Abstract
Many reactions within the cell occur only in specific intracellular regions. Such local reaction networks give rise to microdomains of activated signaling components. The dynamics of microdomains can be visualized by live cell imaging. Computational models using partial differential equations provide mechanistic insights into the interacting factors that control microdomain dynamics. The mathematical models show that, for membrane-initiated signaling, the ratio of the surface area of the plasma membrane to the volume of the cytoplasm, the topology of the signaling network, the negative regulators, and kinetic properties of key components together define microdomain dynamics. Thus, patterns of locally restricted signaling reaction systems can be considered an emergent property of the cell.
Homeostasis and regulated state change in the living cell require many processes to function in a coordinated fashion. Such multitasking is precisely orchestrated (1). For this, the cell integrates information from many extracellular signals in a timely and specific manner to evoke physiological processes. Complex networks of interacting pathways and regulatory mechanisms control and modulate information flow (2). Such complex systems cannot be intuitively understood. Mathematical modeling has served as a powerful analytical and predictive tool to understand how signals evoke responses and identify emergent properties of networks (3) that create an ensemble of output patterns that control cellular response machines. In this review, we describe how computational models help us understand the biochemical mechanisms underlying spatially restricted signaling events within cells.
Functional Specificity from Spatially Restricted Signaling
Organelles often spatially constrain cellular functions. Electrical activity occurs at the plasma membrane, ATP production in the mitochondria, and transcription in the nucleus. It is increasingly recognized that even when such organelle-based barriers do not exist, much of the biochemistry in the cell functions in a spatially restricted manner. The best examples of such spatially restricted biochemical reaction systems come from cell signaling pathways. Many receptors that evoke diverse cellular responses activate a relatively smaller number of shared intracellular signaling pathways, raising the question of how signaling specificity is achieved. One mechanism for achieving response specificity is the spatial separation of signaling reactions. The notion that signaling was spatially restricted was first proposed for the cAMP system by Brunton et al. (4) over 20 years ago. Compartmentalization was skeptically received then, given the popularity of the fluid mosaic membrane model (5) at the time and the prevalent idea that diffusible molecules would be impossible to segregate.
An example of spatially restricted signaling is found in cardiac myocytes, where elevation of cAMP levels regulates contractility, electrical excitability, and carbohydrate metabolism in a hormone-specific manner. For instance, β2-adrenergic stimulation leads to an increase in myocyte contractibility due to the PKA2-dependent modulation of L-type Ca2+ channels and troponin I (6), an actin-binding protein that regulates the myosin ATPase activity via tropomyosin (7). In contrast, other ligands such as prostaglandin E1 can induce similar temporal increases in cAMP but are incapable of regulating cardiac muscle contraction. These data suggested that cells spatially and functionally compartmentalize intracellular signaling. Advances in live cell imaging, new fluorescent probes, and use of fluorescence resonance energy transfer for the visualization of biochemical reactions have shown that, in the live cell, spatially localized signaling is a common phenomenon. Using these approaches, it has been shown in several cell types that increases in cAMP levels are local (8, 9).
Local signaling can provide context specificity to cellular responses to the MAPK pathway. Activation of Ras can lead to differentiation or proliferation of PC12 cells. How these two opposing cellular events can result from activation of the same pathway has been explained by correlating the response with duration of MAPK activation (10), which in turn depends on the location of Ras activation. Ras is activated at two sites within the cell: the plasma membrane and the Golgi membrane (11). Activation of Ras at the Golgi is prolonged and leads to differentiation, whereas brief plasma membrane activation leads to proliferation. The determinant of the location of Ras activation is the length of the intracellular Ca2+ signal, as a prolonged Ca2+ signal leads to translocation of the Ras guanine nucleotide exchange factor RasGRP1 to the Golgi membrane, where it activates Ras (11).
During maturation, the fate of thymocytes is determined either by their ability to recognize themselves (negative selection) or by the presence of a functional T-cell receptor (positive selection). Both processes are dependent on the Ras-MAPK pathway. Location of Ras activation determines whether a thymocyte will undergo negative or positive selection (12). Positive selecting ligands induced the recruitment of RasGRP1 to the Golgi membrane, leading to Ras activation at the Golgi, whereas negative selecting ligands promoted Ras activation at the plasma membrane. Selective activation of Ras in the Golgi, but not the plasma membrane, has been linked to T-cell activation (13).
