et al., 2003a). These efforts delivered a concise picture of a ~20-polypeptide particle, the 20S editosome, which contains activities for mRNA cleavage, U-insertion or deletion, and religation. The 20S particle, however, does not fully reflect the complexity of the editing interactome. Enzymatic reactions essential for the editing process also include gRNA 3′ uridylylation by RNA editing TUTase 1 (RET1) (Aphasizhev et al., 2002; Aphasizhev et al., 2003c).
Several proteins that bind to gRNA and/or mRNA in mitochondrial extract have been identified. The α2β2 complex of the mitochondrial RNA binding proteins 1 and 2 (MRP1/2) promotes annealing of complementary RNAs and interacts with the 20S editosome and RET1 (Allen et al., 1998; Aphasizhev et al., 2003b; Blom et al., 2001; Koller et al., 1997; Köller et al., 1994; Muller et al., 2001; Schumacher et al., 2006). The dual RNAi knockdown of MRP1/2, however, affected only two edited mRNAs and some never-edited transcripts (Vondruskova et al., 2005). Similar observations were made in cells depleted of the 16 kDa RNA binding protein (RBP16), which reportedly binds the gRNA’s oligo[U] tail (Hayman and Read, 1999; Pelletier et al., 2000; Pelletier et al., 2001; Pelletier and Read, 2003). REAP-1 protein, originally identified by binding to pre-edited mRNA, apparently plays a general role in mitochondrial RNA stability (Hans et al., 2007). A more selective effect, manifested by the decreased steady-state levels of edited mRNAs, was induced by repression of TbRGG1 (Hashimi et al., 2008). Guide RNA binding by RGG1 has not been established although its affinity to poly[U] RNA was demonstrated (Vanhamme et al., 1998). To conclude, the protein factors essential for gRNA maintenance in an organelle where numerous nuclease activities are present, including those possibly specific for gRNAs (Ryan et al., 2006), remained unknown.
We have reported earlier that the MRP1/2 complex from L. tarentolae, three polypeptides of ~50, 52, and 55 kDa (associated proteins 1, 2 and 3, (AP1-3)) and gRNAs co-fractionate through tandem affinity purification and glycerol gradient sedimentation as ~30S particle (Aphasizhev et al., 2003b). After subsequent identification of AP1-3 by mass spectrometry, we noticed an unexpected link: the orthologs of AP1 and 2 were also present in the T. brucei mRNA polyadenylation (KPAP1) complex (Etheridge et al., 2008). AP3, which contains a signature domain of the Nudix hydrolase superfamily, was not detected in the KPAP1 complex. We therefore sought to assess the RNA-mediated and RNA-independent interactions between gRNA binding, gRNA uridylylation, mRNA polyadenylation, and editing complexes, and to determine the functions of AP1-3 proteins. Here, we show that homologous proteins AP1 and AP2 form a α2β2-type particle, which is a core component of the gRNA binding complex (GRBC). These proteins have been renamed GRBC1 and 2, respectively. The heterotetramer directly binds gRNAs and is essential for stability of these molecules in mitochondria. AP3 is apparently required for the maintenance of edited mRNAs, and hence is re-named mitochondrial edited mRNA stability factor 1 (MERS1). GRBC interacts with RET1, MRP1/2, MERS1 and the polyadenylation complexes via RNA. Several common subunits associate with the GRBC, MERS1 and KPAP1 complexes.
Results
Interactions of RNA Processing Complexes in Mitochondria of L. tarentolae
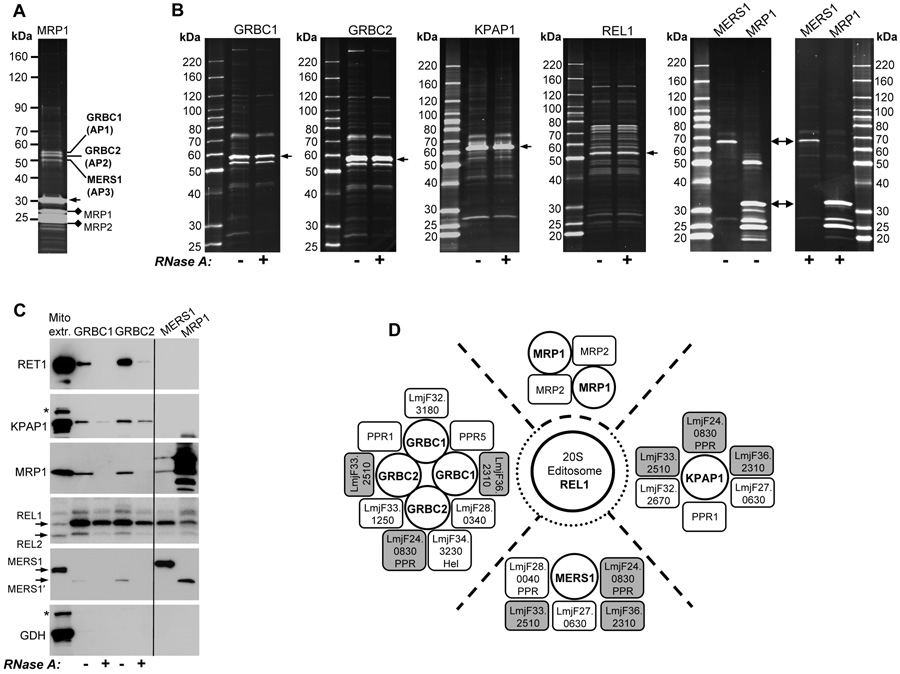
Three protein bands that co-purified with TAP-tagged L. tarentolae MRP1 (Aphasizhev et al., 2003b) were separated on a 8–16% SDS polyacrylamide gel (SDS-PAGE) and identified by mass spectrometry (Figure 1A, Table S1). The GRBC1 and 2 proteins show a high degree of similarity but no discernable functional motifs (Figure S1). These proteins were also detected in the T. brucei polyadenylation complex (Etheridge et al., 2008) and in the putative mitochondrial RNA binding complex 1 (put-MRB 1) (Panigrahi et al., 2007). Protein motif analysis delineated a NUDIX (nucleoside diphosphate linked moiety X) hydrolase domain in the C-terminal region of MERS1 (Figure S1).
Figure 1. RNA-mediated and RNA-independent Interactions of Editing and Polyadenylation Complexes in Mitochondria of L. tarentolae.
(A) Identification of GRBC1, 2 and MERS1. TAP-purified MRP1/2 complex was sedimented on 10–30% glycerol gradient. The ~30S peak fraction was separated on SDS-PAGE and stained with Sypro Ruby. Tagged MRP1 is shown by arrow. Individual bands (AP1, 2 and 3) were excised and digested with trypsin. Peptide sequences (Table S1) were matched to corresponding proteins by Blast searches of the L. major genome.
(B) Purification of GRBC1, GRBC2, KPAP1, REL1, MERS1 and MRP1 complexes. (+): The extract was treated with RNase A during IgG binding step of the TAP procedure. Protein profiles of the final fractions are shown.
