Abstract
Eukaryotic flagella from organisms such as Trypanosoma brucei can be isolated and their protein components identified by mass spectrometry. Here we used a comparative approach utilizing two-dimensional difference gel electrophoresis and isobaric tags for relative and absolute quantitation to reveal protein components of flagellar structures via ablation by inducible RNA interference mutation. By this approach we identified 20 novel components of the paraflagellar rod (PFR). Using epitope tagging we validated a subset of these as being present within the PFR by immunofluorescence. Bioinformatic analysis of the PFR cohort reveals a likely calcium/calmodulin regulatory/signaling linkage between some components. We extended the RNA interference mutant/comparative proteomic analysis to individual novel components of our PFR proteome, showing that the approach has the power to reveal dependences between subgroups within the cohort.
The eukaryotic cilium/flagellum is a multifunctional organelle involved in an array of biological processes ranging from cell motility to cell signaling. Many cells in the human body, across a range of tissues and organs, produce either single or multiple, motile or nonmotile cilia where they perform diverse biological processes essential for maintaining human health. This diversity of function is reflected in an equally diverse range of pathologies and syndromes that result from ciliary/flagellar dysfunction via inherited mutations. This diversity is a reflection of the molecular complexity, both in components and in protein interactions of this organelle (1, 2).
The canonical eukaryotic flagellum displays a characteristic “9 + 2” microtubular profile, where nine outer doublet microtubules encircle two singlet central pair microtubules, an arrangement found in organisms as diverse as trypanosomes, green algae, and mammals. Although this 9 + 2 microtubule arrangement has been highly conserved through eukaryotic evolution, there are examples where this standard layout has been modified, including the “9 + 0” layout of primary cilia and the “9 + 9 + 2” of many insect sperm flagella. In addition to this highly conserved 9 + 2 microtubule structure, flagella and cilia show a vast range of discrete substructures, such as the inner and outer dynein arms, nexin links, radial spokes, bipartite bridges, beak-like projections, ponticuli, and other microtubule elaborations that are essential for cilium/flagellum function. Cilia and flagella can also exhibit various extra-axonemal elaborations, and although these are often restricted to specific lineages, there is evidence that some functions, such as metabolic specialization, provided by these diverse structures are conserved (3, 4). Examples of such extraaxonemal elaborations include the fibrous or rod-like structures in the flagellum of the parasite Giardia lamblia (5), kinetoplastid protozoa (6, 7), and mammalian sperm flagella, along with extra sheaths of microtubules in insect sperm flagella (8).
Several recent studies have set out to determine the protein composition of the flagellum and demonstrated the existence of both an evolutionarily conserved core of flagellum/cilium proteins and a large number of lineage-restricted components (9–13). Although these approaches provide an invaluable catalogue of the protein components of the flagellum, they provide only limited information on the substructural localization of proteins and do not address either the likely protein-protein interactions or the function of these proteins within the flagellum. To address these issues, the protein composition of some axonemal substructures (radial spoke complexes; for example see Ref. 14) has been determined by direct isolation of these structures, and a number of complexes have been resolved by the use of co-immunoprecipitation of indicator proteins (for example see Refs. 15 and 16). In addition the localization and function of a number of flagellar proteins have been investigated by detailed analysis of mutant cell lines (particularly of Chlamydomonas reinhardtii) that exhibit defined structural defects within the assembled axoneme. Early studies employed two-dimensional PAGE to compare the proteomic profile of purified flagella derived from C. reinhardtii mutants and wild type cells (17–22) that showed numerous proteomic differences in the derived profiles. The available technology did not allow identification of the individual proteins within the profiles. Recent proteomic advances offer the opportunity for this identification. For instance the comparative proteomic technique isotope coded affinity tagging has been used to identify components of the outer dynein arm (23). This technique utilizes stable isotope tagging to quantify the relative concentration of proteins between two samples.
Trypanosomatids are important protozoan parasites whose flagellum is a critical organelle for their cell biology and pathogenicity. Their experimental tractability also provides opportunities for generic insights to the eukaryotic flagellum. They are responsible for a number of devastating diseases of humans and other mammals, including commercially important livestock, in some of the poorest areas of the world (24–26). All kinetoplastids build a flagellum that contains an extra-axonemal structure termed the paraflagellar rod (PFR).3 In the case of the African trypanosome Trypanosoma brucei brucei, this consists of a complex subdomain organization of a proximal, intermediate, and distal domain as well as links to specific doublets of the axoneme and a structure known as the flagellum attachment zone (FAZ) by which the flagellum is attached to the cell body for much of its length (6, 7). The PFR is required for cell motility (27, 28) and serves as a scaffold for metabolic and signaling enzymes (3, 29, 30). We have previously shown that the presence of this structure is essential for the survival of the mammalian bloodstream form of the parasite both in vitro (in culture) (12) and in vivo (in mice) (31) as part of a wider requirement for motility in this life cycle stage (12, 32, 33).
Two major protein components of the PFR (PFR1 and PFR2) have been identified (34–38) along with several minor PFR protein components (3, 29, 30, 39–43). The availability of RNAi techniques in T. brucei allowed the generation of the inducible mutant cell line snl2 (44), in which RNAi-mediated ablation of the PFR2 protein causes the specific loss of both the distal and intermediate PFR subdomains (see Fig. 1A). After RNAi induction cells become paralyzed but remain viable (44). Our laboratory (3) has previously identified two PFR-specific adenylate kinases by comparing two-dimensional SDS-PAGE gels of purified flagella from induced and noninduced snl2 cells. These proteins cannot be incorporated into the PFR after PFR2 ablation.
FIGURE 1.
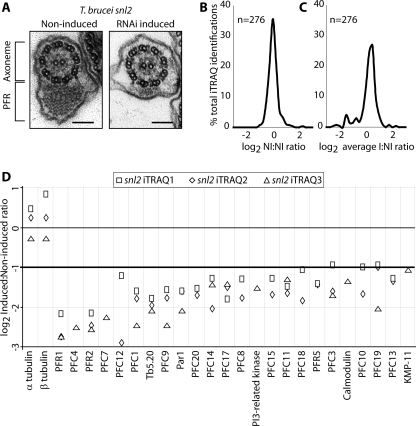
A, electron microscopy images (prepared as described previously (12)) of T. brucei snl2 noninduced and RNAi-induced flagellar transverse sections shows the loss of a large part of the PFR structure. Bar, 100 nm. B, frequencies (resolution 0.25) of log2 protein abundance ratios of noninduced to noninduced samples from quadruplex iTRAQ. C, averaged frequencies (resolution 0.25) of log2 protein abundance ratios of induced to noninduced samples from quadruplex iTRAQ. D, log2 protein abundance ratios of induced to noninduced samples from all iTRAQ experiments for all proteins that show at least a 2-fold decrease after RNAi induction of snl2. α- and β-tubulin show a less than 2-fold change as expected. The results of individual sample pairs are graphed separately as per key.
The ability to ablate PFR2 and hence disable assembly of a major portion of the PFR affords an opportunity to apply advanced proteomic approaches to identify additional PFR proteins. In this present study we have used two complementary proteomic approaches, two-dimensional fluorescence difference gel electrophoresis (DIGE) (45) and isobaric tags for relative and absolute quantitation (iTRAQ; Applied Biosystems), to investigate PFR+ and PFR–flagella to define 30 components of these two PFR subdomains. We have also conducted a bioinformatic analysis of amino acid motifs present in this protein cohort to gain insights into the possible functions of novel proteins and used epitope tagging approaches to confirm the PFR localization of a test set of identified proteins. We then asked whether it was possible to combine comparative proteomics with further analysis of RNAi mutant trypanosomes to provide detailed information on the individual interactions and assembly dependences within the novel PFR components we had identified. By iterating the subtractive proteomic analysis with novel putative PFR proteins, we were able to reveal the existence of distinct PFR protein dependence relationships and provide intriguing new insight into regulatory processes potentially operating within the trypanosome flagellum. Finally, this study establishes the mutant/proteomic combination as a powerful enabling approach for revealing dependences within subcohorts of the flagellar proteome.
