Abstract
INSM1 is a zinc finger transcription factor that plays an important role in pancreatic β-cell development. To further evaluate its role in cell fate determination, we investigated INSM1 effects on cell cycle function. The cyclin box of cyclin D1 is essential for INSM1 binding. Competitive pull-down and co-immunoprecipitation revealed that INSM1 binding to cyclin D1 interrupts its association with CDK4 and induces hypophosphorylation of the retinoblastoma protein. An inducible Tet-on system was established in Cos-7 and Panc-1 cells. Using serum starvation, we synchronized the cell cycle and subsequently induced cell cycle progression by serum stimulation. Comparison of the INSM1 induction group with the noninduced control group, INSM1 ectopic expression causes cell cycle arrest, whereas the INSM1-mediated cell cycle arrest could be reversed by cyclin D1 and CDK4 overexpression. The proline-rich N-terminal portion of INSM1 is required for cyclin D1 binding. Mutation of proline residues abolished cyclin D1 binding and also diminished its ability to induce cell cycle arrest. Cellular proliferation of Panc-1 cells was inhibited by INSM1 overexpression demonstrated by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay, soft agar colony formation, as well as tumor growth in a nude mouse model. Taken together, we provide evidence to support that INSM1 binds to cyclin D1, interrupts cell cycle signaling, and inhibits cellular proliferation.
INSM12 (formerly IA-1) is a zinc finger transcription factor originally isolated from a human insulinoma subtraction library (1). Functional analysis of INSM1 revealed that multiple downstream target genes and upstream regulatory proteins of the INSM1 gene are closely associated with pancreatic endocrine cell differentiation (2–4). Global deletion of the INSM1 gene was embryonic lethal and resulted in abnormal β-cell development (5) as well as impaired sympatho-adrenal lineage development (6). The expression patterns of INSM1 in both humans and rodents are restricted to the fetal neuroendocrine system and silenced in adult tissues (1, 7–9). However, the INSM1 gene is reactivated in tumors of neuroendocrine origin, suggesting that de-differentiation events occur in neuroendocrine tumors that mimic normal embryonic development. Embryonic development requires the generation of cells in appropriate numbers and the timely acquisition of specialized cell functions (10). Specialization occurs gradually over multiple rounds of cell division, with the end point being a nondividing terminally differentiated cell. Usually, cell fate determination is controlled in part by external signals and/or differentiation factors that govern cells to either enter or exit the cell cycle. Our previous study revealed a direct interaction between INSM1 and cyclin D1 (11). Therefore, we hypothesize that INSM1 could not only function as a transcription factor in regulating its downstream target genes but also plays an important role in switching between cellular proliferation and differentiation pathways.
In the present study, the cyclin box of cyclin D1 was identified to be essential for INSM1 binding. Conversely, a proline-rich N-terminal region (amino acids 43–58) in INSM1 facilitates binding to cyclin D1. The cyclin box is known to be the main binding site for cyclin-dependent kinase 4 (CDK4) (12). Competitive immunoprecipitation of cyclin D1 with INSM1 or CDK4 demonstrated that INSM1 could interrupt CDK4 binding, subsequently induced Rb protein hypophosphorylation, and cell cycle arrest. We employed an INSM1 Tet-on system in Cos-7 and Panc-1 cells. Induction of INSM1 in these cells results in cell cycle arrest and inhibition of cellular proliferation.
EXPERIMENTAL PROCEDURES
Cell Lines and Chemicals—An African green monkey kidney cell line (Cos-7) and a human pancreatic carcinoma cell line (Panc-1) were obtained from ATCC and maintained according to the ATCC recommendation. βTC-1 cells were kindly provided by Dr. E. H. Leiter (Jackson Laboratory, Bar Harbor, ME). Antibodies to CDK4 (C-22), cyclin D1, phospho-Rb780, and Rb were purchased from BD Biosciences (Palo Alto, CA). Anti-GFP was purchased from Clontech (Palo Alto, CA). A mouse anti-INSM1 monoclonal antibody (6-1-1) was generated against the C-terminal peptide of the INSM1 protein. Anti-FLAG and anti-actin antibodies were purchased from Sigma. Anti-hemagglutinin antibody was obtained from BIOSOURCE (Camarillo, CA).
Plasmids Constructs—For the yeast two-hybrid screen, the INSM1 or cyclin D1 bait plasmid was constructed by subcloning the full-length INSM1 or cyclin D1 cDNA into a pGBKT7 vector (Clontech), and different fragments of cyclin D1 or INSM1 cDNA were subcloned into a pGADT7 vector (Clontech). For pull-down experiments, full-length cyclin D1 was cloned into a prokaryotic expression vector pET41a(+) (Novagen). For the Tet-on system, the INSM1 cDNA was cloned into pTRE-BI-AcGFP vector (Clontech). The INSM1 mutant was constructed by synthesis of mutated oligonucleotides overlapping with the wild type INSM1. PCR amplification of mutated sequence was subcloned to replace the wild type sequence. The mutant sequence was verified by sequence analysis. Rat insulin I promoter (rInIp) driven luciferase vector was constructed as described (13).
Yeast Two-hybrid Screen—Bait pGBKT7-INSM1 or pGBK7-cyclin D1 and a series of pGADT7-cyclin D1 or pGADT7-INSM1 vectors containing various cyclin D1 or INSM1 fragments were co-transformed into competent AH109 yeast cells according to the manufacturer's protocol (Clontech). The positive clones were selected for their ability to grow in quadruple selection plate (Ade–/His–/Leu–/Trp–) with 5-bromo-4-chloro-3-indolyl-a-d-galactopyranoside (X-a-Gal).
Construction of Recombinant Adenovirus—Recombinant adenoviruses were generated using the AdEasy XL adenoviral vector system (Stratagene) as previously described (11). Briefly, a 3× FLAG tag was subcloned in frame with the C terminus of the human INSM1 cDNA into the pCMV-shuttle vector. Full-length CDK4 cDNA was also subcloned into pCMV-shuttle vector. The recombinant shuttle vector was electroporated into BJ5183-AD-1 cells, and the smaller colonies present on the kanamycin plate (the smaller size of the colonies represents recombinant clones) were selected. The clones produce 4.5- and/or 3-kb bands after PacI enzyme digestion, representing that the recombination took place either at the origins of replication or between the left arms. After selecting the correct recombinant clones, they were transfected into AD293 cells to generate adenovirus. For titering the adenovirus, the adeno X rapid titer kit was used to stain the virally infected cells with anti-hexon antibody (Clontech).