(C) Interactions of GRBC with MERS1, KPAP1, MRP1/2, RET1 and 20S editosome. Equal protein loading from TAP-purified samples was verified by staining of the nitrocellulose membrane (Figure S2A), which was sequentially probed with polyclonal antibodies against RET1, KPAP1, MRP1, MERS1 and GDH. RNA ligases were visualized by self-adenylation. Cross-reacting bands in mitochondrial extract are depicted by asterisks.
(D) Schematic representation of RNA-mediated and RNA-independent interactions between GRBC, MERS1, MRP1/2, the 20S editosome and polyadenylation complexes. The cut off for inclusion of individual proteins into this model was arbitrary set at three or more peptide matches. The interaction was considered RNA-mediated if the RNase treatment caused more than a 50% drop in the number of unique peptides. See Table S2 and S3 for a complete list of detected proteins and Table S4 for the common contaminants found in all preparations. Proteins that were TAP-tagged are encircled. Common subunits are shaded in grey. RNA mediated contacts between complexes are shown by dashed lines; transient interactions are indicated by dotted lines. The proximity of polypeptides within distinct complexes reflects their co-purification under conditions used; it does not imply actual protein-protein interactions. With the exception of MRP1-2 and GRBC1-2, these contacts remain to be identified.
To determine the composition of GRBC1, 2, and MERS1-containing complexes and assess their protein-protein or RNA-mediated interactions with MRP1/2, the polyadenylation particle, and the 20S editosome, the L. major genes indicated in Figure 1B were expressed in L. tarentolae with a C-terminal TAP tag (Puig et al., 2001). All complexes were isolated under uniform conditions in the presence or absence of RNase A. Final fractions were digested with trypsin and analyzed by LC-MS/MS mass spectrometry (Table S2). The compositions of GRBC1 and 2 complexes were remarkably similar and included MRP1/2, MERS1 and several components of the polyadenylation complex such as pentatricopeptide repeat (PPR) proteins. Polypeptides overlapping with put-MRB1 complex included a putative DExH RNA helicase, proteins with Zn-finger and RRM motifs and four hypothetical proteins. In addition, three PPR proteins and polypeptides bearing no recognizable motifs were detected (Table S2). The high peptide yield of GRBC1 and 2 in reciprocal purifications was unaffected by RNAse treatment. In agreement with protein profiles, this illustrates that GRBC1 and 2 are the major subunits of the GRBC.
The 20S editosome components, RET1 and KPAP1 proteins were not detected by mass spectrometry in GRBC1 and 2 pull-downs. In a cross-tagging experiment, only two peptides from GRBC1 and a single GRBC2-derived peptide were found in the 20S editosome indicating that these are distinct complexes. To establish whether editing, gRNA uridylylation (RET1) and annealing (MRP1) and mRNA polyadenylation complexes interact with the GRBC, we used more sensitive radiolabeling and immunochemical methods. Purified fractions were incubated with [α-32P]ATP to self-adenylate editing ligases, separated by SDS-PAGE and sequentially probed for RET1, KPAP1, MRP1 and MERS1 on immunoblots (Figure 1C). The mitochondrial glutamate dehydrogenase served as negative control. Consistent with the data in Table S2, the MRP1/2-GRBC-MERS1 co-purification was eliminated by RNase treatment. Interestingly, it appears that MERS1 co-purifying with GRBC or MRP1/2 (MERS1′) undergoes proteolysis. RET1 is associated with GRBC, as demonstrated by Western blotting and TUTase activity (not shown), but these contacts are also RNA-dependent. RNase A treatment reduced but did not abolish GRBC-KPAP1 or GRBC-20S editosome interaction (Figure 1C). This implies either transient protein-protein interactions or participation of an RNase A-resistant structure, e.g. poly(A) or double-stranded RNA. RET1, MRP1/2 and MERS1 were not detected in the purified 20S editosome (Table S3, (Aphasizhev et al., 2003a)) further characterizing these RNA binding/processing entities as distinct protein particles engaged in transient or RNA-mediated contacts. A summary of interacting complexes is shown in Figure 1D.
Two Major Subunits of GRBC Form a Stable Complex
Mitochondrial extracts from Leishmania cells expressing TAP-tagged GRBC2 were mock-treated or incubated with RNase A at 0.1 mg/ml for 1h at 4 °C and fractionated on a 10–30% glycerol gradient. Each fraction was separated by 8–16% PAGE under native and denaturing conditions. Affinity-tagged GRBC2 was detected with peroxidase anti-peroxidase (PAP) reagent while the 20S editosome and MRP1 were monitored to verify the fractionation efficiency (Figure 2A). Two discrete GRBC2-containing particles of apparent masses ~200 and ~400 kDa, as determined by native gel migration, were observed to sediment in fractions 2 and 3 of the gradient (10–12S region). The higher-molecular weight complexes were detectable in fractions 4–8 (12–20S region) upon overexposure (not shown). Similar experiments with tagged GRBC1 produced virtually identical sedimentation and gel migration patterns (Figure S2B) indicating that these two proteins may interact directly.
Figure 2. GRBC1 and 2 Form a Complex.
(A) Two-dimensional size-fractionation of the mitochondrial extract. Mitochondrial fraction was purified from L. tarentolae cells expressing TAP-tagged GRBC2. Mitochondrial extract (~1 mg of total protein) was fractionated on 10–30% glycerol gradient. Fractions (0.7 ml) were collected from the top, separated on native or denaturing gradient PAGE, and probed for RNA editing ligases (adenylation), tagged protein and MRP1. Catalase [11S], thyroglobulin [19S] and E. coli 30S ribosome subunit were used as sedimentation standards.
(B) RNase A sensitivity of GRBC2 complexes. Mock-treated and RNase-treated extracts were fractionated as in A. Fractions 2 and 3 were separated on 8–16% native PAGE and probed for TAP-tagged GRBC2.
(C) Reconstitution of the GRBC1/2 particle. Respective genes were cloned into T7 RNA polymerase-driven expression vector and coupled transcription-translation was performed in the presence of [35S] methionine. Reactions were separated on 8–16% native gradient gel alongside gradient fraction #3 (from panel A) and native size markers. The tagged GRBC2 and radiolabeled proteins were detected by Western blotting and exposure to the phosphor storage screen, respectively. Probable oligomeric states corresponding to the observed apparent molecular masses are denoted by arrows.
RNase treatment did not affect the sedimentation profile of the GRBC2 particle but led to a re-distribution of signal between the 200 and 400 kDa bands. Quantitative Western blotting of fraction 3 from untreated and RNase-treated gradients showed a change in the upper to lower band percentage ratio from 60/40 to 20/80, respectively (Figure 2B). This suggests an RNA-dependent oligomerization, but also indicates the presence of a tightly bound RNA which is partially resistant to RNase. A similar collapse of the 400 kDa into the 200 kDa band was observed upon multiple cycles of freeze-thawing (not shown).