EXPERIMENTAL PROCEDURES
Cell Culture—Procyclic T. brucei cells were cultured at 28 °C in SDM-79 medium supplemented with 10% (v/v) heat-inactivated fetal calf serum as previously described (46). For induction of RNAi doxycyclin was added to the medium to a final concentration of 1 μg ml–1.
Vector Construction—200–800 bp from the open reading frame of the gene of interest (open reading frame product) and 200–300 bp of the sequence immediately upstream of the gene of interest (untranslated region product) were amplified by PCR from genomic DNA with the addition of appropriate restriction endonuclease recognition sequences (supplemental text). Open reading frame products were inserted into p2T7-177 (47) between SpeI and XhoI sites, and open reading frame and untranslated region products were inserted into pENT6 BTyYFP (48) between SpeI and BamHI sites.
Transfection—Purified linearized plasmid DNA was used to transfect logarithmically growing cultures of procyclic form T. brucei by electroporation (3 × 100 μs pulses of 1700V). Transfected cells were selected by the addition of 10 μg ml–1 Blasticidin (pENT6 BTyYFP derivatives) and/or 5 μg ml–1 Phleomycin (p2T7-177 derivatives) to the growth medium.
Preparation of Flagella—Procyclic form T. brucei were first treated with PEME (100 mm PIPES, pH 6.9, 2 mm EGTA, 1 mm MgSO4, 0.1 mm EDTA) + 1% Nonidet P-40 and then PEME + 1 m NaCl in the presence of protease inhibitors, DNaseI, and RNaseA. Insoluble material consisting of components of the axoneme, PFR, and a number of other flagellar associated structures but not the flagellar membrane or other soluble components such as IFT particles, was either used immediately or stored for short periods at –20 °C. One-dimensional SDS-PAGE and Western blotting were performed using standard protocols.
Analysis by DIGE—Paired protein samples were labeled with Cy Fluors for DIGE (GE Healthcare) and pooled according to the manufacturer's protocols. Immobilised pH gradient strips were rehydrated in the presence of the samples for 20 h before first dimension focusing (50 μA strip–1 current limit; 10–500 V gradient for 4 h; 500–8000 V gradient for 5 h; 8000 V hold for 6 h). Second dimension separation was performed using SDS-PAGE (1 Watt gel–1 for 1 h and then 13 Watt gel–1 for 4–5 h). Spots were visualized on a Typhoon scanner (GE Healthcare) and analyzed using DeCyder software (GE Healthcare). Spots of interest (criteria in main text) were excised, and proteins identification was performed as below.
Tryptic Digests and MALDI—Two-dimensional gel spots were excised and in-gel digested with trypsin. Briefly gel pieces were washed twice in 25 mm ammonium bicarbonate (Fluka) in 50% acetonitrile (Sigma), dehydrated with an acetonitrile wash, and reduced in 10 mm dithiothreitol (Fluka) for 30 min, before being washed again and dehydrated prior to alkylation using 55 mm iodoacetamide (Fluka) for 60 min. Gel pieces were digested with 200 ng of trypsin at 37 °C overnight. Peptides were acidified using 1 μl of trifluoroacetic acid (Fluka) and extracted with a wash of 0.1% trifluoroacetic acid in 50% acetonitrile and a wash of 0.1% trifluoroacetic acid in 100% acetonitrile. Supernatants were pooled and dried in a SpeedVac (Thermo). Peptides were purified using a home-made C18 purification tips. Peptides were spotted using α-cyano-hydoxycinnamic matrix and analyzed on an Applied Biosystems 4800 MALDI-TOF-TOF. Data were searched using MASCOT (MatrixScience) against an in-house curated T. brucei data base containing trypsin and human keratin. Tolerance was set at 50 ppm for MS and 0.1 Da for MS/MS. Carbamidomethylation of cysteine was set as a fixed modification, and methionine oxidation was set as a variable modification. Positive identifications were accepted with a confidence interval of 99% or greater and two unique peptides.
iTRAQ and Liquid Chromatography MALDI—iTRAQ was performed as per manufacturer's recommendations and labeled peptides purified on an SCX cartridge (Applied Biosystems). The iTRAQ-labeled peptides were fractionated by C18 reverse phase HPLC using a Dionex U3000 nano-HPLC coupled to a Probot spotting robot. A 100-min gradient was used, and fractions were spotted, along with MALDI matrix, directly onto the MALDI target at 15-s intervals. The liquid chromatography run was analyzed on an Applied Biosystems 4800 MALDI-TOF-TOF mass spectrometer, and the data were analyzed using GPS Explorer (Applied Biosystems) and MASCOT. Tolerance was set at 50 ppm for MS and 0.1 Da for MS/MS. Positive identifications were accepted with a confidence interval of 99% or greater and two unique peptides.
Immunofluorescence—Cells were settled onto glass slides and extracted by the addition of 1% Nonidet P-40 in PEME. Cytoskeletons were fixed in methanol and then labeled with BB2 (49) (Ty epitope) and L6B3 (50) (FAZ). Labeling was visualized with 488 fluor-conjugated α-mouse IgM (Invitrogen) or 594 fluor-conjugated α-mouse IgG1 (Invitrogen). The slides were mounted in Vectashield mounting medium with 4′,6′-diamino-2-phenylindole (Vector Laboratories Inc) and examined on a Zeiss Axioplan 2 microscope.
Bioinformatics—BLAST (51) alignments were performed either by using software and data bases available via the Sanger Institute at GeneDB or using an in-house BLAST program available via NCBI with genomes downloaded from JGI or NCBI. For reciprocal BLAST an e-value maximum of 1 e–10 was used, and BLAST results were processed using custom Perl scripts and Excel spreadsheets. MEME searches (52, 53) were parameterized to find any number of repetitions between 6 and 300 amino acids with no limitation on the number of motifs and an entry p value cut-off of 1 e–5. WebLogo was used to generate sequence logos (54). Hidden Markov models were generated as previously described (55).
RESULTS
Comparative Proteomic Analysis of the snl2 RNAi Mutant Cell Line Identifies Known and Putative PFR Components—We have used two complementary comparative proteomic techniques, iTRAQ and DIGE, to identify proteins that are absent from flagella purified from the snl2 induced cells but present in noninduced flagellum samples.