Tet-on System—Clones expressing the rtTA (the reverse tetracycline transactivator) were obtained by transfection of Cos-7 and Panc-1 cells with the pTet-On vector according to the protocol (Clontech). Selected rtTA positive clones were cultured in complete medium (Dulbecco's modified Eagle's medium supplemented with 5% Tet-free fetal bovine serum, 2 mm glutamine, 100 international units/ml penicillin, and 100 μg/ml streptomycin, and 0.6 mg/ml G418). Selected clones were then co-transfected with either mutant or wild type pTRE-BI-AcGFP-INSM1 and pTK-hygromycin vector using Lipofectamine 2000 (Invitrogen). Transfected cells were double selected with hygromycin (0.2 μg/ml) and G418 (0.6 μg/ml). The resistant clones were expanded and tested for the induction of INSM1 and GFP expression upon the addition of 0.2 μg/ml doxycycline by reverse transcription-PCR and Western blot analysis. Two individual clones for Cos-7 and Panc-1 were selected with no basal INSM1 expression in the absence of doxycycline and high levels of INSM1 with doxycycline treatment using Western blot analysis. For further experiments, these clones were maintained in complete medium containing 0.3 μg/ml of G418 and 0.1 μg/ml of hygromycin.
Recombinant Protein Expression and Purification—Expression construct pET41a(+)-cyclin D1 was transformed into BL21(DE3) competent cells (Novagen). GST and recombinant GST-cyclin D1 proteins were expressed and purified by glutathione-agarose beads (Sigma) according to the manufacturer's instruction.
Pull-down Experiments—Recombinant GST-tagged cyclin D1 or GST were incubated with cell lysates infected with either Ad-INSM1 or Ad-CDK4 and glutathione-agarose beads in 500 μl of binding buffer (PBS, 1% bovine serum albumin, 0.4% Triton X-100, and 2 mm dithiothreitol) at 4 °C for overnight. The beads were washed four times with 1 ml of binding buffer, resolved by SDS-PAGE, and analyzed via Western blot. GST was used as a negative control for nonspecific binding.
Examination of Cell Cycle Progression—Tet-on selected Cos-7 cells were induced to express INSM1 with doxycycline for 24 h and subjected to serum deprivation overnight to synchronize the cell cycle. After starvation, the cells were stimulated with 10% serum for 1, 4, and 24 h. Cell cycle progression was examined by Western blot and fluorescence-activated cell sorter analysis.
Co-immunoprecipitation—Tet-on selected Cos-7 cells were seeded in a 100-mm dish and subjected to serum deprivation overnight to synchronize the cell cycle and stimulated with 10% serum for different time points before harvesting for co-immunoprecipitation and Western blot analysis. The cell lysates were prepared using protein binding buffer (20 mm Tris/HCl, pH 7.5, 150 mm NaCl, 0.5% Nonidet P-40, 0.1% (v/v), Triton X-100, and 1 mm phenylmethylsulfonyl fluoride). For co-immunoprecipitation, 300 μg of cell lysate was precleared with recombinant protein G-agarose and incubated at 4 °C for 2 h with an anti-cyclin D1 antibody or IgG control, followed by incubation overnight at 4 °C with recombinant protein G-agarose beads. The beads were washed five times with binding buffer, separated by 10% SDS-PAGE, and transferred onto a nitrocellulose membrane (Invitrogen). Cyclin D1 complexed with INSM1 or CDK4 was detected by Western blot analysis (1:1000) with anti-INSM1 or anti-CDK4 antibody, respectively. The horseradish peroxidase-conjugated anti-mouse secondary antibody (1:4000) and chemiluminescent substrate (Pierce) were used for detection of INSM1, CDK4, or cyclin D1 protein.
siRNA Knockdown of INSM1 Restores Cyclin D1 and CDK4 Interaction—Control siRNA or INSM1 siRNA targeting the human INSM1 gene sequence, 5′-GGGATCTGCTTAAAGTTTTAA-3′, were purchased from Ambion (Austin, TX). Tet-on selected Cos-7 cells were transfected with either control or INSM1 siRNA (100 nm) using Lipofectamine 2000 (Invitrogen). The transfected cells were induced by doxycycline for 48 h and collected for Western blot and co-immunoprecipitation analyses.
Transient Transfection and Insulin Promoter Assays—The rat insulin promoter-driven luciferase vector (1.0 μg), rInIp (I: –410/+50 bp), was transfected into a 70% confluent βTC-1 cell line in the presence or absence of INSM1 wild type (0.5 μg) or mutant (0.5 μg). An empty pcDNA3 vector was also included to ensure that each transfection had an equal amount of DNA. A cytomegalovirus β-galactosidase construct (0.5 μg) was used as an internal control to normalize transfection efficiency. Twenty-four hours post-transfection, the cells were washed with PBS and collected for luciferase and β-galactosidase assays (Promega). Each experiment was performed at least four times, and the results represent the averages of four times ± S.E. The wild type rat insulin I promoter activity was designated as 100%.
MTT Assay—Tet-on selected Panc-1 cells were seeded into a 96-well plate (1 × 104 cells/well) with Dulbecco's modified Eagle's medium. After doxycycline treatment for 24 h and after Panc-1 cells were subjected to serum starvation, cell viability under serum stimulation at various time points was determined by MTT assay. Briefly, 10 mm of MTT reagent was added to each well for an additional 4 h. The blue MTT formazan precipitate was dissolved in 100 μl of Me2SO and measured on a multi-detection microplate reader model KC4 (Bio-Tek) at the absorbance wavelength of 570 nm and reference wavelength of 630 nm.
Soft Agar Assay—Tet-on selected Panc-1 cells were mixed with culture medium containing 0.33% agarose with or without doxycycline and plated on a presolidified layer of the same medium containing 0.5% agarose in 35-mm Petri dish. An additional 1 ml of culture medium with or without doxycycline was added after solidification to the top layer, and cells were incubated for 2 weeks at 37 °C. The plates were stained with crystal violet (Invitrogen), and colony numbers were counted (defined as ≥50 cells).