The calculated molecular masses of tagged GRBC1 and 2 proteins lacking predicted mitochondrial importation signals are ~52 and 57 kDa, respectively. However, the smallest particles observed in gradient fractions from both extracts had a virtually identical apparent mass of 200–210 kDa. We next asked if this particle represents a GRCB1-GRBC2 oligomer. Individual proteins expressed in bacteria were insoluble (not shown) and could only be purified as a complex from Leishmania. For that reason, GRBC1 and 2 were synthesized separately or co-synthesized in an in vitro coupled transcription/translation system. The reaction mixtures were centrifuged to remove insoluble proteins and ribosomes and separated under native (Figure 2C) and denaturing conditions (Figure S2C). Individual GRBC2 was soluble, while ~80% of GRBC1 was removed by centrifugation (Figure S2C). In the case of co-synthesis, a complex of ~200 kDa was detected on a gradient native gel. MERS1 did not associate with either protein and migrated at ~55 kDa in close agreement with its calculated mass. As a control, fraction 3 from the untreated glycerol gradient was run alongside and probed for the GRBC2-TAP fusion (Figure 2C). Based on the apparent molecular masses, individual GRBC2 may form a dimer or a tetramer, but in the presence of GRBC1, it readily assembles into a particle with a molecular mass of a heterotetramer. Thus, GRBC1 and 2 form a stable complex. In mitochondria, this particle is engaged in higher-order RNA-mediated and RNA-independent associations.
GRBC1, 2 and MERS1 are Essential for Parasite Viability and the Editing Process
The roles of GRBC1, 2 and MERS1 in mitochondrial RNA processing were examined by inducible RNAi knockdown in the insect (procyclic) form of T. brucei. Upon RNAi induction in all three cell lines, a severe growth inhibition at 72–96 hr was followed by elimination of live parasites in the culture (Figure 3A). Three representative mitochondrial mRNAs were analyzed by “poisoned” primer extension at early time-points (Figure 3B). The abundance of never-edited cytochrome oxidase subunit 1 (CO1) and cis-edited subunit 2 (CO2) mRNAs remained unchanged. The edited form of the pan-edited subunit 3 (CO3) mRNA was reduced by ~85% in RNA isolated from GRBC1 and 2 RNAi cells and by ~60% in MERS1 RNAi cells. Remarkably, the abundance of pre-edited CO3 mRNA increased in GRBC1 or 2-depleted parasites, congruent with results observed in RNAi knockdowns of several 20S editosome’s components (for example (Trotter et al., 2005)) or RET1 RNAi (Aphasizhev et al., 2002). Conversely, the level of CO3 pre-edited mRNA did not change during MERS1 RNAi.
Figure 3. Effects of GRBC1, 2 and MERS1 Repression on Cell Viability and the Abundance of Mitochondrial RNAs.
(A) Growth kinetics of RNAi cell lines after mock induction (closed squares) or addition of tetracycline (open diamonds).
(B) “Poisoned” primer extension analysis of mitochondrial mRNAs. DNA oligonucleotides were hybridized with total RNA isolated at indicated time points of RNAi induction. Reverse transcription was performed in the presence of ddGTP and reaction products were separated on 10% polyacrylamide/urea gel. P - pre-edited mRNA; E - edited mRNA. Only extension products are shown, 5′ labeled primers are omitted. The decrease in abundance was calculated assuming the mitochondrial mRNA/β -tubulin mRNA ratio in mock-induced cells as 100%.
(C) Quantitative RT-PCR analysis of mitochondrial mRNAs, 12 S rRNA and RNAi-targeted transcripts. The RNA levels were normalized to β-tubulin mRNA. P – pre-edited mRNA; E - edited form. Error bars represent the standard deviation from at least three replicates. The thick line at “1” stands for no change in mRNA’s relative abundance with bars above or below representing an increase or decrease, respectively.
To establish the general nature of the observed RNAi effects, the change in relative abundance of six pre-edited, corresponding edited and three never-edited mRNAs was determined by quantitative RT-PCR (Figure 3C). Uniformly, the 12S ribosomal RNA and never-edited mRNAs remained unaltered. All edited transcripts showed a 75–95% decline in abundance. In agreement with primer extension data, most pre-edited mRNAs increased by 2–5 fold in GRBC1 and 2 RNAi; the same RNAs were unaffected or increased by ~50% in the MERS1 knockdown cells. The pre-edited mRNAs are relatively stable in mitochondria and typically accumulate if not processed by editing (Etheridge et al., 2008), suggesting that the repression of either GRBC1 or 2 expression has a direct impact on editing. On the contrary, the pre-edited mRNAs did not accumulate in MERS1 RNAi cells signifying an unaffected editing process, but accelerated decay of edited transcripts.
Editing Complexes Remain Functional in GRBC1, 2 and MERS1-Depleted Mitochondria
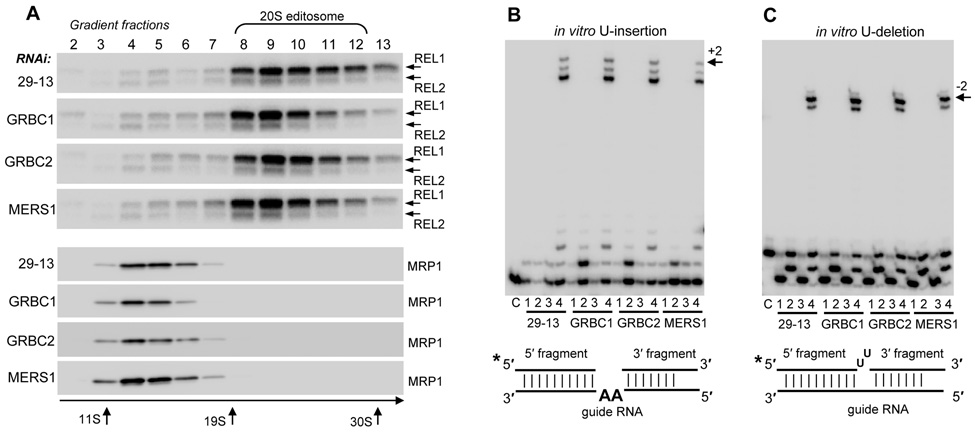
Because the edited mRNAs declined in parasites lacking GRBC1, 2 and MERS1, we next investigated if the integrity and activity of the 20S editosome, MRP1/2 and RET1 complexes were compromised. Mitochondrial fractions were purified from the T. brucei parental cell line (29–13) and RNAi cells at ~68 hours post-induction. The extracts were fractionated on 10–30% glycerol gradients and each fraction was tested for the 20S editosome and MRP1/2 (Figure 4A). Glycerol gradient sedimentation patterns are sensitive to even minor alterations in the editing complexes (Kang et al., 2006; Aphasizhev et al., 2003c), yet no apparent differences were observed. Sedimentation patterns of RET1 and MERS1 were also unaffected (not shown). Peak fractions (#9) from all gradients showed virtually identical U-insertion (Figure 4B) and U-deletion (Figure 4C) editing activities on a pre-cleaved mRNA substrate (Igo et al., 2000). These data confirm that the 20S editosome, MRP1/2, RET1, MERS1 and GRBC are likely to be discrete complexes - knockdown of an integral subunit often leads to the destabilization of the respective particle (Law et al., 2007; Vondruskova et al., 2005). To conclude, the GRBC1, 2 and MERS1 RNAi knockdowns did not compromise the activity, abundance or integrity of RNA editing complexes.