Cdc42 activation is also spatially delimited (14). In a fibroblast spreading on a fibronectin-coated surface, Cdc42 regulates Arp2/3 nucleation of actin at the leading edge. Visualization of Cdc42 activation showed that the region of the cell perimeter undergoing rapid spreading had an enhanced accumulation of active Cdc42, whereas filopodia had little or no active Cdc42 (14). These observations led to the concept of “microdomains” defined as functional regions of regulated local increases in concentrations of activated signaling components.
Intracellular Microdomains
What is a microdomain, and how are its dynamics regulated? A microdomain is a region of micron/submicron dimensions within which increases in levels of signaling components are controlled by the relative rates of its production, diffusion, and destruction. The number of reactions underlying a microdomain can range from at least two, as in the simplest case of Ca2+, to tens, as in the case of MAPK microdomains. A microdomain has two defining characteristics: a point at which the concentration of the activated signaling component is the highest and a gradient that defines the change in concentration of the active component from the highest point to the level present in the surrounding milieu. The slope of the gradient defines the boundaries of the microdomains. Both these characteristics are dynamic, i.e. they are changing continually with time. Hence, all microdomain views are snapshots in time, and steady-state assumptions are mostly invalid.
The idea of signaling gradients was first articulated by Turing (15) more than 50 years ago in the context of developmental biology, where extracellular morphogen gradients provide positional cues for a developing tissue. These gradients can be produced by the local synthesis of a diffusible molecule and its slow degradation across space-creating gradients that may be several microns long. Mechanisms underlying morphogen gradients have been widely studied experimentally and computationally (16, 17). Analysis of intracellular gradients lagged behind both theoretically and experimentally. Since its discovery (18, 19), the use of green fluorescent protein (20) and similar probes to track proteins and interactions in real time within live cells has accelerated progress.
Among the first intracellular microdomains to be visualized were those for Ca2+ at the intracellular terminus of Ca2+ channels because the speed of conductance of Ca2+ channels is several orders of magnitude higher than the diffusion coefficient of Ca2+ (21). Ca2+/calmodulin feedback inhibitory mechanisms (22) tightly control the buildup of Ca2+ within these microdomains. Dissipation of these microdomains is due not only to diffusion but also to multiple sequestration mechanisms involving the endoplasmic reticulum stores and mitochondria. Several other types of spatiotemporal patterns of intracellular calcium regulation have been described (23).
Diffusible second messengers (24, 25), activated small GTPases (14, 26–29), and protein kinases (8, 9, 30) exist within microdomains. Factors underlying the formation and dissipation of these microdomains are complex. An integration of experimental data and mathematical modeling is needed to provide a mechanistic understanding. Even though experimental approaches yield physical evidence for the existence of microdomains, they offer only limited insight into origins. Mathematical models can complement the empirical approaches by probing possible mechanistic bases of the observations. Simulation-derived hypotheses can then be validated experimentally. The combination of experiments and mathematical modeling has highlighted a number of factors that contribute to the formation of microdomains. These factor are cell shape, topology of the reaction network, diffusion coefficients, and reaction kinetics (intrinsic properties of the components involved). To further complicate matters, the factors are inter-dependent. Thus, for a mechanistic understanding, it is necessary to determine how interdependence among these factors controls microdomain dynamics.
Computational Approaches to Modeling Spatially Restricted Biochemical Reactions
Models of spatially restricted biochemical reactions can be built using ODE-based compartmental models or PDE-based spatial models. ODE models describe the rate of change in the amount of molecules over time, and PDE models describe the rate of change in the amount of a molecule over time and space (36). Both approaches yield predictive models. Compartmental models can be easily solved using commercial simulators such as MATLAB. Modeling and running numerical simulations of realistic spatial models are more challenging, and there are very few simulators that biological researchers can use. Virtual Cell (37) is a freely available simulator useful for developing simulations from cell imaging data using both ODE and PDE models.