Using iTRAQ, we analyzed three independent sample pairs, each consisting of a noninduced and 72-h RNAi-induced purified flagella. Two sample pairs were analyzed in a quadruplex experiment using four iTRAQ labels, whereas the remaining sample pair was analyzed in a duplex iTRAQ experiment utilizing two of the available labels. In total, 239 proteins were identified in these samples, of which 53% were present in our recent T. brucei flagellum proteome (12). An advantage of using a quadruplex design for two of the pairs is the ability to obtain abundance ratios between the noninduced samples of each pair. When plotted as a frequency distribution of log2 ratios, this shows a near symmetrical distribution with 98% of log2 ratios falling between –1 and +1 (i.e. a less than 2-fold change in either direction) (Fig. 1B). A plot of log2 ratios of the two RNAi-induced samples reveals a similar distribution (not shown). When average log2 ratios of the RNAi-induced to noninduced samples are plotted in the same way, a shoulder is observed on the distribution for values of log2 ratio less than –1 (Fig. 1C). With reference to the ratio distribution obtained by comparison of the two noninduced samples, we defined our proteins of interest as those with a log2 ratio of less than –1 in either iTRAQ experiment (i.e. a greater than 2-fold reduction after PFR2 ablation). In cases where ratios passed this test in one experiment and failed in the other, the ratio generated by the highest number of peptides was accepted. If this occurred between samples in the quadruplex iTRAQ, we applied a stringent approach and did not classify the protein as of interest. The portfolio of PFR candidates generated by this approach consists of 24 proteins, and results from each sample pair are plotted separately in (Fig. 1D).
We also performed a comparative analysis on the snl2 mutant using DIGE, another established proteomic technology, in three experiments using two independent paired samples (noninduced and 72 h RNAi-induced) in each experiment. The resulting gels were analyzed using DeCyder software, and spots were selected on the basis of a fold change in spot volume greater than two (consistent with the criteria applied to the iTRAQ results) (Fig. 2). Spots that exhibited this fold modulation were excised from the gels and subjected to tandem MS protein identification. In total 62 spots were sequenced, and 36 proteins were identified. In cases where multiple identifications were forthcoming from a single spot, we again applied a conservative criterion and did not classify these as proteins of interest because we cannot be sure of the specific contribution of each protein to the reduction in spot volume. It is likely, however, that at least some of these excluded 20 proteins are bona fide PFR components and await further investigation for confirmation. Although one specific group of spots did show an increase in spot volume after induction, no protein identifications were forthcoming from MS/MS analysis. The observation that several spots increased in abundance as a result of PFR2 ablation may be due to differences in post-translational modification altering the mobility of proteins in either one or both of the electrophoresis dimensions. There is no evidence of any other spots increasing in abundance or appearing as a consequence of PFR2 ablation, so it is likely that the majority of the changes observed are due to the absence of proteins in the sample. In total 16 proteins were identified as PFR candidates in this screen, 10 of which were also identified by iTRAQ.
FIGURE 2.
Two-dimensional DIGE analysis of snl2 noninduced and induced flagella. The gels were analyzed using Decyder software (GE Healthcare), and spots that show a greater than 2-fold decrease in volume are marked.
In summary these two proteomic approaches identified 30 proteins as PFR candidates (Table 1) of which 20 are novel. These novel proteins are named here as paraflagellar rod proteome components (PFCs) 1–20. Two proteins in the data set have existing annotations but have not previously been associated with the PFR. KMP-11 has been shown to be differentially expressed during the life cycle of several kinetoplastids and has been localized to the flagellum (56, 57). Tb11.01.6300 is annotated as a PI3K-related kinase by homology, and our analyses of the predicted domain architecture and size of the protein are consistent with this automated annotation (see below). 15 proteins have been identified as PFR components by previous studies, and eight of these proteins are present in our data set. These are the major PFR proteins PFR1 and PFR2 (34–38), PAR1 (39, 42), PFR5 (43), Tb5.20 (40), calmodulin (30), and, as mentioned above, the PFR adenylate kinases ADKA and ADKB (3).
TABLE 1.
Summary of PFR candidates identified in this analysis
PFCs and known PFR proteins are identified by a reduction in protein abundance following inducible RNAi against PFR2. Accession numbers relate to the T. brucei genome project. Relative abundance of proteins is shown as a log2 of the ratio of spot volumes (DIGE) or peak areas of reporter ions (iTRAQ) between RNAi-induced and noninduced samples.
| Accession number | Name | DIGE peptides for identification | Log2 average induced:noninduced DIGE ratio | iTRAQ quadruplex peptides | iTRAQ duplex peptides | Log2 average induced:noninduced ratio |
|---|---|---|---|---|---|---|
| Tb09.211.4513 | KMP-11 | 10 | -1.06 | |||
| Tb10.26.0680 | PFC16 | 7 | -2.79 | |||
| Tb10.389.0100 | PFC20 | 2 | -1.61 | |||
| Tb10.61.1260 | PFC15 | 6 | -1.47 | |||
| Tb10.6k15.0140 | PFC19 | 16 | -1.42 | 3 | 2 | -1.24 |
| Tb10.6k15.0810 | PFC14 | 27 | -2.24 | 7 | 2 | -1.56 |
| Tb10.6k15.1510 | PFC18 | 2 | -1.40 | |||
| Tb11.01.3000 | PFC17 | 4 | -1.38 | 3 | -1.57 | |
| Tb11.01.4623 | Calmodulin | 3 | -1.34 | |||
| Tb11.01.5100 | Par1 | 38 | -2.78 | 8 | 6 | -1.75 |
| Tb11.01.6300 | PI3K-related kinase | 2 | -1.52 | |||
| Tb11.01.6510 | PFC9 | 3 | -1.88 | |||
| Tb11.01.6740 | Tb5.20 | 4 | 2 | -1.94 | ||
| Tb11.02.2350 | PFC12 | 2 | -1.83 | |||
| Tb927.2.2160 | PFC11 | 24 | -1.11 | 3 | 2 | -1.47 |
| Tb927.2.3660 | PFC10 | 2 | -1.30 | |||
| Tb927.2.4330 | PFR5 | 3 | -1.42 | |||
| Tb927.2.5660 | ADKA | 15 | -3.38 | -1.63 | ||
| Tb927.2.950 | PFC13 | 2 | -1.31 | |||
| Tb927.3.3750 | PFC7 | 8 | -2.84 | 2 | -2.25 | |
| Tb927.3.3770 | PFC6 | 20 | -1.84 | 2 | -0.66 | |
| Tb927.3.4290 | PFR1 | 35 | -2.72 | 28 | 30 | -2.53 |
| Tb927.6.3670 | PFC8 | 2 | -1.51 | |||
| Tb927.6.4140 | PFC4 | 2 | -2.51 | |||
| Tb927.7.1920 | PFC5 | 14 | -1.77 | |||
| Tb927.8.1550 | PFC3 | 33 | -2.13 | 9 | 8 | -1.38 |
| Tb927.8.3790 | PFC2 | 9 | -2.63 | -2.45 | ||
| Tb927.8.4970 | PFR2 | 38 | -2.72 | 27 | 32 | -2.38 |
| Tb927.8.6660 | PFC1 | 14 | -1 | 9 | 2 | -1.90 |
| Tb10.70.7330 | ADKB | 13 | -2.12 |
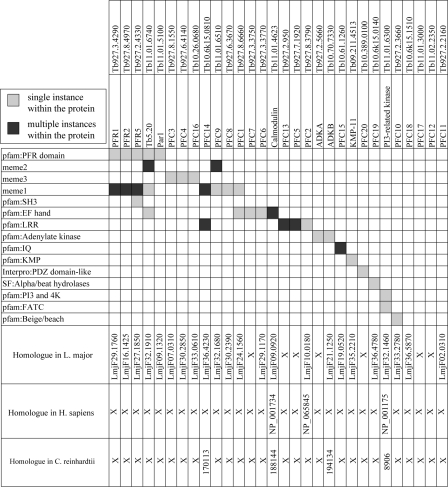
Bioinformatic Analysis of PFR Proteins Reveals Known and Novel Motifs—Because the PFR is an extra-axonemal structure specific to trypanosomes and related protozoa, it might be expected that many of the PFR proteins will be restricted to this lineage. Indeed in silico analysis using a reciprocal BLASTP methodology (Fig. 3) revealed that 25 of the proteins identified are either restricted to T. brucei or have a corresponding gene in the Leishmania major genome but cannot be found in either the Homo sapiens or C. reinhardtii genomes. However, homologues were detected in either H. sapiens, C. reinhardtii, or both for five of these proteins. In some cases, for example calmodulin, this may be as a result of other functions in the cell, but it may also give an insight into conserved flagellar functions, albeit built into variable flagellar structures (4).