Figure 4. Depletion of GRBC1, 2 and MERS1 does not Affect RNA Editing Complexes.
(A) The effect of GRBC1, 2 or MERS1 repression on the integrity of the 20S editosome and MRP1/2 complex. Mitochondrial extracts from respective RNAi cell lines were sedimented on 10–30% glycerol gradients and 0.6 ml fractions were collected. Each fraction was tested for REL1 and REL2 editing ligases, and MRP1.
(B) U-insertion editing activity in peak gradient fractions. RNA substrates for the pre-cleaved editing assay were assembled as follows 1) 5′ fragment; 2) 5′ fragment + “guide” RNA; 3) 5′ fragment + 3′ fragment; 4) fully assembled substrate for +2 addition, 5′ fragment + 3′ fragment + “guide RNA”. The position of +2 edited products is shown by arrow.
(C) U-deletion editing activity in peak gradient fractions. RNA substrates for the pre-cleaved editing assay were assembled as in (B).
mRNA Polyadenylation is Not Affected by GRBC1, 2 or MERS1 RNAi
Polyadenylation differentially influences the stability of pre-edited and edited mRNAs in mitochondrial extracts (Kao and Read, 2005) and determines the abundance of edited and never-edited transcripts in vivo (Etheridge et al., 2008). Because GRBC interacts with the polyadenylation complex (Figure 1), we next analyzed if the decline of edited transcripts in GRBC1, 2 and MERS1-depleted cells was caused by inhibition of mRNA polyadenylation. The short (~20 nts) poly(A) tails are found in both pre-edited and edited mRNA species (Read et al., 1994). These structures, although not required for the stability of pre-edited mRNAs, are essential for the maintenance of the edited transcripts (Etheridge et al., 2008). Therefore, the pre-edited mRNAs represent a convenient target for analyzing polyadenylation defects.
Total RNA was separated on a 6% polyacrylamide/ 8M gel and sequentially probed for pre-edited, partially-edited (5′ region unedited) and fully-edited forms of RPS12 mRNA. As seen in Figure 5A (pre-edited panel), the short A-tails (ST) remained intact and pre-edited mRNAs accumulated in GRBC1 and 2 RNAi cells, increasing by approximately four-fold. This indicates a disabled editing process. Conversely, the abundance of pre-edited mRNA did not change in MERS1 RNAi cells, suggesting that pre-edited RNA was consumed by editing. The fully-edited mRNAs bearing a short A-tail and long (A/U) tail (Etheridge et al., 2008) and the partially-edited transcripts were effectively reduced in all RNAi cell lines. Quantitation of Northern blotting for never-edited CO1 mRNA in the same RNA samples revealed no major differences among GRBC1, 2 and MERS1 knockdowns (Figure 5B). Sedimentation of KPAP1 complexes was not affected by GRBC1 or 2 RNAi (not shown). Collectively, these data illustrate operative polyadenylation process in GRBC1, 2 and MERS1 RNAi cells.
Figure 5. mRNA Polyadenylation in Mitochondria Lacking GRBC1, 2 or MERS1.
(A) Northern blotting of pre-edited, partially edited and fully-edited RPS12 mRNAs. RNA was isolated from GRBC1, 2 and MERS1 mock-induced (−) or tetracycline induced (+) cells and separated on 6% acrylamide/urea gel. [dT]: RNA was treated with RNase H in the presence of 18-mer [dT]. (−): RNase H treatment in the absence of oligo[dT]. 12S rRNA: mitochondrial large subunit rRNA. ST: short (A)-tail. LT: long tail of never-edited mRNAs. A/U: long (A/U) tail of fully-edited mRNAs. NA: non-adenylated transcript.
(B) Northern blotting of the never-edited CO1 mRNA. Total RNA from parental cell line (29–13) and induced RNAi cells was separated on 1.8% agarose formaldehyde gel and probed for CO1 mRNA, cytosolic 18S and mitochondrial 12S rRNAs.
Loss of gRNAs in GRBC1 or 2-depeleted Mitochondria
The repression of GRBC1, 2 and MERS1 abolished editing of all tested mRNAs, except the cis-guided insertion of four Us into cytochrome oxidase subunit 2 (CO2) transcript (Benne et al., 1986). Combined with data demonstrating intact transcription, RNA editing and polyadenylation processes, this observation led us to inquire if guide RNAs were affected. The overall gRNA abundance and size distribution were assayed via 5' labeling of primary transcripts with guanylyltransferase in the presence of [α-32P]GTP (Blum et al., 1990). As a control for the loss of oligo[U] tails in gRNAs (Aphasizhev et al., 2003c), RNA isolated from RET1 RNAi cells was also subjected to G-labeling. The overall gRNA abundance decreased by ~70% and ~50% in GRBC1 and 2 RNAi cells, respectively. A minor increase was seen upon repression of MERS1 (Figure 6A).
Figure 6. Binding to GRBC is Essential for the gRNA Stability.
(A) Analysis of total gRNA abundance. RNA was isolated from purified mitochondria (kRNA), parental cell line (29–13), RET1, GRBC1, 2 and MERS1 RNAi cells at ~ 65 hr post-induction. Guide RNAs were 5′ labeled with [α-32P]GTP in the presence of vaccinia virus guanylyltransferase and separated on 10 % polyacrylamide/8M urea gel. The unidentified cytosolic RNA labeled with [α-32P]GTP was used as a loading control. Quantitation area is shown by rectangle. The ratio of gRNA vs. cytoplasmic RNA in mock-induced cells was used as 100%; the percentage of total gRNA remaining is provided under RNAi (+) lines.
(B) Northern blotting of individual gRNAs. RNAs from the parental cells (29–13) and indicated cell lines with induced RNAi were separated on 12% polyacrylamide/8M urea gels and sequentially hybridized with oligonucleotide probes. Splice leader (SL) and 9S ribosomal RNAs were used as loading controls.
(C) Total gRNA labeling in RNA isolated from MRP1/2 RNAi cells.
(D) Northern blotting of individual gRNAs in MRP1/2 RNAi cells. RNA was isolated from the parental cells (29–13) and cell lines with induced RNAi.
(E) gRNA association with affinity-purified editing complexes. RNA was extracted from purified GRBC1, 2 and MRP1/2 complexes and the 20S editosome (Figure S3C), G-labeled and analyzed as in (A).