Compartmental models disregard details of spatial organization and the movement of molecules. The steps in setting up a compartmental model are shown in supplemental Fig. S1. Spatial information is abstracted to two entities, volume and surface area, and within each compartment, all molecules are assumed to be evenly distributed (i.e. fast diffusion). Even with these simplifying assumptions, compartmental models provide valuable insight into complex phenomena. A multicompartment model of myocyte function integrating mitochondrial biochemical reactions with electrical activity (ion channel function) in the plasma membrane and excitation-contraction in the sarcolemma provided valuable insight into how feedback mechanisms between mitochondrial oxidative phosphorylation reactions (cellular biochemistry) and the electrical and mechanical activities (cell physiology) drive metabolic regulation of cardiac function (38).
Spatial models deal with cell shape explicitly. To understand the regulatory insights spatial models provide, it is useful to know how these models are developed and computed. The steps involved in setting up a model with a realistic geometry in Virtual Cell are shown in supplemental Fig. S1. The reactions are set up as an ODE compartmental model, and then compartments are mapped onto a spatial geometry from a microscopic image. Mapping involves placing the components in the appropriate location at the appropriate concentration and defining their diffusion coefficients. Numerical simulations are then carried out using a finite volume regular grid solver, and results can be displayed as color-coded concentrations within the geometry, allowing for a visually accessible comparison between experiments and simulations.
Insights from Spatial Models
Spatial PDE models of cellular reactions are few because the diffusion coefficients of few cellular components are known. Although diffusion coefficients can be calculated from molecular weights, it is likely that molecular crowding will affect diffusion within the cell (39–41). With fluorescence recovery after photobleaching experiments, it is becoming feasible to obtain direct measures of diffusion of cellular components. Systematic variation of the diffusion parameter can be used to identify computationally parameters that are likely to yield experimentally observed patterns of microdomain dynamics. Despite these limitations, spatial models have yielded the first mechanistic insights. Spatial models have shown that microdomain dynamics is a systems level property arising from the interplay of several variables. Mathematical models have identified spatial information as an entity that is transmitted through a signaling network separately from information regarding activity states. Here, we describe spatial models of microdomains in myocytes and neurons to highlight the insight obtained from these models.
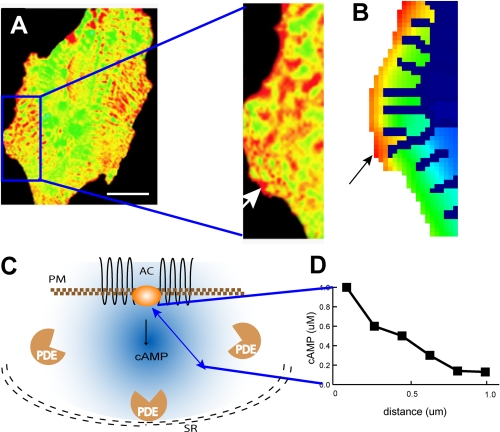
An experimentally observed cAMP microdomain from the studies of Zaccolo and Pozzan (9) is shown in Fig. 1A. Results from a spatial model that simulate these observations are shown Fig. 1B. To understand what a computational model tells us, consider the schematic in Fig. 1C. A microdomain depends on the localization of a key upstream component, in this case, adenylyl cyclase, which is an intrinsic membrane protein. Whereas cAMP-degrading phosphodiesterases are located largely in the cytoplasm, some phosphodiesterase isoforms are anchored to the endoplasmic reticulum or other membranes through anchoring proteins. The spatial separation of cAMP production and degradation results in a microdomain. The spatial gradient of cAMP is shown in Fig. 1D. In the myocyte, there is an extensive, specialized endoplasmic reticulum that forms a physical barrier for cAMP diffusion and thus defines the observed shape of the microdomains. Although contributing to microdomain characteristics, such a barrier is not essential.
FIGURE 1.
cAMP microdomains in cardiac myocytes. A, experimental demonstration of the cAMP microdomain in a study by Zaccolo and Pozzan (9) reprinted with permission from AAAS. The fluorescence resonance energy transfer ratio images are color-coded, with green signifying basal concentrations of cAMP and yellow to red signifying high concentrations of cAMP. Scale bar = 10 μm. The white arrow in the enlarged image points to a cAMP microdomain (∼1 μm). B, simulation in Virtual Cell of receptor-triggered cAMP increases using the cell shape traced from the myocyte image in A. The black arrow shows the corresponding cAMP microdomain in same region as in A. C, schematic diagram of factors controlling cAMP microdomains in myocytes: plasma membrane (PM) localization of adenylyl cyclase (AC), separation of production and degradation (phosphodiesterase (PDE)) activities for cAMP, and diffusion hindrance by intracellular organelles such as the sarcoplasmic reticulum (SR, depicted as the black double dashed line). D, microdomain characteristics are defined by the highest concentration point within the microdomain and the resulting gradient that extends to the periphery and defines the microdomain boundary. These are represented as a function of distance from the site of production. Arbitrary numbers are used for illustrative purposes.