FIGURE 3.
Domain and motif architecture of the PFR proteins identified using Pfam and Interpro data bases and MEME (see text for details). BLAST analysis reveals that 25 proteins of the 30 identified in our proteome are trypanosomatid specific, whereas the remaining five are found in either or both Chlamydomonas and human.
We subsequently analyzed the domain and motif architecture of proteins present in our data set using the motif analysis tool MEME (52, 53). This analysis identified numerous domains, many of which correspond to previously predicted Pfam domains (Fig. 3). As previously reported the PFR domain (PF05149) was identified in PFR1, PFR2, PFR5, and PAR1 (43), but we also detected an additional novel occurrence of this domain in Tb5.20. A motif corresponding to the EF hand domain (PF00036) was detected in five proteins (Tb5.20, PFC1, PFC7, PFC6, and calmodulin) and one corresponding to the leucine-rich repeat domain (PF00560) was present in four proteins (PFC13, PFC14, PFC2, and PFC5). As expected, motifs consistent with adenylate kinase signatures were detected for ADKA and ADKB. A Pfam analysis of the data set also identified two IQ calcium-independent calmodulin binding motifs (PF00612) in PFC15, a Beige/BEACH domain (PF02138) in PFC10, as previously reported (43), an SH3 (PF00018) domain in PFR5, and a phosphatidylinositol 3- and 4-kinase (PF00454) and FATC (PF02260) domain in Tb11.01.6300, consistent with the automatic PI3K-related kinase annotation.
In addition to these known domains, MEME also revealed the presence of three novel motifs named here meme 1–3. meme1 is a variable 15-amino acid motif that is present in eight proteins within this data set (PFR1, PFR2, PFR5, Tb5.20, PFC1, PFC9, PFC8, and PFC14) (supplemental Fig. S1A). Although the domain is present in four proteins that also carry the PFR domain, the extent of meme1 does not coincide with any part of this larger domain. Interrogation of the whole T. brucei genome using a hidden Markov model (generated from the alignment of meme1) identified only nine proteins, eight of which were identified in our PFR proteome (supplemental Fig. S1B). Interestingly, the additional protein (Tb10.70.4370) has previously been identified in our T. brucei flagellar proteome (12) and appears to be trypanosomatid-specific. meme2 is a short variable motif of 11 amino acids that is repeated 53 times in Tb5.20 and 10 times in PFC9 (supplemental Fig. S1C). Interestingly, when a hidden Markov model generated from the alignment of meme2 was used to query the predicted proteins in the T. brucei genome, the only additional protein identified was TbI2 (41), a known PFR protein that contains this motif 19 times. meme3 is a 21-amino acid motif present in PFC4, PFC16, and PFC3 (supplemental Fig. S1D). A hidden Markov model generated from the alignment of this motif did not identify any additional proteins when used to search the T. brucei genome.
PFC Proteins Localize to the PFR—The portfolio of proteins generated in this analysis contains eight proteins previously proposed as PFR components. These initial descriptions have come from a number of kinetoplastids (3, 30, 38, 40, 42) and are supported by variable levels of evidence. Where necessary the annotation of these proteins in the T. brucei data set has been inferred from bioinformatics using the TriTryp genome projects (58–60). The presence of these PFR proteins in this data set (representing over 25% of the identifications) is comforting and shows that this RNAi mutant/comparative proteomic method is capable of identifying genuine PFR components and supports the annotation of PFR proteins previously identified only by bioinformatics. To validate the remaining proteins in the data set, we selected seven novel proteins that are representative of the methodologies used to identify them (PFC5 and PFC16: identified only by DIGE; PFC15 and PFC4: identified only by iTRAQ; PFC3, PFC11, and PFC14: identified by both methods) for subcellular localization by epitope tagging and immunofluorescence microscopy. We also used this epitope tagging strategy to localize PFR2 and PAR1, proteins that have previously been shown to localize to the PFR. Transgenic cell lines were generated in which one of the endogenous copies of the gene of interest carried the in-frame coding sequence for the Ty epitope tag (49) immediately downstream of the start codon. Trypanosome cells were fixed and assayed by immunodouble labeling using antibodies against the Ty epitope tag and the FAZ (50). In T. brucei, the PFR lies alongside the axoneme from a point after the flagellum exits the flagellar pocket, beyond the start point of the FAZ, to a point beyond the region of attachment to the cell body. In all cases the tagged protein localized in a portion of the flagellum (Fig. 4A) with a labeling pattern consistent with the PFR (Fig. 4B). All of the proteins were distributed along the length of the flagellum as either a continuous or punctate line. Including the known PFR proteins in our data set, we now have strong evidence for PFR localization of 50% of the proteins identified (and 100% of those tested), suggesting that this is a robust data set that contains a very high proportion of bona fide PFR proteins.
FIGURE 4.
A, Ty epitope tagging of endogenous loci of seven PFC proteins. All seven tagged proteins localize to the flagellum with a pattern consistent with the PFR. B, Ty epitope tagging of endogenous loci of the known PFR components PFR2 and PAR1 exemplify a PFR localization. Green, Ty tagged protein; magenta, FAZ; blue, 4′, 6′-diamino-2-phenylindole. Bar, 2 μm. Arrow, distal extent of the FAZ; arrowhead, start point of the Ty signal.