(F) UV-induced gRNA crosslinking with affinity-purified complexes. The major crosslink in GRBC1 and 2 fractions is depicted by the arrow (left panel). Sypro Ruby-staining of protein fractions subjected to cross-linking is shown on the right panel. TEV protease, which was observed to crosslink to RNA non-specifically, and bovine serum albumin (BSA) were included as controls. Note equal crosslinking efficiency of the gRNA and RPS12 mRNA fragment with TEV protease.
(G) gRNA stability in T. brucei cells depleted of GRBC1. After 42 h of GRBC1 RNAi induction, Actinomycin D and ethidium bromide were added to inhibit transcription, and 5 × 107 cells were collected at indicated time points. Total RNA was separated on 10% polyacrylamide/8M urea gel, transferred onto membrane and sequentially probed for mitochondrial gRNAs and 9S ribosomal RNA, and nuclearly encoded SL RNA. Nuclearly encoded tRNACys(GCA), which is imported into mitochondria (J. Alfonzo, personal communication), was used as normalization standard. The decrease in relative abundance was calculated assuming the gRNA/tRNA ratio in mock-induced cells at the time of Actinomycin D addition as 100%.
To further validate these results for specific gRNAs and also to examine if gRNAs encoded in minicircles (gRPS12[64], gCyb[560]) and maxicircles (gMurf2[II]) are equally affected, we probed total RNA for the species shown in Figure 6B and Figure S3A. In agreement with G-labeling results, the abundance of all gRNAs tested decreased by 70–95%, depending on the species but not the gene location. These results indicate that GRBC1 and 2, but not MERS1, are required for gRNA maintenance in the mitochondria of trypanosomes.
gRNAs were unaffected in RNAi knockdowns of RBP16 (Pelletier and Read, 2003) and RGG1 (Hashimi et al., 2008), but the possible role of MRP1/2 in gRNA maintenance has not been established. The dual RNAi knockdown of mitochondrial RNA binding proteins 1 and 2 eliminated editing of Cyb, had a moderate effect on RPS12 mRNA, and decreased the abundance of never-edited CO1 mRNA ((Vondruskova et al., 2005), Figure S3B). To examine the possible role of MRP1/2 in gRNA biogenesis or stability, we analyzed the effect of dual MRP1 and 2 knockdown on the gRNA population. Total gRNA (Figure 6C) and specific gRNAs participating in the editing of transcripts most impacted by MRP1/2 RNAi (Figure 6D), were largely unaffected as compared to the parental cell line (29–13) or KPAP1 RNAi.
gRNA Binding to GRBC
To determine if gRNAs are associated with GRBC and MERS1, RNA was extracted from the tandem affinity-purified fractions and G-labeled with guanylyltransferase (Figure 6E). Purified MRP1/2, which binds gRNA with high affinity (Aphasizhev et al., 2003b; Schumacher et al., 2006) served as a positive control. The 20S editosome ((Aphasizhev et al., 2003a), Figure S3C) was also included in the analysis. All fractions were adjusted for the bait protein and laced with a 24-mer RNA tracer to control the uniformity of RNA isolation. Although gRNAs from GRBC1 and 2 samples were degraded to some degree, the co-purification shows that under the experimental conditions used (150 mM KCl, 1 mM EDTA) gRNAs are stably associated with GRBC. The reason for gRNA’s partial degradation is unclear as we did not detect any nuclease activity in the purified complexes (not shown). gRNAs were not observed in the MERS1 fraction.
Sequence analysis of GRBC1 and 2 proteins did not reveal similarity to any known RNA binding motif. However, several components of the complex have the potential to bind RNA, e.g., pentatricopeptide repeat proteins, Zn-finger subunit and a putative RNA helicase. To elucidate which specific protein is responsible for gRNA binding, we performed short UV-induced RNA-protein crosslinking. The gND7[506] was synthesized by in vitro transcription with T7 RNA polymerase in the presence of [α-32P]ATP. The 3′ fragment of edited RPS12 mRNA of approximately the same length but with a different predicted secondary structure (Figure S4) was used as a control. In a label-transfer experiment, TAP-purified fractions were UV-crosslinked with radioactive RNAs, treated with RNase A, separated on an SDS gel (Figure 6F), and transferred to a nitrocellulose membrane. The membrane was stained with Sypro Ruby to assure concurrence between the radioactive signals (Figure 6F, left panel) and the protein bands (Figure 6F, right panel). The major crosslinks in GRBC1 and 2 TAP fractions juxtapose well with GRBC1/2 bands, but unambiguous identification of the crosslinked polypeptide was precluded by similar gel migration. The signal intensity, nonetheless, was virtually identical in GRBC1 and 2 complexes as was the peptide yield for GRBC1 (Table S2). GRBC1 may have crosslinked more efficiently than GRBC2 although its affinity-purified fraction contained less gRNA (Figure 6E), suggesting that both homologous proteins are involved in RNA binding. MRP1/2 predictably generated a strong crosslinking signal with gRNA.
GRBC is Essential for gRNA Stability
GRBC1 or 2 RNAi abrogated minicircle- and maxicircle-encoded gRNAs but had no effect on the synthesis and processing of the precursor which yields rRNA, pre-edited and never-edited pre-mRNAs. The direct binding of gRNA argues for GRBC’s role in regulating gRNA stability. Still, it is possible that gRNA transcription was selectively impaired. To gain further mechanistic insight, we have developed a gRNA stability assay. The time of GRBC1 RNAi induction was optimized to achieve more than 90% depletion of the targeted mRNA while retaining sufficient amount of gRNAs for Northern blotting detection. After blocking mitochondrial transcription with a combination of Actinomycin D and ethidium bromide, the change in gRNA relative abundance was monitored over eight hours period of time. The nuclearly encoded tRNACys, which is imported into the mitochondria, was found to be sufficiently stable to serve as a normalization control. As seen in Figure 6G, the stability of individual gRNAs varies in mockinduced RNAi cells but ultimately decreases upon repression of GRBC1. Hence, we concluded that GRBC acts as a gRNA-stabilizing factor in the mitochondria of trypanosomes.
Discussion
Previous studies of guide RNA binding proteins have produced a substantial amount of data on polypeptides that function in editing and apparently in other processes (reviewed in (Pelletier et al., 2007)). Here, we identified two homologous proteins, GRBC1 and GRBC2, and a protein bearing the NUDIX domain, MERS1. We show that these proteins are essential for the viability of Trypanosoma brucei and are critical for the RNA editing process. In a remarkable parallel to MRP1/2 RNA chaperones, GRBC1 and 2 are engaged in protein-protein interactions forming a ~200 kDa heterotetramer, which directly binds gRNA. This particle has been reconstituted in vitro opening an opportunity for structural examination of this RNA binding module. Upon overexpression in L. tarentolae, unassociated GRBC1 or 2 were not detected, suggesting that individual proteins are unstable in the mitochondria. The GRBC1/2 particle forms a higher-order complex with several polypeptides designated as the gRNA binding complex, GRBC.