Nude Mice Tumor Model—Eight-week NU/NU strain nude mice were purchased from Charles River Laboratory. For measurement of tumor growth, 1 × 107 Panc-1 cells were injected subcutaneously into the flank of nude mice. Either doxycycline (20 mg/kg, intraperitoneally) or PBS was administered daily 72 h after Panc-1 cell injection. After 6 weeks, the tumors from both groups were retrieved and weighed. Tumor weight was derived from the average of four tumors.
Statistical Analysis—Statistical analysis was performed using one-way analysis of variance with a Bonferroni adjustment or a Student's unpaired t test. The data are expressed as the means ± S.E.
RESULTS
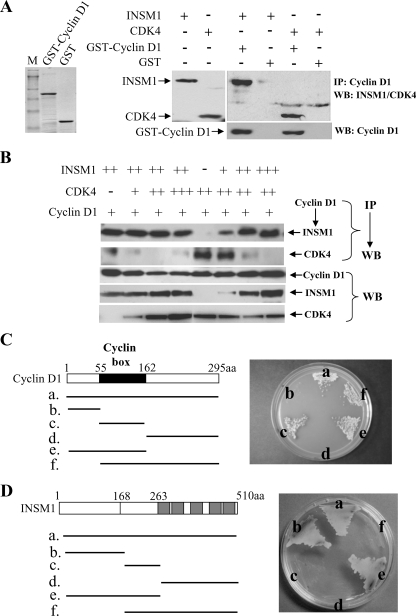
INSM1 Interaction with Cyclin D1 Disrupts CDK4 Binding—Our previous studies demonstrated that INSM1 could suppress NeuroD/β2 and insulin gene expression via the recruitment of cyclin D1 and histone deacetylase-3 (11, 13). The physical interaction between INSM1 and cyclin D1 was verified by co-immunoprecipitation. To further elucidate which region of cyclin D1 and INSM1 are responsible for the physical interaction, we determined the sequence of cyclin D1 essential for INSM1 binding and vise versa. We first verified whether recombinant cyclin D1 is capable of binding to INSM1 and CDK4. The expression of INSM1 and CDK4 in adenoviral infected Cos-7 cells was measured by Western blot analysis. Our in vitro GST pull-down experiment showed that cyclin D1 could pull-down INSM1 and CDK4 effectively as compared with the GST control (Fig. 1A). We assessed the cyclin D1 binding affinities for INSM1 and/or CDK4 using an in vitro competitive pull-down experiment (Fig. 1B). Using a fixed INSM1 concentration and gradually increasing amounts of CDK4, a cyclin D1 pull-down experiment showed that INSM1 is the dominant component associated with cyclin D1. Similarly, a gradual increase in INSM1 concentration competes against a fixed amount of CDK4 in binding to cyclin D1. These results suggest that in the presence of INSM1, the cyclin D1-INSM1 complex interferes with normal cyclin D1-CDK4 interaction. We further mapped the INSM1-binding domain in the cyclin D1 protein using a yeast two-hybrid system. The INSM1 bait vector and six different fragments of cyclin D1 were co-transformed into AH109 competent yeast cells and selected on a quadruple selection plate (Ade–/His–/Leu–/Trp–) with X-α-Gal. As shown in Fig. 1C, only the b and d fragments fail to grow on the selection plate. The vector b and d fragments do not contain the cyclin box sequence (amino acids 55–162), suggesting that the cyclin box of cyclin D1 is essential for INSM1 binding. Consistent with our pull-down result, it is known that CDK4 also binds to the cyclin box in cyclin D1 (12). Inversely, we also performed the yeast two-hybrid experiment to map the cyclin D1-binding domain in the INSM1 protein. As shown in Fig. 1D, the a, b, and e fragments are essential for cyclin D1 binding, suggesting that the N-terminal portion of the INSM1 (amino acids 1–168), which contains a proline-rich segment between amino acids 43 and 58 could contribute to the protein-protein interaction. Therefore, we hypothesize that the interaction between INSM1 and cyclin D1 could disrupt CDK4 binding during the normal cell cycle.
FIGURE 1.
Interaction of INSM1, CDK4, and cyclin D1. A, GST and GST-cyclin D1 were expressed in Escherichia coli and purified by glutathione affinity column. Cos-7 cells were infected with Ad-INSM1 or Ad-CDK4, and cellular extracts were subjected to Western blot (WB) analysis. Pull-down experiments were conducted using recombinant GST-cyclin D1 or GST mixed with INSM1 or CDK4 extract and demonstrated a specific interaction of cyclin D1 with either INSM1 or CDK4. IP, immunoprecipitation. B, competitive pull-down experiments using either GST-cyclin D1 or GST with various concentrations of cell extract infected with Ad-INSM1 or Ad-CDK4 showed preferential binding INSM1 to CDK4 by cyclin D1. C, schematic diagram shows the full-length cyclin D1 and various fragments of cyclin D1. The cyclin D1 sequences in pGADT7 vector were co-transformed with pGBKT7-INSM1 bait vector for the yeast two-hybrid screen. Only fragments b and d failed to grow on the quadruple selection plate (Ade–/His–/Leu–/Trp–) with X-α-Gal, indicating that the cyclin box is essential for INSM1 and cyclin D1 binding. D, inversely, full-length INSM1 and various fragments of INSM1 in pGADT7 vector were co-transformed with pGBKT7-cyclin D1 bait vector for the yeast two-hybrid screen. Only fragments a, b, and e grow on the quadruple selection plate (Ade–/His–/Leu–/Trp–) with X-α-Gal, indicating that the N-terminal portion of INSM1 (amino acids 1–168) is essential for cyclin D1 binding.