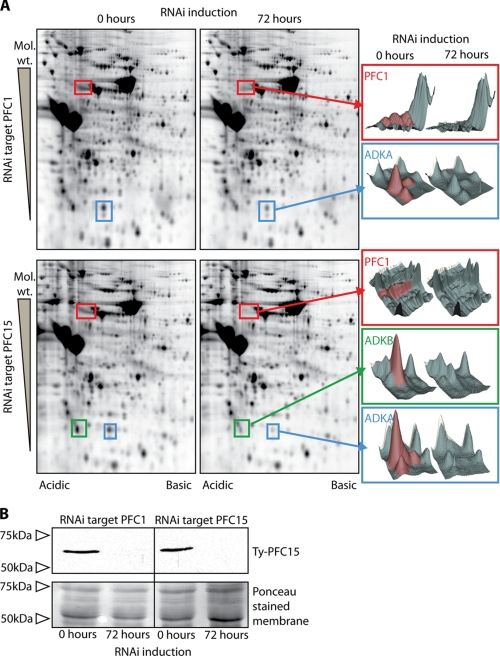
Comparative Proteomics and RNAi Identifies Subgroups, Dependences, and Interactions within the Cohort of PFR Proteins—The presence of calmodulin and the calcium and calmodulin recognition domains in the PFC proteins is indicative of a calcium-regulated system operating within the PFR. To investigate interactions of components within this potential calcium signaling pathway, we conducted RNAi/comparative proteomic analyses using DIGE against two cryptic proteins with predicted domains involved in calcium signaling; PFC1 (EF-hand calcium-binding domain) and PFC15 (IQ-calmodulin-binding domain). A number of spots showed volume reductions following RNAi-mediated ablation of PFC1 and PFC15, and by reference to snl2 DIGE gels (Fig. 2), the identity of the corresponding proteins was determined to be PFC1, ADKA, and ADKB. ADKA spot volumes decreased significantly as a result of RNAi against either PFC1 (log2 ratio ADKA, –0.84) or PFC15 (log2 ratio ADKA, –2.22). PFC1 spot volume was reduced as effectively by PFC15 RNAi (log2 ratio PFC1, –1.50) as it was by PFC1 RNAi (log2 ratio PFC1, –1.53); however, the effect on ADKB spot volume in these RNAi backgrounds differed with a significant reduction only observed after PFC15 RNAi (PFC1 RNAi log2 ratio: –0.37, PFC15 RNAi log2 ratio, –1.25) (Fig. 5A). Although the reasons for this are not immediately clear, it may suggest a role for other proteins in this complex or transport into the flagellum/PFR as a factor. PFC15 has not been detected in DIGE analyses, possibly because of its highly basic nature (predicted pI 10.4). To determine the fate of PFC15 in these RNAi cell lines, we tagged one of the endogenous copies of the gene with a Ty epitope in both PFC1 and PFC15 RNAi backgrounds. RNAi induced and noninduced detergent-extracted pellets derived from each cell line were compared by Western blotting using an antibody that recognizes the Ty epitope. This revealed that, as expected, Ty-PFC15 is readily detectable in noninduced samples and is not present after PFC15 RNAi. However, this analysis also showed that the Ty-PFC15 protein is not correctly assembled into the flagellum after RNAi against PFC1 (Fig. 5B). DIGE analyses using these tagged RNAi cell lines reproduced the previous result for untagged cell lines (data not shown). Overall these results show the interdependency of PFC1 and PFC15 and suggest a possible role for calcium regulation of adenylate kinase function in the PFR. In contrast to the severe motility consequences following ablation of PFR2 and the gross reduction in the PFR structure (28), RNAi against either PFC1 or PFC15 did not obviously affect the motility of the cells under culture conditions (data not shown), as similarly reported previously following RNAi ablation of both ADKA and ADKB (3).
FIGURE 5.
A, two-dimensional DIGE analysis of PFC1 and PFC15 noninduced and induced flagella. The gels were analyzed using Decyder software (GE Healthcare), which was used to generate three-dimensional representations of the spots that show a change in volume after induction. In both RNAi environments, significant reductions in volume were seen for spots corresponding to PFC1 and ADKA. An equally significant volume decrease was observed for ADKB after PFC15 RNAi, but this was not observed after PFC1 RNAi. B, Western blot showing the absence of Ty epitope-tagged PFC15 from the detergent-resistant fraction following RNAi against PFC15 and against PFC1. Ponceau-stained membrane is shown as a loading control.
DISCUSSION
Our aim in this work was to establish a method that combines RNAi ablation of proteins of interest with cutting edge comparative proteomics techniques to generate proteomes for flagellar substructures and provide additional information about protein-protein interactions within these substructures. We have tested this protocol on the well characterized T. brucei PFR mutant snl2 and have identified 30 proteins as components of the PFR. Furthermore, we have been able to iterate the process with novel PFR proteins to define a subset of interdependent components within the cohort. Whether the detected dependences are due to interactions in the final PFR structure or are a result of the process of transporting proteins to the flagellum remains to be determined. There are many advantages to the use of T. brucei for studies of this type. Reverse genetics approaches are well advanced, and the availability of a completed and well annotated genome (Ref. 58; hosted by the Sanger Institute) with a near total absence of introns greatly facilitates the construction of vectors for RNAi, overexpression, and epitope tagging, as well as protein identification by mass spectrometry. In the course of this study we have been able to rapidly turn novel protein identifications into localizations and RNAi phenotypes that have allowed us to target specific cohorts of interacting proteins within the larger framework of the PFR. Most importantly for general future use, our reiteration of this RNAi mutant/proteomic approach at the level of individual proteins (PFC1 and PFC15) shows it to have high sensitivity in revealing subcohort protein dependences.
Previously Identified PFR Components—Previous studies have identified 15 proteins as PFR components in Trypanosome or Leishmania species, based either on interactions, localization, bioinformatics, or a combination of these approaches (3, 29, 30, 39–43, 61). Eight of these previously identified PFR proteins were identified in this screen along with an additional 20 previously hypothetical proteins and two annotated proteins that have not previously been identified as PFR components (KMP-11 and PI3K-related kinase). Because PFR2 was the RNAi target in the snl2 cell line, it was expected that the level of PFR2 protein would be significantly reduced, and this was indeed the case, both by DIGE and iTRAQ analyses. The other major component of the PFR, PFR1 is also reduced, and although it is difficult to resolve these two proteins using two-dimensional DIGE, the average log2 ratios of induced to noninduced samples detected by iTRAQ are –2.4 (PFR1) and –2.6 (PFR2), which is consistent with a near stoichiometric loss of these two proteins after RNAi. Four other proteins have previously been given the soubriquet of PFR (or PAR) (39, 42, 43), and of these PAR1 and PFR5 are both present in our data set.
Calmodulin has been shown to localize to the proximal and distal domains of the PFR as well as to the fibers attaching the PFR to the axoneme (30). Consistent with this, calmodulin was identified by iTRAQ as being reduced following RNAi ablation of the PFR structure in the snl2 cell line. There is evidence that calmodulin interacts directly with one of the major PFR components (30), and several novel proteins described in our analysis have pfam motifs predicted as calmodulin- or calcium-binding domains. In the original proteomic analysis of the snl2 mutant (3), two novel adenylate kinases were identified as PFR components. These two proteins, designated ADKA and ADKB, have an unusual N-terminal extension that is both necessary and sufficient to localize these proteins to the PFR. Interestingly, neither ADKA nor ADKB were observed by iTRAQ within our criteria, but both were identified in the DIGE comparisons and are included in the final data set on this basis. Similarly a number of proteins were detected only by iTRAQ, which supports the use of both comparative methods to more fully explore the samples. The final previously known PFR component detected in this screen is the repetitive protein known as Tb5.20 (40). This protein was isolated from a cDNA library using a complex antisera raised against T. brucei cytoskeletons, and specific antibodies to Tb5.20 localize along the whole length of the PFR.
Seven proteins that have previously been proposed as PFR components are not in our final data set: γ-tubulin, PAR4, PFR5, TbI2, TbI17, PDEB1, and PDEB2 (29, 41–43, 61). Their absence could be due to sampling variations, low protein abundance, masking by other proteins in the case of DIGE, or physical properties that may be refractory to MS identification. However, such absences might indicate discreet localization in the PFR substructures that are not ablated by RNAi against PFR2 such as the proximal domain or the links to the axoneme and FAZ (28).
Novel PFR Components—Two proteins identified in our data set have a pre-existing annotation but have not previously been identified as PFR components. KMP-11 is a conserved membrane protein of kinetoplastids that is mainly associated with the developmental form present in the insect vector where it has been localized to the flagellum and flagellar pocket (56, 57). KMP-11 is currently exciting interest because of its immunological properties (62), and a recent examination of the KMP-11 RNAi phenotype in T. brucei has suggested a role for this protein in regulating basal body segregation with additional consequences for nuclear and cell division (63). Interestingly, a feature of this phenotype was the inability of cells to correctly assemble the FAZ filament in the procyclic but not the bloodstream form. A FAZ is still made in the snl2 mutant as evidenced by the attachment of flagella to the cell body, and so this suggests that KMP-11 may have a complex localization within the PFR such that only a portion of the protein is lost after PFR2 ablation. Tb11.01.6300 is annotated in the T. brucei genome as a PI3K-related kinase by homology, and our analysis of the predicted physical properties of the protein would support this. The family of PI3K-related kinases do not phosphorylate lipids but instead have a Ser/Thr protein kinase activity (64). We have not yet determined a function for this protein in the PFR, but a number of proteins identified in this analysis migrate on two-dimensional gels with a multi-spot pattern that suggests a role for protein phosphorylation in the PFR (Fig. 2).