We have confirmed the previously reported association of the GRBC with the mitochondrial polyadenylation complex in T. brucei and partial sensitivity of these contacts to RNase treatment (Etheridge et al., 2008). The overlap between GRBC and KPAP1 complexes in closely related species was expected; of particular interest is the presence of two shared pentatricopeptide repeat proteins, LmjF18.0010 (PPR1) and LmjF24.0830. MERS1 forms a discrete assembly with five polypeptides of which three are shared with GRBC and KPAP1 complexes. All interactions of MRP1/2 appear to be RNA-mediated. PPR5 was detected in GRBC, KPAP1 and MRP1/2 preparations but its repression reportedly leads to the loss of ribosomal RNAs (Pusnik et al., 2007). Further studies are required to elucidate specific functions of each protein identified in GRBC.
Biochemical evidence points out that MRP1/2 and RBP16 are likely to be involved in gRNA use by the editing machinery at the gRNA-mRNA annealing step. The RNAi repression of RGG1 led to a decline of edited mRNAs but the mechanism is less clear. In any event, depletion of these proteins had no effect on the gRNA population (Figure 6, (Hashimi et al., 2008; Pelletier and Read, 2003)). We demonstrate that repression of GRBC1 or 2 by RNAi causes a loss of gRNAs and decreases the abundance of all tested edited mRNAs with the exception of cis-edited CO2. The mRNA editing, polyadenylation, gRNA 3′ processing and annealing complexes remained unaffected, supporting the conclusion that selective inhibition of editing was caused by the loss of gRNAs. Measurements of gRNA turnover show a faster decay of these molecules in the GRBC1-depleted cells. This finding directly implicates binding to GRBC as essential for gRNA protection against nucleolytic degradation. Interestingly, low levels of several non-uridylylated gRNAs were detected in RET1-depleted cells whereas no decay intermediates were observed in GRBC1 or 2 RNAi knockdowns. These results indicate that 3′ uridylylation by RET1 is also required for gRNA maintenance, but probably occurs after gRNA’s association with GRBC. The mode of action for MERS1 is distinctly different; upon analysis of editing, polyadenylation and gRNA processing in MERS1-depleted cells, we conclude that MERS1 is most likely required for the stability of edited mRNAs.
GRBC associates with the polyadenylation and editing machineries but the RNAi knockdowns of components integral to each specific assembly, e.g., GRBC1 or 2, REL1, RET1, MERS1, MRP1/2 or KPAP1 had no discernable effects on interacting complexes, although often compromised respective resident particles. This suggests a model in which distinct entities containing common subunits interact transiently or via an RNA component, rather than through shared polypeptides. For example, hybridization of the gRNA’s U-tail with a short A-tail found in all pre-mRNAs may cause GRBC-KPAP1 association. Coherent with accumulating evidence for RET1’s involvement into mRNA 3′ end processing (Etheridge et al., 2008; Ryan and Read, 2005), this may have an additional benefit of tethering RET1 to the 3′ end of mRNA for subsequent addition of the long A/U tail. These interactions may be facilitated by RNA binding factors such as PPR1, a protein required for the long tail formation in both never-edited and fully-edited mRNAs (Pusnik et al., 2007). Based on data reported here and recent findings in the field, we propose a model of RNA processing in the mitochondria of trypanosomes (Figure 7). Although speculative and only partially inclusive, the model does not contradict existing knowledge and has a certain predictive power.
Figure 7. Model for RNA Processing in the Mitochondria of Trypanosomes.
Two transcript types are produced by the mitochondrial RNA polymerase from maxicircle and minicircle DNA: multi-cistronic precursor RNA and gRNAs, respectively. A few gRNAs are transcribed from maxicircles as independent units. The precursor is processed into rRNAs, which are uridylylated, and into pre-mRNAs of two kinds: never-edited (contain open reading frame) and pre-edited (must be edited prior to translation). Both kinds undergo addition of ~20 As by KPAP1; the 3′ adenylation is required for the stability of never-edited RNAs but is dispensable for the maintenance of pre-edited transcripts. The primary gRNA transcripts are stabilized via binding to GRBC and 3′ uridylylation by RET1. The GRBC-gRNA-RET1 complex associates with the polyadenylation complex bound to A-tailed pre-edited and never-edited mRNAs. Partial RNase A resistance of GRBC-KPAP1 contacts agrees with the mRNA’s A-tail, or a hybrid formed by gRNA’s U-tail and mRNA’s A-tail, as being possible bridging elements. This association brings essential factors to the 3′ ends of both mRNA types: PPR1 (Pusnik et al., 2007; Mingler et al., 2006) and, possibly, RET1, for addition of long tails to never-edited mRNAs, and gRNAs for editing of pre-edited transcripts. The ~ 20 bp-long gRNA-mRNA “tail-tail” hybrid is thermodynamically more stable that most gRNA-mRNA “anchor” hybrids, and may be more efficient in the initial tethering of these molecules. gRNA’s “anchor” sequence, which is typically 5–10 nucleotides-long, is then engaged in specific hybridization with pre-edited mRNA by MRP1/2 chaperone and/or RBP16. The gRNA binding generates a substrate for editing endonucleases and initiates the 20S editosome recruitment. Upon progression of editing beyond the first block, the mRNA is subjected to surveillance process. A short A-tail in partially-edited mRNAs is a required cis-element for mRNA stability; the RNAi knockdown of MERS1 (this study) and RGG1 (Hashimi et al., 2008) put these proteins forward as potential trans-acting stabilizing factors. Finally, upon completion of the editing process, the long A/U tail is added to the fully-edited mRNAs concluding mRNA maturation and marking translationally-competent molecules.
Experimental Procedures
Cell Cultures and RNAi
L. tarentolae cells were transfected with pTAP-based plasmids (Aphasizhev et al., 2003a), and maintained in brain heart infusion medium supplemented with 100 mg/L of Geneticine (G418, Sigma). Ten liter batches were grown to 250 × 106 cells/ml in a Biostat C10 Fermentor (Sartorius). Mitochondria were isolated on Renografin density gradients (Pelletier et al., 2007). Clonal inducible RNAi cell lines were obtained by transfecting p2T7-177-based (Wickstead et al., 2002) plasmids into procyclic T. brucei strain 29–13 (Wirtz et al., 1999). RNAi was induced with 10 µg/ml of tetracycline. For RNA analysis and mitochondrial isolation cells were collected at 48–55 hours post induction unless otherwise indicated (Aphasizhev and Aphasizheva, 2007). Details of plasmids construction and oligonucleotides are provided in the Supplemental Data.
Affinity Purification and Mass Spectrometry
Tandem affinity purification was performed using highly-enriched mitochondria (3–4 g of wet weight, 150–200 mg of total protein) as previously described (Pelletier et al., 2007). Unless otherwise noted, KCl was maintained at 150 mM and EDTA was at 1 mM during extract preparation and the IgG chromatography step. For RNase treatment, RNase A was added to the 50 µg/ml. The details of protein identification are provided in Supplemental Data.