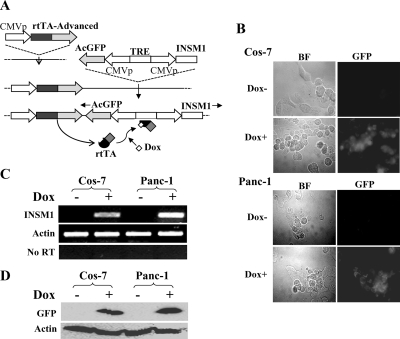
To further study the functional effects of INSM1 during the normal cell cycle, we established a doxycycline inducible Tet-on system in Cos-7 and Panc-1 cells. Fig. 2 shows a schematic diagram of how to establish the Tet-on system (from Clontech.). We chose a double reporter gene system that could simultaneously express GFP and the gene of interest (INSM1). Both INSM1 and GFP were clearly shown to be induced by doxycycline treatment.
FIGURE 2.
Establishment of the INSM1 Tet-on system in Cos-7 and Panc-1 cells. A, flow chart of establishing the INSM1 Tet-on system. B, selected Cos-7 or Panc-1 Tet-On clones were treated with doxycycline (Cos-7, 0.2 μg/ml; Panc-1, 2 μg/ml) or without doxycycline for 24 h, GFP signals were examined under a fluorescence microscope. BF, bright field. C, reverse transcription-PCR of INSM1 and actin control. From left to right, each lane represents Dox-treated (+ or –) Cos-7 or Panc-1 cells. D, Western blot analysis of GFP. GFP was detected using anti-GFP antibody (1:1000). Anti-actin was used as loading control.
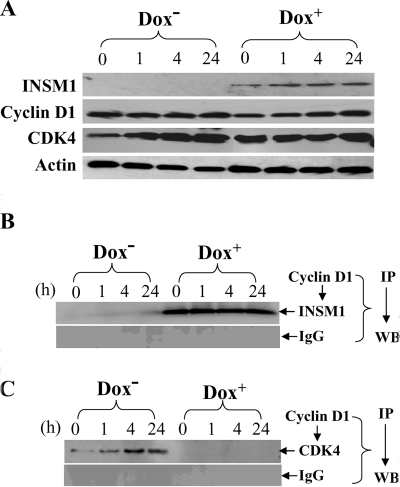
To test whether INSM1 competes with CDK4 binding to cyclin D1 in vivo, we first induced INSM1 expression for 24 h with doxycycline. On the next day, the cells were synchronized in the G0 phase via serum starvation. On the third day, the cells were stimulated to re-enter the cell cycle via regular serum containing medium. The serum stimulated cells were harvested at different time points: 0, 1, 4, and 24 h, and then subjected to competitive binding for either INSM1 or CDK4 to cyclin D1. Fig. 3A shows the expression patterns of INSM1, cyclin D1, and CDK4. Co-immunoprecipitation was performed using cyclin D1 antibody or control IgG and then immunoblotted with an anti-INSM1 (Fig. 3B) or an anti-CDK4 antibody (Fig. 3C). The INSM1 protein was precipitated by the cyclin D1 antibody in the Dox+ group, whereas no INSM1 was detected in the Dox– group. For CDK4, the CDK4 protein precipitated by the cyclin D1 antibody gradually increased in the Dox– group from 0 to 24 h in contrast to the Dox+ group where no CDK4 protein was co-precipitated because of the interference by INSM1. The result suggested that during the progression of the cell cycle, overexpression of INSM1 disrupts cyclin D1-CDK4 binding.
FIGURE 3.
Ectopic expression of INSM1 competes with CDK4 for cyclin D1 binding. Tet-on selected Cos-7 cells were synchronized by serum starvation and stimulated with 10% serum at various time points. A, cell extracts derived from Dox+ or Dox– group was subjected to Western blot (WB) analysis for INSM1, cyclin D1, CDK4, and actin control. Co-immunoprecipitation (IP) of cyclin D1 with INSM1 (B) or CDK4 (C) was performed using Dox+ or Dox– Cos-7 cells. After 24 h Dox+ treatment, the INSM1 and cyclin D1 interaction interrupts CDK4 binding to the cyclin D1. Normal IgG was used as a control.
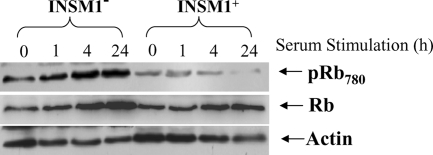
INSM1 Blocks Rb Protein Phosphorylation—During the normal cell cycle, the cyclin D1·CDK4 complex is critical for CDK4 activation that subsequently results in Rb protein phosphorylation and cell cycle progression. If the interaction between INSM1 and cyclin D1 disrupts cyclin D1-CDK4 binding, the subsequent cyclin D1 downstream signaling pathway should be blocked. Thus, we investigated the phosphorylation state of Rb protein when INSM1 was induced by doxycycline. In the INSM1 negative Cos-7 cells, the phosphorylated Rb (pRb780), and the total Rb (Rb) proteins were gradually increased after serum stimulation. However, in the INSM1 positive group, a dramatic decrease in phosphorylated Rb protein was observed; in contrast the total Rb protein detected was still similar to the INSM1 negative group. These results indicate that INSM1 binding to cyclin D1 could inhibit the CDK4 activity (Fig. 4).
FIGURE 4.
Ectopic expression of INSM1 suppresses Rb protein phosphorylation. Tet-on selected Cos-7 cells were treated with doxycycline for 48 h. An equal amount of cell lysate was separated on a 10% SDS-PAGE and Western blotted with anti-phospho-Rb780 or anti-Rb antibody (1:1000). Anti-actin was used as a control. Phospho-Rb780 protein was dramatically reduced in the Dox-treated group as compared with untreated Cos-7 cells without a significant change in the total Rb protein detected.