We have also identified 20 proteins previously annotated as conserved hypothetical as components of the PFR and have verified seven of these by immunolocalization at the light microscope level. We used various bioinformatic strategies as an initial screen to probe for possible functions for these novel proteins and identified a number of interesting patterns. Six PFR proteins, including previously known components and representing 20% of the data set, have domains associated with calcium sensing, and taken together with the previously published interactions of calmodulin with PFR1/2 (30), this suggests an important role for calcium regulation in the PFR. We have also identified a new domain designated meme1 that appears to be largely PFR-specific. Of the nine proteins that could be found to contain meme1, eight are in this data set, and we would predict that the ninth, which we have previously shown to be a trypanosomatid-specific component of the flagellum (12), is also present in the PFR, perhaps in one of the structures less affected by PFR2 ablation. Given that the distribution of meme1 is restricted in the genome, we would predict that this motif has a role in specific protein-protein interactions of the PFR or assembly into or transport or recruitment to this structure, similar to that already established for the N-terminal extension of ADKA and ADKB (3). Finally, an intriguing domain found in this bioinformatic analysis is the Beige/BEACH pfam domain of PFC10. In humans a protein containing this domain is implicated in Chediak-Higashi syndrome, an autosomal recessive disease likely resulting from abnormalities in vesicular transport. However, to our knowledge, none of the pathology associated with this syndrome is likely to be caused as a result of flagella/cilia dysfunctions (65).
Dependence Subgroups Provide Clues about the Role of the PFR in the Regulation of Flagellar Motility—In this work we have demonstrated a reciprocal dependence relationship between two novel PFR proteins: PFC1 and PFC15. These two proteins were chosen for further study because domain predictions suggested a role in a potential PFR calcium signaling network as also suggested by the localization of calmodulin to the PFR and the finding that it interacts with the major PFR components (30). Intriguingly, we have also shown that the PFC1/PFC15 relationship involves the previously identified PFR-specific adenylate kinases ADKA and ADKB (3). Roles for calcium signaling and adenine nucleotides (in addition to the role of ATP as an energy source) in the regulation of flagellar and dynein arm function are well established, and our results may point toward these two systems being linked in the trypanosome PFR. We hypothesize that adenylate kinase function in the PFR responds to changes in calcium concentration to regulate adenine nucleotide homeostasis in the flagellar compartment. This could function to directly regulate the activity of dynein arms (66, 67) or perhaps provide and/or limit substrates for calcium-regulated cyclic nucleotide signaling pathways that have been described in the flagellum and shown to influence the mode of flagellar motility (68, 69). This could then provide a mechanism for calcium-regulated control of flagellar waveform. Flagellar wave reversal, changes in wave form, and regulation of microtubule sliding as a response to changes in calcium concentration have been described in a number of organisms, including trypanosomes (70–81). Recent work from our group demonstrated the switching from flagellar to ciliary waveform in three species closely related to T. brucei (82), and our unpublished observations suggest that this is also a feature of motility in T. brucei. Calcium regulation is an important factor in the hyperactivation of mammalian sperm that involves changes in the flagellar beat (83). Substantial evidence points to the central pair complex and radial spokes as key transducers of calcium signals to the dynein arms in C. reinhardtii (84), and calmodulin has been localized to both of these structures (15, 16). Analysis of C. reinhardtii mutants suggests that the outer dynein arms control the beat frequency of the flagellum, whereas the inner dynein arms are responsible for the shape of the waveform (85). In trypanosomes, however, it appears that beat frequency can be maintained in the absence of outer dynein arms, although the direction of wave propagation is reversed (33). This highlights differences in the regulation of flagellar motility between these organisms, another example being the fixed central pair position of trypanosomes compared with the rotating central pair of Chlamydomonas.
Regulation of adenylate kinase function by calcium has previously been reported in other organisms, including in the flagellum of sea urchin sperm (86–88), and two adenylate kinases have been localized to the fibrous sheath of mouse sperm flagella (89), a structure to which a number of intriguing parallels can be drawn to the PFR (4), suggesting that this could be a more general feature of flagellar beat regulation in other eukaryotes.
Forward View—Several trypanosomatids are the causative agents of devastating parasitic disease in man. In Africa, T. brucei species are responsible for African trypanosomiasis or sleeping sickness, and in central and South America Chagas disease is the result of infection with Trypanosoma cruzi. No vaccines are currently available, and existing drug treatments are associated with high toxicity and, increasingly, drug resistance (24–26). The paraflagellar rod is a specific feature of all of these organisms, and work over several years has shown that, in model systems, vaccination with PFR proteins can confer total immunity to subsequent challenge with T. cruzi (90) or more limited protection against Leishmania species (91). In this work we have presented a list of PFR proteins, many of which are conserved among trypanosomatids but are also restricted to this lineage. Further work is needed to confirm any of these as possible vaccine candidates, although recent studies on one, KMP-11, have shown promising results (62, 92).
Recent work from our group has shown that the mammalian bloodstream form of T. brucei is exquisitely sensitive to loss of the PFR as a result of RNAi ablation of PFR2 whereby mice are able to completely clear a normally lethal challenge by this parasite (31). RNAi mutants affecting axonemal components give a similar phenotype in the bloodstream form (12, 32, 33), suggesting that impaired motility is the major factor in this phenotype and not a specific effect of PFR ablation. However, the restricted evolutionary distribution of the PFR structure compared with the more conserved components of the axoneme makes this a particularly valuable result from the viewpoint of therapeutic potential.
Supplementary Material
Author's Choice—Final version full access.
This work was supported by the Wellcome Trust, the Human Frontier Science Program, and the EP Abraham Trust (to KG). This work was also supported by the Engineering and Physical Sciences Research Council (EPSRC) and the Biotechnology and Biological Sciences Research Council (BBSRC). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental methods and supplemental figure S1.
Footnotes
The abbreviations used are: PFR, paraflagellar rod; RNAi, RNA interference; FAZ, flagellum attachment zone; MS, mass spectrometry; MS/MS, tandem MS; DIGE, difference gel electrophoresis; iTRAQ, isobaric tags for relative and absolute quantitation; PIPES, 1,4-piperazinediethanesulfonic acid; MALDI, matrix-assisted laser desorption ionization; TOF, time-of-flight; HPLC, high pressure liquid chromatography; PFC, paraflagellar rod proteome component; PI3K, phosphatidylinositol 3-kinase.