Many protein kinases and phosphatases have anchors and scaffolds that create local signaling hubs. Prominent among these are A-kinase anchoring protein for PKA, microtubule affinity-regulating kinases for protein kinases C, JNK-interacting proteins for JNK, and MP1 for MAPK. Microdomains within the plasma membrane such as lipid rafts and caveolae (31) also promote spatial organization and separate extracellular signals spatially before their transduction. Certain signaling proteins such as lipid-linked non-receptor tyrosine kinases appear to be preferentially localized to these intramembrane structures (32). Intuitively, one would assume that such anchors are essential for microdomains; however, mathematical analyses indicate they are not essential.
When the signal originates at the membrane and is degraded in the cytoplasm, cell shape or the ratio of the surface area of the plasma membrane to the cytoplasmic volume (surface/volume ratio (S/V)) becomes important. The S/V of thin protrusions like filapodia or dendrites is larger than that of thicker structures such as the cell body of a neuron. Models predict and experiments show that, in regions of high S/V ratio, elevation of cAMP is enhanced (33), as is activation of Cdc42 and protein kinase C (34, 35). Because both amplitude and duration of a signal are related to the numbers (moles) of signaling molecules along a pathway, increasing the surface area of the plasma membrane relative to the volume of the cytoplasm decreases the volume within which the signaling molecule can diffuse and the gradient dissipate. In most cases, as the concentration (molecules/volume) stays constant, increasing the S/V ratio increases the signal source. This results in a buildup of the activated signaling component and the resultant microdomain. Although computational analyses indicate that anchors and scaffolds are not required for the formation of microdomains, the role of signaling complexes in microdomain formation and dynamics has not yet been extensively studied. An intriguing hypothesis is that cytoplasmic anchors may help override some of the spatial constraints that low S/V areas impose on microdomains. This hypothesis can be computationally addressed.
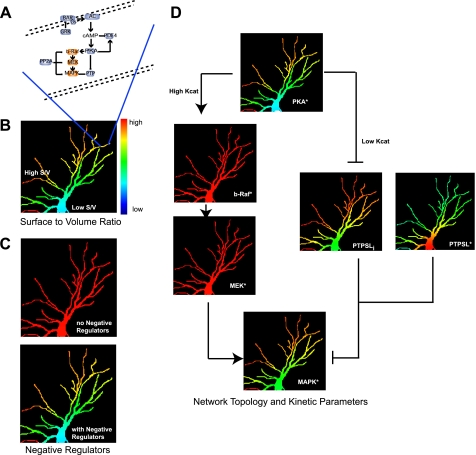
Computational analyses show that S/V ratio is only one of several factors regulating microdomains. Negative regulators also play a critical role. Dependence of cAMP microdomains on the activity of phosphodiesterases has been shown (9) as depicted in Fig. 1C. At its core, this phenomenon is analogous to Turing's original description of a point source with a diffusing signal and negative regulator resulting in a gradient, and a signaling version of this phenomenon was described (42). The toy graph in Fig. 1D shows this gradient. From S/V ratio consideration, the notion arises that thin, elongated regions of a cell (such as filopodia or dendrites in neurons) are favored for the formation of microdomains. This is shown in the simulations in Fig. 2. Here, a cAMP-PKA-MAPK network in a neuronal dendritic arbor was modeled (Fig. 2A). Although the receptor, Gs, and adenylyl cyclase are uniformly distributed throughout the cell, the thin dendrites (high S/V ratio) show the highest levels of PKA activation (Fig. 2B). Thus, cell shape becomes a determinant of microdomain characteristics (33, 34). When the phosphodiesterase is fully inhibited, then irrespective of cell shape, PKA activation is uniformly elevated (Fig. 2C). These observations indicate that the biochemistry of the system can override the physical constraints.
FIGURE 2.