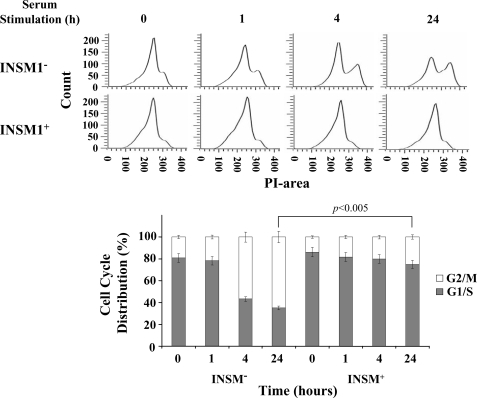
INSM1 Ectopic Expression Induces Cell Cycle Arrest—Using the Tet-on system, we induced INSM1 expression in a similar experimental setting during serum starvation followed by stimulation at various time points. Propidium iodide staining and flow cytometric analysis is shown in Fig. 5. In these flow cytometric histograms, the cell cycle curve was composed of a G1/S peak and a G2/M peak. At 0 h, the cell cycle curve was mainly made up of a single G1/S peak. The cells were then stimulated to re-enter the cell cycle by the return of serum-containing media. From 0 to 24 h, in the absence of doxycycline the G1/S peak decreased while the G2/M peak increased. While in the presence of doxycycline, the majority of the cells remained in the G1/S peak and had no obvious G2/M peak. We observed that at 0 h, almost 80–90% of the cells in both nontreated and Dox-treated groups were blocked at the G1/S phase. Following serum stimulation, the cells from the uninduced group normally progressed with an accompanying increase of G2/M cells from 20 to 70%, whereas almost 80% of cells in the INSM1 induced group at 24 h were blocked at the G1/S phase. It implicated that ectopic expression of INSM1 caused cell cycle arrest at the G1/S phase.
FIGURE 5.
INSM1 ectopic expression induces cell cycle arrest. Cos-7 cells were cultured with or without 0.2 μg/ml of doxycycline for 24 h. The cells were synchronized with serum-free medium and subsequently stimulated with 10% serum for 0, 1, 4, or 24 h. The cells were subjected to propidium iodide (PI) staining and flow cytometric analysis. The cell cycle distribution revealed that less G2/M phase cells were present during INSM1 ectopic expression as compared with the control group, suggesting that cell cycle arrest occurs during INSM1 expression.
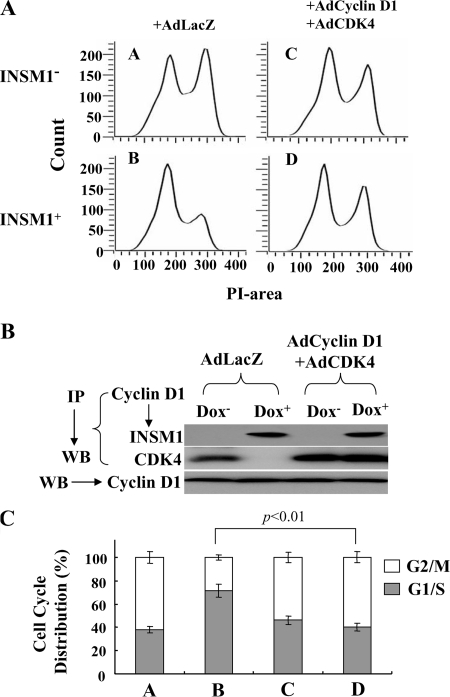
To further clarify that the interruption of cyclin D1 and CDK4 complex formation contributed to cell cycle arrest, cyclin D1 and CDK4 were overexpressed in Tet-on Cos-7 cells via adenovirus infection. Flow cytometric analysis showed nearly 70% of the cells were blocked at the G1/S phase in the presence of INSM1. In contrast, overexpression of cyclin D1 and CDK4 in the presence of INSM1 resulted in more than 60% of cells entering the G2/M phase by 24 h (Fig. 6, A and C). Co-immunoprecipitation of cyclin D1 with INSM1 or CDK4 indicated that overexpression of cyclin D1·CDK4 in the presence of INSM1 can maintain cyclin D1·CDK4 complex formation in addition to the cyclin D1·INSM1 complex (Fig. 6B). These results demonstrate that excess cyclin D1·CDK4 can reverse the cell cycle arrest caused by INSM1.
FIGURE 6.
Cyclin D1·CDK4 can reverse the cell cycle arrest induced by INSM1. A, Cos-7 cells ectopically expressing INSM1 were synchronized by serum starvation and then stimulated by adding 10% serum for 24 h. Ad-LacZ or Ad-cyclin D1·CDK4 was added during the serum stimulation. Cell cycle progression was measured by propidium iodide (PI) staining using flow cytometric analysis. Overexpression of cyclin D1·CDK4 could reverse the cell cycle arrest caused by INSM1. B, co-immunoprecipitation (IP) of cyclin D1 revealed that overexpression of cyclin D1 and CDK4 can restore the cyclin D1·CDK4 complex even in the presence of INSM1. C, percentage of cell cycle distribution. WB, Western blot.
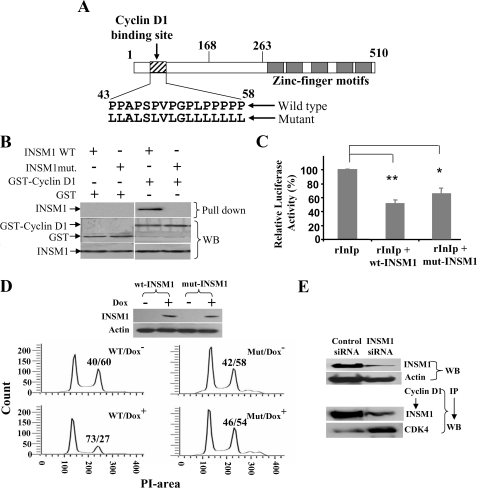
INSM1 Binding to the Cyclin D1 Is Essential to Induce Cell Cycle Arrest—In Fig. 1D, we have shown that the N-terminal portion of the INSM1 (amino acids 1–168) is essential for cyclin D1 binding. Within this region, there is a stretch of proline-rich sequence (amino acids 43–58) that we suspect may contribute directly to the cyclin D1 binding. Therefore, we constructed an INSM1 mutant by replacing all proline with leucine residues (Fig. 7A). A pull-down experiment showed indeed that the INSM1 mutant is no longer capable of binding to cyclin D1 (Fig. 7B). When we tested the INSM1 mutant for its transcriptional repressor activity using a rat insulin promoter as a target gene, only a slight decrease in repressor activity was observed (Fig. 7C). We further established a mutant INSM1 Tet-on cell line in Cos-7 cells. Both wild type and mutant INSM1 Tet-on cell lines were verified for wild type and mutant INSM1 expression induced by doxycycline using Western blot analysis. However, the INSM1 mutant lost its ability to induce cell cycle arrest because of its inability to bind to the cyclin D1 (Fig. 7D). The mutant INSM1 expression even at 24 h failed to inhibit G2/M phase (54%) conversion in contrast to the wild type INSM1, which inhibited G2/M phase to 27%. We also used siRNA against INSM1 to knock down the INSM1 expression in the Cos-7 Tet-on system. As shown in Fig. 7E, INSM1 expression was suppressed by over 80% as compared with the control siRNA. When the INSM1 reduced, the cyclin D1 and CDK4 interaction was increased greatly because the amount of INSM1 was decreased by the siRNA. Overall, these experiments strongly support that cyclin D1 binding to INSM1 can interrupt CDK4 binding and subsequently induce cell cycle arrest.