Gradient Fractionation, Native Gel Electrophoresis, in vitro Editing and Western Blotting
Mitochondrial extract was prepared as per TAP purification and spun on a 10–30% glycerol gradient containing 25 mM HEPES (pH 7.6), 150 mM KCl, 1 mM EDTA and 1 mM CHAPS for 20 h in a SW41 rotor at 111,000 g. Sedimentation values were calculated using catalase (11.5S), thyroglobulin (19.3S) and 30S bacterial ribosome subunits. Self-adenylation of RNA ligases was carried out with 5 µCi of [α-32P]ATP per 10 µl of gradient fraction followed by electrophoresis on an 8–16% Tris-glycine gel for 16 h at 4 °C, and tank-transfer for 10 h. For analysis under denaturing conditions, the reactions were separated on SDS gels and blotted onto nitrocellulose membrane. Quantitative chemiluminescent images were acquired with LAS-4000 digital analyzer (Fujifilm) and automatically superimposed with native high molecular mass markers (GE Healthcare). The pre-cleaved in vitro editing reactions were performed as described (Aphasizhev and Aphasizheva, 2007).
In vitro Transcription/Translation
Genes lacking sequences corresponding to the predicted mitochondrial importation peptides were cloned into a pET15b vector (Novagen) and expressed in the TNT T7 Quick coupled transcription/translation system (Promega) as recommended by manufacturer. For co-synthesis, 0.5 µg of each plasmid was added to a 20 µl reaction and incubated for 90 min at 30 °C. The reaction mixture was centrifuged for 30 min at 250,000g and was loaded onto 8–16% native Tris-glycine gel, which was run and electroblotted as described for glycerol gradient fractions.
RNA Analysis and UV crosslinking
Real-time quantitative PCR, “poisoned” primer extensions, mRNA Northern blotting and gRNA G-labeling were performed as described earlier (Etheridge et al., 2008). For RNA isolation from affinity purified complexes, the amount of tagged subunit was adjusted to ~ 1 µg. Details of gRNA stability assay and UV crosslinking experiments are described in Supplemental Data.
Supplementary Material
Acknowledgments
We thank Larry Simpson for his unconditional support at the initial stages of this project and members of our laboratory for reading the manuscript. We appreciate Juan Alfonzo’s advice on mitochondrial tRNA detection, and helpful suggestions from the reviewers. This work was supported by NIH grants AI064653 and AI073810 to RA, and GM74830 to LH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
James Weng, Department of Microbiology and Molecular Genetics, School of Medicine, University of California, Irvine, CA 92697, USA..
Inna Aphasizheva, Department of Microbiology and Molecular Genetics, School of Medicine, University of California, Irvine, CA 92697, USA..
Ronald D. Etheridge, Department of Microbiology and Molecular Genetics, School of Medicine, University of California, Irvine, CA 92697, USA.
Lan Huang, Department of Physiology & Biophysics, School of Medicine, University of California, Irvine, CA 92697, USA..
Xiaorong Wang, Department of Physiology & Biophysics, School of Medicine, University of California, Irvine, CA 92697, USA..
Arnold M. Falick, Howard Hughes Medical Institute Mass Spectrometry Laboratory and Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA.
Ruslan Aphasizhev, Department of Microbiology and Molecular Genetics, School of Medicine, University of California, Irvine, CA 92697, USA..
Reference List
- Adler BK, Harris ME, Bertrand KI, Hajduk SL. Modification of Trypanosoma brucei mitochondrial rRNA by posttranscriptional 3' polyuridine tail formation. Mol. Cell. Biol. 1991;11:5878–5884. doi: 10.1128/mcb.11.12.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TE, Heidmann S, Reed R, Myler PJ, Goringer HU, Stuart KD. Association of guide RNA binding protein gBP21 with active RNA editing complexes in Trypanosoma brucei. Mol Cell Biol. 1998;18:6014–6022. doi: 10.1128/mcb.18.10.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I. RNA Editing Uridylyltransferases of Trypanosomatids. Methods Enzymol. 2007;424:51–67. doi: 10.1016/S0076-6879(07)24003-0. [DOI] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I. Terminal RNA uridylyltransferases of trypanosomes. Biochim. Biophys. Acta. 2008;1779:270–280. doi: 10.1016/j.bbagrm.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003a;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Nelson RE, Simpson L. A 100-kD complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA. 2003b;9:62–76. doi: 10.1261/rna.2134303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Simpson L. A tale of two TUTases. Proc. Natl. Acad. Sci. U. S. A. 2003c;100:10617–10622. doi: 10.1073/pnas.1833120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Sbicego S, Peris M, Jang SH, Aphasizheva I, Simpson AM, Rivlin A, Simpson L. Trypanosome Mitochondrial 3' Terminal Uridylyl Transferase (TUTase): The Key Enzyme in U-insertion/deletion RNA Editing. Cell. 2002;108:637–648. doi: 10.1016/s0092-8674(02)00647-5. [DOI] [PubMed] [Google Scholar]
- Benne R, Van den Burg J, Brakenhoff J, Sloof P, Van Boom J, Tromp M. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- Bhat GJ, Souza AE, Feagin JE, Stuart K. Transcript-specific developmental regulation of polyadenylation in Trypanosoma brucei mitochondria. Mol. Biochem. Parasitol. 1992;52:231–240. doi: 10.1016/0166-6851(92)90055-o. [DOI] [PubMed] [Google Scholar]
- Blom D, Burg J, Breek CK, Speijer D, Muijsers AO, Benne R. Cloning and characterization of two guide RNA-binding proteins from mitochondria of Crithidia fasciculata: gBP27, a novel protein, and gBP29, the orthologue of Trypanosoma brucei gBP21. Nucleic Acids Res. 2001;29:2950–2962. doi: 10.1093/nar/29.14.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: "Guide" RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- Carnes J, Trotter JR, Peltan A, Fleck M, Stuart K. RNA Editing in Trypanosoma brucei requires three different editosomes. Mol. Cell Biol. 2008;28:122–130. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge RD, Aphasizheva I, Gershon PD, Aphasizhev R. 3' adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 2008;27:1596–1608. doi: 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans J, Hajduk SL, Madison-Antenucci S. RNA-editing-associated protein 1 null mutant reveals link to mitochondrial RNA stability. RNA. 2007;13:881–889. doi: 10.1261/rna.486107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimi H, Zikova A, Panigrahi AK, Stuart KD, Lukes J. TbRGG1, an essential protein involved in kinetoplastid RNA metabolism that is associated with a novel multiprotein complex. RNA. 2008;14:970–980. doi: 10.1261/rna.888808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman ML, Read LK. Trypanosoma brucei RBP16 is a mitochondrial Y-box family protein with guide RNA binding activity. J. Biol. Chem. 1999;274:12067–12074. doi: 10.1074/jbc.274.17.12067. [DOI] [PubMed] [Google Scholar]