Multiple factors regulate microdomain dynamics and the flow of spatial information. A, schematic of the spatially specified network topology used in the simulations. B–D, simulations of microdomains of signaling components in a dendritic arbor of a neuron. In all cases the signal originates at the plasma membrane. B, S/V ratios regulate microdomains. Regions of high S/V ratios show increased levels of activated PKA (PKA*) compared with regions with low S/V ratios. C, cytoplasmic negative regulators control microdomain characteristics. Upper panel, uniform activation of PKA when the phosphodiesterase is inhibited; lower panel (same image as in A), PKA microdomains when phosphodiesterase is active. D, network topology and kinetic parameters control the transfer of microdomain characteristics (i.e. spatial information) from upstream to downstream components. These simulations show that kinetic parameters are crucial factors in determining the transfer of microdomain characteristics. In the model, PKA has a high Kcat for b-Raf, which leads to extensive activation of b-Raf and loss of microdomains. In contrast, PKA has a low Kcat for PTPSL, resulting in microdomains of phosphorylated PTPSL (PTPSLi) that are similar to those of PKA. The regulation of PTP by PKA* leads to two populations of phosphatase activity, PKA*-inhibited activity (PTPSLi) and the active non-phosphorylated enzyme (PTPSL*). Areas with a high concentration of PKA* have low PTP activity and consequently high phospho-MAPK. Areas with a low concentration of PKA* have high PTP activity and low phospho-MAPK. This spatial and activity heterogeneity of PTPSL results in recapitulating the PKA* microdomains as MAPK* microdomains.
Although S/V ratios, cell shape, and negative regulators explain the dynamics of microdomains of signaling molecules such as cAMP, Ca2+, and even some small GTPases (34), these two factors do not explain the gradients in downstream components such as MAPKs. The computational analyses indicate that network topology is also important (Fig. 2D). The hierarchy of the negative regulators within the network topology determines microdomain characteristics of the downstream component. Both the upstream cAMP phosphodiesterase and downstream PTP control the MAPK microdomain. This control is evident only when S/V conditions are high; thus, cell shape becomes an overall constraint within which the network topology can be altered to determine downstream microdomains. These computational studies showed that the microdomain characteristics (i.e. gradient) of PKA are transmitted to MAPK through PTP but not through b-Raf and MEK. Nevertheless, the magnitude of the increase in MAPK activity results from b-Raf and MEK activation (Fig. 2D). Thus, information regarding position of the activated component (i.e. spatial information) is handled differently from information regarding activity state. These two types of information can be transmitted through different paths within a signaling network (Fig. 2D). This insight, originally obtained from the computational model, was experimentally verified by ablating the key component (PTP) utilized for spatial information transfer. When PTP was ablated, information regarding activation state was still transmitted, but the spatially restricted activation of MAPK was lost, indicating that information regarding activity state is distinct from spatial information (33).
Separation of spatial information from activity information raises an interesting question: what are the characteristics within a pathway that allow for the flow of spatial information? Mathematical simulations reveal that the relationship between the kinetic parameters of the upstream and downstream components is an important determinant. If the upstream enzyme extensively activates (high Kcat) the downstream component (e.g. PKA → b-Raf) (Fig. 2D), the microdomain characteristics of the upstream component will not be preserved in the downstream component. Inversely, a low Kcat reaction such as the phosphorylation of PTP by PKA results in the phosphorylated form of PTP exhibiting microdomains similar to those of upstream PKA* (Fig. 2D). The non-homogenous distribution of PKA* results in differing ratios of active (non-phosphorylated PTPSL*) to inactive (phosphorylated PTPSLi), allowing for the formation of activated MAPK1/2 microdomains (Fig. 2D). Thus, the microdomains of PKA* are recapitulated in the microdomains of phospho-MAPK. The extent to which the signal controls the negative regulator becomes a critical factor. Too much active PTP in all locations would result in a near-complete loss of phospho-MAPK, and too much PTP inhibition would result in high levels of phospho-MAPK and in the dissipation of the microdomain. This is a Goldilocks relationship in which the kinetic parameters and network topology need to be tuned just right for the spatial information to be transmitted. The balance between kinetic parameters and network topology works only if the overall S/V ratios are in the correct range. For spatial information to flow through the cell, the cell shape, signaling network topology, and kinetic characteristics of the signaling components need to be balanced. Hence, spatial information flow is an emergent property that arises from the functional organization of the cell within an optimized shape.