References
- 1.Fliegauf, M., Benzing, T., and Omran, H. (2007) Nat. Rev. Mol. Cell. Biol. 8 880–893 [DOI] [PubMed] [Google Scholar]
- 2.Marshall, W. F. (2008) J. Cell Biol. 180 17–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pullen, T. J., Ginger, M. L., Gaskell, S. J., and Gull, K. (2004) Mol. Biol. Cell 15 3257–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oberholzer, M., Bregy, P., Marti, G., Minca, M., Peier, M., and Seebeck, T. (2007) Trends Parasitol. 23 71–77 [DOI] [PubMed] [Google Scholar]
- 5.Elmendorf, H. G., Dawson, S. C., and McCaffery, J. M. (2003) Int. J. Parasitol. 33 3–28 [DOI] [PubMed] [Google Scholar]
- 6.Bastin, P., Matthews, K. R., and Gull, K. (1996) Parasitol. Today 12 302–307 [DOI] [PubMed] [Google Scholar]
- 7.Maga, J. A., and LeBowitz, J. H. (1999) Trends Cell Biol. 9 409–413 [DOI] [PubMed] [Google Scholar]
- 8.Werner, M., and Simmons, L. W. (2008) Biol. Rev. Camb. Philos. Soc. 83 191–208 [DOI] [PubMed] [Google Scholar]
- 9.Ostrowski, L. E., Blackburn, K., Radde, K. M., Moyer, M. B., Schlatzer, D. M., Moseley, A., and Boucher, R. C. (2002) Mol. Cell. Proteomics 1 451–465 [DOI] [PubMed] [Google Scholar]
- 10.Pazour, G. J., Agrin, N., Leszyk, J., and Witman, G. B. (2005) J. Cell Biol. 170 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith, J. C., Northey, J. G., Garg, J., Pearlman, R. E., and Siu, K. W. (2005) J. Proteome Res. 4 909–919 [DOI] [PubMed] [Google Scholar]
- 12.Broadhead, R., Dawe, H. R., Farr, H., Griffiths, S., Hart, S. R., Portman, N., Shaw, M. K., Ginger, M. L., Gaskell, S. J., McKean, P. G., and Gull, K. (2006) Nature 440 224–227 [DOI] [PubMed] [Google Scholar]
- 13.Liu, Q., Tan, G., Levenkova, N., Li, T., Pugh, E. N., Jr., Rux, J. J., Speicher, D. W., and Pierce, E. A. (2007) Mol. Cell. Proteomics 6 1299–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang, P., Diener, D. R., Yang, C., Kohno, T., Pazour, G. J., Dienes, J. M., Agrin, N. S., King, S. M., Sale, W. S., Kamiya, R., Rosenbaum, J. L., and Witman, G. B. (2006) J. Cell Sci. 119 1165–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wargo, M. J., Dymek, E. E., and Smith, E. F. (2005) J. Cell Sci. 118 4655–4665 [DOI] [PubMed] [Google Scholar]
- 16.Dymek, E. E., and Smith, E. F. (2007) J. Cell Biol. 179 515–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luck, D., Piperno, G., Ramanis, Z., and Huang, B. (1977) Proc. Natl. Acad. Sci. U. S. A. 74 3456–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piperno, G., Huang, B., and Luck, D. J. (1977) Proc. Natl. Acad. Sci. U. S. A. 74 1600–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, B., Piperno, G., and Luck, D. J. (1979) J. Biol. Chem. 254 3091–3099 [PubMed] [Google Scholar]
- 20.Adams, G. M., Huang, B., Piperno, G., and Luck, D. J. (1981) J. Cell Biol. 91 69–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piperno, G., Huang, B., Ramanis, Z., and Luck, D. J. (1981) J. Cell Biol. 88 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piperno, G., and Luck, D. J. (1982) Prog. Clin. Biol. Res. 80 95–99 [DOI] [PubMed] [Google Scholar]
- 23.Wirschell, M., Pazour, G., Yoda, A., Hirono, M., Kamiya, R., and Witman, G. B. (2004) Mol. Biol. Cell 15 2729–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Souza, W. (2007) Microbes Infect. 9 544–545 [DOI] [PubMed] [Google Scholar]
- 25.Checchi, F., and Barrett, M. P. (2008) Br. Med. J. 336 679–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simarro, P. P., Jannin, J., and Cattand, P. (2008) PLoS Med. 5 e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santrich, C., Moore, L., Sherwin, T., Bastin, P., Brokaw, C., Gull, K., and LeBowitz, J. H. (1997) Mol. Biochem. Parasitol. 90 95–109 [DOI] [PubMed] [Google Scholar]
- 28.Bastin, P., Sherwin, T., and Gull, K. (1998) Nature 391 548. [DOI] [PubMed] [Google Scholar]
- 29.Oberholzer, M., Marti, G., Baresic, M., Kunz, S., Hemphill, A., and Seebeck, T. (2007) FASEB J. 21 720–731 [DOI] [PubMed] [Google Scholar]
- 30.Ridgley, E., Webster, P., Patton, C., and Ruben, L. (2000) Mol. Biochem. Parasitol. 109 195–201 [DOI] [PubMed] [Google Scholar]
- 31.Griffiths, S., Portman, N., Taylor, P. R., Gordon, S., Ginger, M. L., and Gull, K. (2007) Eukaryot. Cell 6 1248–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ralston, K. S., and Hill, K. L. (2006) PLoS Pathog. 2 e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Branche, C., Kohl, L., Toutirais, G., Buisson, J., Cosson, J., and Bastin, P. (2006) J. Cell Sci. 119 3443–3455 [DOI] [PubMed] [Google Scholar]
- 34.Hyams, J. S. (1982) J. Cell Sci. 55 199–210 [DOI] [PubMed] [Google Scholar]
- 35.Russell, D. G., Newsam, R. J., Palmer, G. C., and Gull, K. (1983) Eur J. Cell Biol. 30 137–143 [PubMed] [Google Scholar]
- 36.Cunha, N. L., De Souza, W., and Hasson-Voloch, A. (1984) J. Submicrosc. Cytol. 16 705–713 [PubMed] [Google Scholar]
- 37.Birkett, C. R., Parma, A. E., Gerke-Bonet, R., Woodward, R., and Gull, K. (1992) Gene (Amst.) 110 65–70 [DOI] [PubMed] [Google Scholar]
- 38.Deflorin, J., Rudolf, M., and Seebeck, T. (1994) J. Biol. Chem. 269 28745–28751 [PubMed] [Google Scholar]
- 39.Saborio, J. L., Manuel Hernandez, J., Narayanswami, S., Wrightsman, R., Palmer, E., and Manning, J. (1989) J. Biol. Chem. 264 4071–4075 [PubMed] [Google Scholar]
- 40.Woodward, R., Carden, M. J., and Gull, K. (1994) Mol. Biochem. Parasitol. 67 31–39 [DOI] [PubMed] [Google Scholar]
- 41.Imboden, M., Muller, N., Hemphill, A., Mattioli, R., and Seebeck, T. (1995) Parasitology 110 249–258 [DOI] [PubMed] [Google Scholar]
- 42.Fouts, D. L., Stryker, G. A., Gorski, K. S., Miller, M. J., Nguyen, T. V., Wrightsman, R. A., and Manning, J. E. (1998) J. Biol. Chem. 273 21846–21855 [DOI] [PubMed] [Google Scholar]
- 43.Clark, A. K., Kovtunovych, G., Kandlikar, S., Lal, S., and Stryker, G. A. (2005) Parasitol. Res. 96 312–320 [DOI] [PubMed] [Google Scholar]