FIGURE 7.
INSM1 proline-rich sequence is required for cyclin D1 binding. A, schematic diagram of the INSM1 mutant. B, GST-cyclin D1 pull-down of wild type and mutant INSM1. GST was used as a control. Either wild type or mutant INSM1 expression vector was transiently transfected into Cos-7 cells. The transfected cell lysate was subjected to a GST-cyclin D1 pull down experiment. C, measurement of transcriptional repressor activity. rInIp driven luciferase vector was transfected alone or with either wild type or mutant INSM1 into β-TC1 cells. Promoter activities were normalized with pCMV-β-galactosidase internal control (**, p < 0.01; *, p < 0.05). D, wild type or mutant INSM1 Tet-on Cos-7 cells were subjected to doxycycline induction. As described above, the cells were synchronized with serum-free medium, and subsequently stimulated with 10% serum for 24 h. Western blot analysis showed the expression of both wild type and mutant INSM1 proteins when treated with doxycycline. The cells were subjected to propidium iodide staining and flow cytometric analysis. A G1/S:G2/M ratio is shown above the G2/M peak. E, siRNA knock down of INSM1 in the Cos-7 Tet-on system. INSM1 siRNA treatment suppressed INSM1 expression and restored cyclin D1 and CDK4 interaction. WB, Western blot; wt or WT, wild type; mut, mutant.
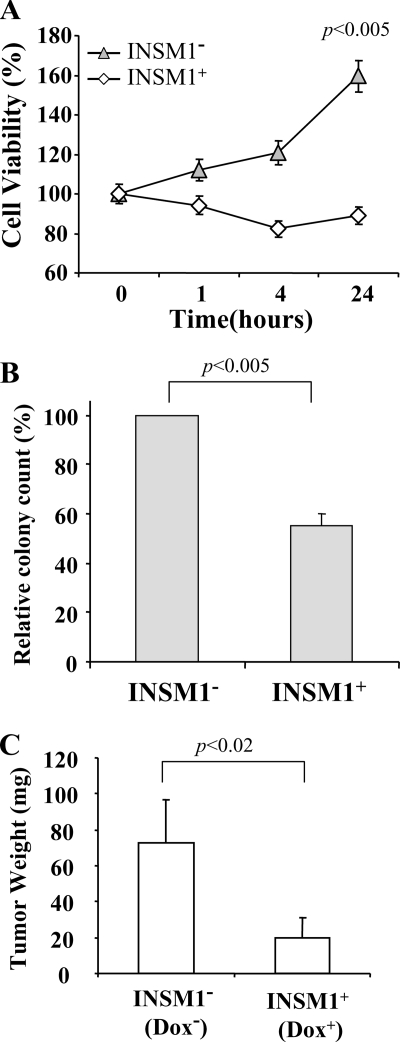
Expression of INSM1 Inhibits Cellular Proliferation—Because ectopic expression of INSM1 induces cell cycle arrest, we speculated that INSM1 could inhibit cellular proliferation in non-neuroendocrine cells. Three experimental procedures were designed to evaluate the effect of INSM1 on cellular proliferation. A MTT assay is the most straightforward measurement of cell viability following INSM1 expression. The Tet-on selected Panc-1 cells were treated with or without doxycycline for 24 h and then subjected to serum starvation and stimulation for various times. Induction of INSM1 inhibited cell proliferation in contrast to the uninduced control cells, which increased to 160% after a 24-h period (Fig. 8A). We further evaluated the effect of ectopic INSM1 expression on colony formation using a soft agar assay. At the beginning, Panc-1 cells were seeded into soft agar as single cells. The number of colonies formed in the INSM1 induced group was significantly lower than that of the uninduced group. In the absence of doxycycline, 50% more colonies were counted than in the Dox-treated group (Fig. 8B). These results implicate that INSM1 expression retards Panc-1 colony formation. Furthermore, we examined Panc-1 cell proliferation in a nude mouse model. Approximately 1 × 107 cells were injected subcutaneously into athymic nude mice on both the left and right hind flanks. Doxycycline (20 mg/kg, intraperitoneally) or PBS was injected daily to the Panc-1-bearing mice for 6 weeks. Doxycycline-induced INSM1 expression inhibited Panc-1 tumor growth by 70% in vivo as compared with the control (Fig. 8C). Collectively, these results support that overexpression of INSM1 in Panc-1 cells inhibits cellular proliferation both in vitro and in vivo.
FIGURE 8.
Ectopic expression of INSM1 inhibits cellular proliferation. A, cell proliferation was measured by a MTT assay. Cell viability was measured after 24 h in Panc-1 cells with or without doxycycline treatment following serum stimulation for various time points. B, the inhibitory effect on cell growth by INSM1 expression in Panc-1 cells was further examined in a soft agar colony formation assay. The relative colony count (%) was determined by averaging 10 separate fields of colonies (colony defined as ≥50 cells). C, an in vivo nude mouse tumor model was employed to evaluate whether INSM1 could delay Panc-1 tumor growth. Approximately 107 Panc-1 cells were injected subcutaneously into the flank of nude mice. Either doxycycline (20 mg/kg, intraperitoneally) or PBS was administered daily 72 h following Panc-1 cell injection. After 6 weeks, the tumors from either group (n = 4) were retrieved and weighed. The result was presented as an average of four tumor weight ± S.E.