- Igo RP, Palazzo SS, Burgess ML, Panigrahi AK, Stuart K. Uridylate addition and RNA ligation contribute to the specificity of kinetoplastid insertion RNA editing. Mol. Cell Biol. 2000;20:8447–8457. doi: 10.1128/mcb.20.22.8447-8457.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X, Gao G, Rogers K, Falick AM, Zhou S, Simpson L. Reconstitution of full-round uridine-deletion RNA editing with three recombinant proteins. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13944–13949. doi: 10.1073/pnas.0604476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CY, Read LK. Opposing effects of polyadenylation on the stability of edited and unedited mitochondrial RNAs in Trypanosoma brucei. Mol. Cell Biol. 2005;25:1634–1644. doi: 10.1128/MCB.25.5.1634-1644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller J, Muller UF, Schmid B, Missel A, Kruft V, Stuart K, Goringer HU. Trypanosoma brucei gBP21. An arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J. Biol. Chem. 1997;272:3749–3757. doi: 10.1074/jbc.272.6.3749. [DOI] [PubMed] [Google Scholar]
- Köller J, Nörskau G, Paul AS, Stuart K, Goringer HU. Different Trypanosoma brucei guide RNA molecules associate with an identical complement of mitochondrial proteins in vitro. Nucleic Acids Res. 1994;22:1988–1995. doi: 10.1093/nar/22.11.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Huang CE, O'Hearn SF, Sollner-Webb B. In Trypanosoma brucei RNA editing, band II enables recognition specifically at each step of the U insertion cycle. Mol. Cell Biol. 2005;25:2785–2794. doi: 10.1128/MCB.25.7.2785-2794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, O'Hearn S, Sollner-Webb B. In Trypanosoma brucei RNA editing, TbMP18 (band VII) is critical for editosome integrity and for both insertional and deletional cleavages. Mol. Cell Biol. 2007;27:777–787. doi: 10.1128/MCB.01460-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingler MK, Hingst AM, Clement SL, Yu LE, Reifur L, Koslowsky DJ. Identification of pentatricopeptide repeat proteins in Trypanosoma brucei. Mol. Biochem. Parasitol. 2006;150:37–45. doi: 10.1016/j.molbiopara.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Muller UF, Lambert L, Goringer HU. Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J. 2001;20:1394–1404. doi: 10.1093/emboj/20.6.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Allen TE, Stuart K, Haynes PA, Gygi SP. Mass spectrometric analysis of the editosome and other multiprotein complexes in Trypanosoma brucei. J. Am. Soc. Mass Spectrom. 2003;14:728–735. doi: 10.1016/S1044-0305(03)00126-0. [DOI] [PubMed] [Google Scholar]
- Panigrahi AK, Ernst NL, Domingo GJ, Fleck M, Salavati R, Stuart KD. Compositionally and functionally distinct editosomes in Trypanosoma brucei. RNA. 2006;12:1038–1049. doi: 10.1261/rna.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Zikova A, Dalley RA, Acestor N, Ogata Y, Anupama A, Myler PJ, Stuart KD. Mitochondrial complexes in trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol. Cell Proteomics. 2007;7:534–545. doi: 10.1074/mcp.M700430-MCP200. [DOI] [PubMed] [Google Scholar]
- Pelletier M, Miller MM, Read LK. RNA-binding properties of the mitochondrial Y-box protein RBP16. Nucleic Acids Res. 2000;28:1266–1275. doi: 10.1093/nar/28.5.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier M, Read LK. RBP16 is a multifunctional gene regulatory protein involved in editing and stabilization of specific mitochondrial mRNAs in Trypanosoma brucei. RNA. 2003;9:457–468. doi: 10.1261/rna.2160803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier M, Read LK, Aphasizhev R. Isolation of RNA Binding Proteins Involved in Insertion/deletion Editing. Methods Enzymol. 2007;424:69–96. doi: 10.1016/S0076-6879(07)24004-2. [DOI] [PubMed] [Google Scholar]
- Pelletier M, Xu Y, Wang X, Zahariev S, Pongor S, Aletta JM, Read LK. Arginine methylation of a mitochondrial guide RNA binding protein from Trypanosoma brucei. Mol. Biochem. Parasitol. 2001;118:49–59. doi: 10.1016/s0166-6851(01)00367-x. [DOI] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Pusnik M, Small I, Read LK, Fabbro T, Schneider A. Pentatricopeptide repeat proteins in Trypanosoma brucei function in mitochondrial ribosomes. Mol. Cell Biol. 2007;27:6876–6888. doi: 10.1128/MCB.00708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read LK, Stankey KA, Fish WR, Muthiani AM, Stuart K. Developmental regulation of RNA editing and polyadenylation in four life cycle stages of Trypanosoma congolense. Mol. Biochem. Parasitol. 1994;68:297–306. doi: 10.1016/0166-6851(94)90174-0. [DOI] [PubMed] [Google Scholar]
- Ryan CM, Kao CY, Sleve DA, Read LK. Biphasic decay of guide RNAs in Trypanosoma brucei. Mol. Biochem. Parasitol. 2006;146:68–77. doi: 10.1016/j.molbiopara.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Ryan CM, Read LK. UTP-dependent turnover of Trypanosoma brucei mitochondrial mRNA requires UTP polymerization and involves the RET1 TUTase. RNA. 2005;11:763–773. doi: 10.1261/rna.7248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Karamooz E, Zikova A, Trantirek L, Lukes J. Crystal structures of T. brucei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell. 2006;126:701–711. doi: 10.1016/j.cell.2006.06.047. [DOI] [PubMed] [Google Scholar]
- Simpson L, Aphasizhev R, Gao G, Kang X. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. RNA. 2004;10:159–170. doi: 10.1261/rna.5170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Trotter JR, Ernst NL, Carnes J, Panicucci B, Stuart K. A deletion site editing endonuclease in Trypanosoma brucei. Mol. Cell. 2005;20:403–412. doi: 10.1016/j.molcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Vanhamme L, Perez-Morga D, Marchal C, Speijer D, Lambert L, Geuskens M, Alexandre S, Ismaïli N, Göringer U, Benne R, Pays E. Trypanosoma brucei TBRGG1, a mitochondrial oligo(U)-binding protein that co-localizes with an in vitro RNA editing activity. J. Biol. Chem. 1998;273:21825–21833. doi: 10.1074/jbc.273.34.21825. [DOI] [PubMed] [Google Scholar]
- Vondruskova E, van den BJ, Zikova A, Ernst NL, Stuart K, Benne R, Lukes J. RNA interference analyses suggest a transcript-specific regulatory role for mitochondrial RNA-binding proteins MRP1 and MRP2 in RNA editing and other RNA processing in Trypanosoma brucei. J. Biol. Chem. 2005;280:2429–2438. doi: 10.1074/jbc.M405933200. [DOI] [PubMed] [Google Scholar]
- Wickstead B, Ersfeld K, Gull K. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol. Biochem. Parasitol. 2002;125:211–216. doi: 10.1016/s0166-6851(02)00238-4. [DOI] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.