Perspective
Many questions regarding the role of scaffolds and anchors in the dynamics of microdomains remain to be addressed. The study by Fuller et al. (43) on the spatiotemporal dynamics of the Aurora B kinase phosphorylation of its substrates during anaphase highlights some of these questions. Interactions with anchors and scaffolds are often regulated by signaling. This regulation introduces the possibility that dynamic scaffolding can play a substantial role in controlling the flow of spatial information. This is an area for future studies where computation and experimentation can be effectively integrated.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM 072853 and Systems Biology Center Grant P50 GM 065085. This is the fourth article of six in the Thematic Minireview Series on Computational Biochemistry: Systems Biology. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: PKA, protein kinase A; MAPK, mitogen-activated protein kinase; ODE, ordinary differential equation; PDE, partial differential equation; JNK, c-Jun N-terminal kinase; S/V, surface/volume; PTP, protein-tyrosine phosphatase; MEK, MAPK/extracellular signal-regulated kinase kinase.
References
- 1.Jordan, J. D., Landau, E. M., and Iyengar, R. (2000) Cell 103 193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhalla, U. S., and Iyengar, R. (1999) Science 283 381–387 [DOI] [PubMed] [Google Scholar]
- 3.Weng, G., Bhalla, U. S., and Iyengar, R. (1999) Science 284 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunton, L., Hayes, J., and Mayer, S. (1981) Adv. Cyclic Nucleotide Res. 14 391–397 [PubMed] [Google Scholar]
- 5.Singer, S. J., and Nicolson, G. L. (1972) Science 175 720–731 [DOI] [PubMed] [Google Scholar]
- 6.Brunton, L., Hayes, J., and Mayer, S. (1979) Nature 280 78–80 [DOI] [PubMed] [Google Scholar]
- 7.Perry, S. V. (1999) Molecular Cell. Biochem. 190 9–32 [PubMed] [Google Scholar]
- 8.Bacskai, B. J., Hochner, B., Mahaut-Smith, M., Adams, S. R., Kaang, B. K., Kandel, E. R., and Tsien, R. Y. (1993) Science 260 222–226 [DOI] [PubMed] [Google Scholar]
- 9.Zaccolo, M., and Pozzan, T. (2002) Science 295 1711–1715 [DOI] [PubMed] [Google Scholar]
- 10.Marshall, C. J. (1995) Cell 80 179–185 [DOI] [PubMed] [Google Scholar]
- 11.Bivona, T. G., Perez De Castro, I., Ahearn, I. M., Grana, T. M., Chiu, V. K., Lockyer, P. J., Cullen, P. J., Pellicer, A., Cox, A. D., and Philips, M. R. (2003) Nature 424 694–698 [DOI] [PubMed] [Google Scholar]
- 12.Daniels, M. A., Teixeiro, E., Gill, J., Hausmann, B., Roubaty, D., Holmberg, K., Werlen, G., Hollander, G. A., Gascoigne, N. R., and Palmer, E. (2006) Nature 444 724–729 [DOI] [PubMed] [Google Scholar]
- 13.Mor, A., Campi, G., Du, G., Zheng, Y., Foster, D. A., Dustin, M. L., and Philips, M. R. (2007) Nat. Cell Biol. 9 713–719 [DOI] [PubMed] [Google Scholar]
- 14.Nalbant, P., Hodgson, L., Kraynov, V., Toutchkine, A., and Hahn, K. M. (2004) Science 305 1615–1619 [DOI] [PubMed] [Google Scholar]
- 15.Turing, A. (1952) Philos. Trans. R. Soc. Lond. B Biol. Sci. 237 37–72 [Google Scholar]
- 16.Eldar, A., Shilo, B. Z., and Barkai, N. (2004) Curr. Opin. Genet. Dev. 14 435–439 [DOI] [PubMed] [Google Scholar]
- 17.Lander, A. D. (2007) Cell 128 245–256 [DOI] [PubMed] [Google Scholar]
- 18.Morin, J. G., and Hastings, J. W. (1971) J. Cell. Physiol. 77 305–312 [DOI] [PubMed] [Google Scholar]
- 19.Morise, H., Shimomura, O., Johnson, F. H., and Winant, J. (1974) Biochemistry 13 2656–2662 [DOI] [PubMed] [Google Scholar]