DISCUSSION
The INSM1 transcription factor has emerged as an important regulator for pancreatic endocrine cell differentiation (4, 5). Extensive biochemical studies have revealed the DNA consensus binding sequence, transcriptional repressor activity, downstream target genes, upstream regulatory protein of INSM1, and its restricted expression pattern (1–3, 9, 14, 15). These results strongly support its role as a differentiation factor during embryonic neuroendocrine cell development. However, the molecular mechanisms that govern cell fate determination are still unclear. In pancreatic endocrine cell differentiation, neurogenin 3 plays a pivotal role in controlling endocrine cell lineage maturation (16–20). Neurogenin 3 activates INSM1 and NeuroD/β2 during the β-cell differentiation pathway (14, 21), whereas INSM1 and NeuroD/β2 counter-regulate each other during β-cell maturation (3, 11). Expression of INSM1 in fetal pancreas development is highly restricted to a very narrow time window. In the pancreas of INSM1 mutant mice, endocrine precursor cells accumulate but fail to differentiate correctly, suggesting that INSM1 is essential for the development of pancreatic β-cells (5). In the embryonic pancreas, rare INSM1-expressing cells were actively cycling, as demonstrated in a bromodeoxyuridine incorporation study (4). INSM1 mutant mice also show a marked change in the terminal differentiation of chromaffin cells and reduced expression of genes whose protein products control catecholamine synthesis and secretion (6). INSM1 has a pan-neurogenic role and promotes the generation and expansion of basal progenitors in the developing mouse neocortex (22). Furthermore, the homolog Ol-insm1b is strongly expressed in a Medaka model during neurogenesis and pancreas organogenesis, where it plays a role in cell cycle exit and down-regulates cell proliferation during development without triggering apoptosis (23). A recent report identified INSM1 as a Sonic Hedgehog-regulated gene in developing cerebellum and medulloblastoma, suggesting that INSM1 may take part in the Hedgehog-induced tumorigenic process (24). These results support that INSM1 not only functions as a transcriptional repressor but also contributes to cellular proliferation and differentiation. Our previous attempts to establish stable INSM1 cell lines other than of neuroendocrine origin repeatedly failed. These observations led us to hypothesize that in non-neuroendocrine cell types, INSM1 could induce cell cycle arrest as a normal process to direct the cell lineage toward the neuroendocrine pathway. Here, we provide evidence to show that the cyclin box of cyclin D1 is essential for INSM1 binding. INSM1 binding to cyclin D1 competes for the same binding site as CDK4. Therefore, binding of INSM1 to cyclin D1 interrupts normal CDK4 interactions and cell cycle signaling including Rb protein phosphorylation. This competition leads to cell cycle arrest and inhibition of cellular proliferation. Inversely, an INSM1 mutant with changes in the N-terminal proline-rich residues failed to bind to cyclin D1 and was incapable of inducing cell cycle arrest in contrast to the wild type INSM1 protein. The INSM1 mutant still maintains most of its transcriptional repressor activity. This can be explained by the transcriptional complex formed by INSM1-cyclin D1-HDAC-3, where any of the two components are tightly bound to each other (11). Disruption of INSM1 and cyclin D1 binding would not totally diminish the trimer complex.
Cyclin D1 is a key regulator governing normal cell cycle progression. Although it has been shown to possess co-repressor activities by its close association with multiple transcription factors and nuclear receptors including INSM1 (11, 25–33), its cell cycle-dependent activity is mainly mediated through binding and activating CDK4. Activation of CDK4 leads to hyperphosphorylation of Rb protein. The phosphorylated Rb protein releases the bound E2F transcription factor and allows the cell cycle to progress. Our previous and current studies demonstrated that INSM1 is the first zinc finger transcription factor involved in cyclin D1 dual cellular functions including both cell cycle-dependent and independent actions (11). The current study demonstrated that overexpression and binding of INSM1 could induce cell cycle arrest and inhibit cellular proliferation of non-neuroendocrine tumor cells. These observations raise an interesting question because INSM1 is readily expressed in neuroendocrine tumors. We speculate that most of the neuroendocrine tumors suffer from Rb protein mutations, which result in abnormal cell cycle regulation and proliferation (34–36). Thus, INSM1 expression could still block Rb protein phosphorylation, but it will not affect its abnormal cell cycle progression. In contrast to normal neuroendocrine cell differentiation, the Rb pathway is intact, and it is essential to exit from the cell cycle before committing into a differentiation pathway.
In summary, INSM1 can interrupt the normal cell cycle. INSM1 binds and competes against CDK4 for binding to cyclin D1. This leads to a block of Rb protein phosphorylation. Blockage of the cell cycle signaling causes cell cycle arrest. Accompanying cell cycle arrest, cellular proliferation is severely inhibited, which could facilitate a switch to the differentiation pathway.
This work was supported, in whole or in part, by National Institutes of Health Grant DK61436 (to M. S. L.). This work was also supported by funds from the Research Institute for Children, Children's Hospital at New Orleans. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: INSM1, insulinoma-associated antigen-1; CDK4, cyclin-dependent kinase 4; Rb, retinoblastoma; GFP, green fluorescence protein; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; X-a-Gal, 5-bromo-4-chloro-3-indolyl-a-d-galactopyranoside; GST, glutathione S-transferase; PBS, phosphate-buffered saline; siRNA, small interfering RNA; Dox, doxycycline.