- 20.Chalfie, M., Tu, Y., Euskirchen, G., Ward, W. W., and Prasher, D. C. (1994) Science 263 802–805 [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Lopez, J. R., Shacklock, P. S., Balke, C. W., and Wier, W. G. (1995) Science 268 1042–1045 [DOI] [PubMed] [Google Scholar]
- 22.Rios, E., and Stern, M. D. (1997) Annu. Rev. Biophys. Biomol. Struct. 26 47–82 [DOI] [PubMed] [Google Scholar]
- 23.Berridge, M. J., and Dupont, G. (1994) Curr. Opin. Cell Biol. 6 267–274 [DOI] [PubMed] [Google Scholar]
- 24.Hirose, K., Kadowaki, S., Tanabe, M., Takeshima, H., and Iino, M. (1999) Science 284 1527–1530 [DOI] [PubMed] [Google Scholar]
- 25.Honda, A., Adams, S. R., Sawyer, C. L., Lev-Ram, V., Tsien, R. Y., and Dostmann, W. R. (2001) Proc. Natl. Acad. Sci. U. S. A 98 2437–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mochizuki, N., Yamashita, S., Kurokawa, K., Ohba, Y., Nagai, T., Miyawaki, A., and Matsuda, M. (2001) Nature 411 1065–1068 [DOI] [PubMed] [Google Scholar]
- 27.Aoki, K., Nakamura, T., and Matsuda, M. (2004) J. Biol. Chem. 279 713–719 [DOI] [PubMed] [Google Scholar]
- 28.Del Pozo, M. A., Kiosses, W. B., Alderson, N. B., Meller, N., Hahn, K. M., and Schwartz, M. A. (2002) Nat. Cell Biol. 4 232–239 [DOI] [PubMed] [Google Scholar]
- 29.Ohba, Y., Kurokawa, K., and Matsuda, M. (2003) EMBO J. 22 859–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ting, A. Y., Kain, K. H., Klemke, R. L., and Tsien, R. Y. (2001) Proc. Natl. Acad. Sci. U. S. A 98 15003–15008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel, H. H., Murray, F., and Insel, P. A. (2008) Annu. Rev. Pharmacol. Toxicol. 48 359–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Filipp, D., Zhang, J., Leung, B. L., Shaw, A., Levin, S. D., Veillette, A., and Julius, M. (2003) J. Exp. Med. 197 1221–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neves, S. R., Tsokas, P., Sarkar, A., Grace, E. A., Rangamani, P., Taubenfeld, S. M., Alberini, C. M., Schaff, J. C., Blitzer, R. D., Moraru, I. I., and Iyengar, R. (2008) Cell 133 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyers, J., Craig, J., and Odde, D. J. (2006) Curr. Biol. 16 1685–1693 [DOI] [PubMed] [Google Scholar]
- 35.Craske, M. L., Fivaz, M., Batada, N. N., and Meyer, T. (2005) J. Cell Biol. 170 1147–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eungdamrong, N. J., and Iyengar, R. (2004) Trends Cell Biol. 14 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loew, L. M., and Schaff, J. C. (2001) Trends Biotechnol. 19 401–406 [DOI] [PubMed] [Google Scholar]
- 38.Cortassa, S., Aon, M. A., O'Rourke, B., Jacques, R., Tseng, H. J., Marban, E., and Winslow, R. L. (2006) Biophys. J. 91 1564–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fulton, A. B. (1982) Cell 30 345–347 [DOI] [PubMed] [Google Scholar]
- 40.Medalia, O., Weber, I., Frangakis, A. S., Nicastro, D., Gerisch, G., and Baumeister, W. (2002) Science 298 1209–1213 [DOI] [PubMed] [Google Scholar]
- 41.Minton, A. P. (2001) J. Biol. Chem. 276 10577–10580 [DOI] [PubMed] [Google Scholar]
- 42.Sick, S., Reinker, S., Timmer, J., and Schlake, T. (2006) Science 314 1447–1450 [DOI] [PubMed] [Google Scholar]
- 43.Fuller, B. G., Lampson, M. A., Foley, E. A., Rosasco-Nitcher, S., Le, K. V., Tobelmann, P., Brautigan, D. L., Stukenberg, P. T., and Kapoor, T. M. (2008) Nature 453 1132–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.