References
- 1.Goto, Y., DeSilva, M. G., Toscani, A., Prabhakar, B. S., Notkins, A. L., and Lan, M. S. (1992) J. Biol. Chem. 267 15252–15257 [PubMed] [Google Scholar]
- 2.Breslin, M. B., Zhu, M., Notkins, A. L., and Lan, M. S. (2002) Nucleic Acids Res. 30 1038–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breslin, M. B., Zhu, M., and Lan, M. S. (2003) J. Biol. Chem. 278 38991–38997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mellitzer, G., Bonne, S., Luco, R. F., Van De Casteele, M., Lenne, N., Collombat, P., Mansouri, A., Lee, J., Lan, M. S., Pipeleers, D., Nielsen, F. C., Ferrer, J., Gradwohl, G., and Heimberg, H. (2006) EMBO J. 25 1344–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gierl, M. S., Karoulias, N., Wende, H., Strehle, M., and Birchmeier, C. (2006) Genes Dev. 20 2465–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wildner, H., Gierl, M. S., Strehle, M., Pla, P., and Birchmeier, C. (2008) Development 135 473–481 [DOI] [PubMed] [Google Scholar]
- 7.Zhu, M., Breslin, M. B., and Lan, M. S. (2002) Pancreas 24 139–145 [DOI] [PubMed] [Google Scholar]
- 8.Lan, M. S., Russell, E. K., Lu, J., Johnson, B. E., and Notkins, A. L. (1993) Cancer Res. 53 4169–4171 [PubMed] [Google Scholar]
- 9.Xie, J. P., Cai, T., Zhang, H., Lan, M. S., and Notkins, A. L. (2002) Genomics 80 54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wildwater, M., The, I., and van den Heuvel, S. (2007) Dev. Cell 12 841–842 [DOI] [PubMed] [Google Scholar]
- 11.Liu, W. D., Wang, H. W., Muguira, M., Breslin, M. B., and Lan, M. S. (2006) Biochem. J. 397 169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zwicker, J., Brusselbach, S., Jooss, K. U., Sewing, A., Behn, M., Lucibello, F. C., and Muller, R. (1999) Oncogene 18 19–25 [DOI] [PubMed] [Google Scholar]
- 13.Wang, H. W., Muguira, M., Liu, W. D., Zhang, T., Chen, C., Aucoin, R., Breslin, M. B., and Lan, M. S. (2008) J. Endocrinol. 198 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breslin, M. B., Wang, H. W., Pierce, A., Aucoin, R., and Lan, M. S. (2007) FEBS Lett. 581 949–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duggan, A., Madathany, T., De Castro, S. C. P., Gerrelli, D., Guddati, K., and Garcia-Anoveros, J. (2008) J. Comp. Neurol. 507 1497–1520 [DOI] [PubMed] [Google Scholar]
- 16.Apelqvist, A., Li, H., Sommer, L., Beatus, P., Anderson, D. J., Honjo, T., Hrabe, D. A., Lendahl, U., and Edlund, H. (1999) Nature 400 877–881 [DOI] [PubMed] [Google Scholar]
- 17.Gradwohl, G., Dierich, A., LeMeur, M., and Guillemot, F. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 1607–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen, J., Heller, R. S., Funder-Nielsen, T., Pedersen, E. E., Lindsell, C., Weinmaster, G., Madsen, O. D., and Serup, P. (2000) Diabetes 49 163–176 [DOI] [PubMed] [Google Scholar]
- 19.Schwitzgebel, V. M., Scheel, D. W., Conners, J. R., Kalamaras, J., Lee, J. E., Anderson, D. J., Sussel, L., Johnson, J. D., and German, M. S. (2000) Development 127 3533–3542 [DOI] [PubMed] [Google Scholar]
- 20.Gu, G., Dubauskaite, J., and Melton, D. A. (2002) Development 129 2447–2457 [DOI] [PubMed] [Google Scholar]
- 21.Huang, H. P., Liu, M., El-hodiri, H. M., Chu, K., Jamrich, M., and Tsai, M. J. (2000) Mol. Cell Biol. 20 3292–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farkas, L. M., Haffner, C., Giger, T., Khaitovich, P., Nowick, K., Birchmeier, C., Paabo, S., and Huttner, W. B. (2008) Neuron 60 40–55 [DOI] [PubMed] [Google Scholar]
- 23.Candal, E., Alunni, A., Thermes, V., Jamen, F., Joly, J. S., and Bourrat, F. (2007) Dev. Biol. 309 1–17 [DOI] [PubMed] [Google Scholar]
- 24.De Smacle, E., Fragomeli, C., Ferretti, E., Pelloni, M., Po, A., Canettieri, G., Coni, S., Di Marcotullio, L., Greco, A., Moretti, M., Di Rocco, C., Pazzaglia, S., Maroder, M., Screpanti, I., Giannini, G., and Gulino, A. (2008) Neoplasia 10 89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skaper, S. X., Rhee, J., Kim, P. S., Novitch, B. G., and Lassar, A. B. (1996) Mol. Cell Biol. 16 7043–7053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inoue, K., and Sherr, C. J. (1998) Mol. Cell Biol. 18 1590–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adnane, J., Shao, Z., Robert-Guroff, M., and P. D. (1999) Oncogene 18 239–247 [DOI] [PubMed] [Google Scholar]
- 28.Ganter, B., Fu, S., and Lipsick, J. S. (1998) EMBO J. 17 255–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knudsen, K. E., Cavence, W. K., and Arden, K. C. (1999) Cancer Res. 59 2297–2301 [PubMed] [Google Scholar]
- 30.Bienvenu, F., Gasean, H., and Coqueret, O. (2001) J. Biol. Chem. 276 16840–16847 [DOI] [PubMed] [Google Scholar]
- 31.Ratineau, C., Petry, M. W., Mutoh, H., and Leiter, A. B. (2002) J. Biol. Chem. 277 8847–8853 [DOI] [PubMed] [Google Scholar]
- 32.Lin, H. M., Zhao, L., and Cheng, S. Y. (2002) J. Biol. Chem. 277 28733–28741 [DOI] [PubMed] [Google Scholar]
- 33.Fu, M., Rao, M., Bouras, T., Wang, C., Wu, K., Zhang, X., Li, Z., Yao, T. P., and Pestell, R. G. (2005) J. Biol. Chem. 280 16934–16941 [DOI] [PubMed] [Google Scholar]
- 34.Mori, N., Yokota, J., Akiyama, T., Sameshima, Y., Okamoto, A., Mizoguchi, H., Toyoshima, K., Sugimura, T., and Terada, M. (1990) Oncogene 5 1713–1717 [PubMed] [Google Scholar]
- 35.Gouyer, V., Gazzeri, S., Brambilla, E., Bolon, I., Moro, D., Perron, P., Benabid, A. L., and Brambilla, C. (1994) Int. J. Cancer 58 818–824 [DOI] [PubMed] [Google Scholar]
- 36.Lubensky, I. A., and Zhuang, Z. (2007) Endocr. Pathol. 18 156–162 [DOI] [PubMed] [Google Scholar]