- 44.Bastin, P., Ellis, K., Kohl, L., and Gull, K. (2000) J. Cell Sci. 113 3321–3328 [DOI] [PubMed] [Google Scholar]
- 45.Unlu, M., Morgan, M. E., and Minden, J. S. (1997) Electrophoresis 18 2071–2077 [DOI] [PubMed] [Google Scholar]
- 46.Brun, R., and Schonenberger. (1979) Acta Trop. 36 289–292 [PubMed] [Google Scholar]
- 47.Wickstead, B., Ersfeld, K., and Gull, K. (2002) Mol. Biochem. Parasitol. 125 211–216 [DOI] [PubMed] [Google Scholar]
- 48.Kelly, S., Reed, J., Kramer, S., Ellis, L., Webb, H., Sunter, J., Salje, J., Marinsek, N., Gull, K., Wickstead, B., and Carrington, M. (2007) Mol. Biochem. Parasitol. 154 103–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bastin, P., Bagherzadeh, Z., Matthews, K. R., and Gull, K. (1996) Mol. Biochem. Parasitol. 77 235–239 [DOI] [PubMed] [Google Scholar]
- 50.Kohl, L., Sherwin, T., and Gull, K. (1999) J. Eukaryot. Microbiol. 46 105–109 [DOI] [PubMed] [Google Scholar]
- 51.Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990) J. Mol. Biol. 215 403–410 [DOI] [PubMed] [Google Scholar]
- 52.Bailey, T. L., and Elkan, C. (1994) Proc. Int. Conf. Intell. Syst. Mol. Biol. 2 28–36 [PubMed] [Google Scholar]
- 53.Bailey, T. L., Williams, N., Misleh, C., and Li, W. W. (2006) Nucleic Acids Res. 34 369–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004) Genome Res. 14 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Devaux, S., Kelly, S., Lecordier, L., Wickstead, B., Perez-Morga, D., Pays, E., Vanhamme, L., and Gull, K. (2007) Mol. Biol. Cell 18 1293–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stebeck, C. E., Beecroft, R. P., Singh, B. N., Jardim, A., Olafson, R. W., Tuckey, C., Prenevost, K. D., and Pearson, T. W. (1995) Mol. Biochem. Parasitol. 71 1–13 [DOI] [PubMed] [Google Scholar]
- 57.Berberich, C., Machado, G., Morales, G., Carrillo, G., Jimenez-Ruiz, A., and Alonso, C. (1998) Biochim. Biophys. Acta 1442 230–237 [DOI] [PubMed] [Google Scholar]
- 58.Berriman, M., Ghedin, E., Hertz-Fowler, C., Blandin, G., Renauld, H., Bartholomeu, D. C., Lennard, N. J., Caler, E., Hamlin, N. E., Haas, B., Bohme, U., Hannick, L., Aslett, M. A., Shallom, J., Marcello, L., Hou, L., Wickstead, B., Alsmark, U. C., Arrowsmith, C., Atkin, R. J., Barron, A. J., Bringaud, F., Brooks, K., Carrington, M., Cherevach, I., Chillingworth, T. J., Churcher, C., Clark, L. N., Corton, C. H., Cronin, A., Davies, R. M., Doggett, J., Djikeng, A., Feldblyum, T., Field, M. C., Fraser, A., Goodhead, I., Hance, Z., Harper, D., Harris, B. R., Hauser, H., Hostetler, J., Ivens, A., Jagels, K., Johnson, D., Johnson, J., Jones, K., Kerhornou, A. X., Koo, H., Larke, N., Landfear, S., Larkin, C., Leech, V., Line, A., Lord, A., Macleod, A., Mooney, P. J., Moule, S., Martin, D. M., Morgan, G. W., Mungall, K., Norbertczak, H., Ormond, D., Pai, G., Peacock, C. S., Peterson, J., Quail, M. A., Rabbinowitsch, E., Rajandream, M. A., Reitter, C., Salzberg, S. L., Sanders, M., Schobel, S., Sharp, S., Simmonds, M., Simpson, A. J., Tallon, L., Turner, C. M., Tait, A., Tivey, A. R., Van Aken, S., Walker, D., Wanless, D., Wang, S., White, B., White, O., Whitehead, S., Woodward, J., Wortman, J., Adams, M. D., Embley, T. M., Gull, K., Ullu, E., Barry, J. D., Fairlamb, A. H., Opperdoes, F., Barrell, B. G., Donelson, J. E., Hall, N., Fraser, C. M., Melville, S. E., and El-Sayed, N. M. (2005) Science 309 416–42216020726 [Google Scholar]
- 59.El-Sayed, N. M., Myler, P. J., Bartholomeu, D. C., Nilsson, D., Aggarwal, G., Tran, A. N., Ghedin, E., Worthey, E. A., Delcher, A. L., Blandin, G., Westenberger, S. J., Caler, E., Cerqueira, G. C., Branche, C., Haas, B., Anupama, A., Arner, E., Aslund, L., Attipoe, P., Bontempi, E., Bringaud, F., Burton, P., Cadag, E., Campbell, D. A., Carrington, M., Crabtree, J., Darban, H., da Silveira, J. F., de Jong, P., Edwards, K., Englund, P. T., Fazelina, G., Feldblyum, T., Ferella, M., Frasch, A. C., Gull, K., Horn, D., Hou, L., Huang, Y., Kindlund, E., Klingbeil, M., Kluge, S., Koo, H., Lacerda, D., Levin, M. J., Lorenzi, H., Louie, T., Machado, C. R., McCulloch, R., McKenna, A., Mizuno, Y., Mottram, J. C., Nelson, S., Ochaya, S., Osoegawa, K., Pai, G., Parsons, M., Pentony, M., Pettersson, U., Pop, M., Ramirez, J. L., Rinta, J., Robertson, L., Salzberg, S. L., Sanchez, D. O., Seyler, A., Sharma, R., Shetty, J., Simpson, A. J., Sisk, E., Tammi, M. T., Tarleton, R., Teixeira, S., Van Aken, S., Vogt, C., Ward, P. N., Wickstead, B., Wortman, J., White, O., Fraser, C. M., Stuart, K. D., and Andersson, B. (2005) Science 309 409–41516020725 [Google Scholar]
- 60.Ivens, A. C., Peacock, C. S., Worthey, E. A., Murphy, L., Aggarwal, G., Berriman, M., Sisk, E., Rajandream, M. A., Adlem, E., Aert, R., Anupama, A., Apostolou, Z., Attipoe, P., Bason, N., Bauser, C., Beck, A., Beverley, S. M., Bianchettin, G., Borzym, K., Bothe, G., Bruschi, C. V., Collins, M., Cadag, E., Ciarloni, L., Clayton, C., Coulson, R. M., Cronin, A., Cruz, A. K., Davies, R. M., De Gaudenzi, J., Dobson, D. E., Duesterhoeft, A., Fazelina, G., Fosker, N., Frasch, A. C., Fraser, A., Fuchs, M., Gabel, C., Goble, A., Goffeau, A., Harris, D., Hertz-Fowler, C., Hilbert, H., Horn, D., Huang, Y., Klages, S., Knights, A., Kube, M., Larke, N., Litvin, L., Lord, A., Louie, T., Marra, M., Masuy, D., Matthews, K., Michaeli, S., Mottram, J. C., Muller-Auer, S., Munden, H., Nelson, S., Norbertczak, H., Oliver, K., O'Neil, S., Pentony, M., Pohl, T. M., Price, C., Purnelle, B., Quail, M. A., Rabbinowitsch, E., Reinhardt, R., Rieger, M., Rinta, J., Robben, J., Robertson, L., Ruiz, J. C., Rutter, S., Saunders, D., Schafer, M., Schein, J., Schwartz, D. C., Seeger, K., Seyler, A., Sharp, S., Shin, H., Sivam, D., Squares, R., Squares, S., Tosato, V., Vogt, C., Volckaert, G., Wambutt, R., Warren, T., Wedler, H., Woodward, J., Zhou, S., Zimmermann, W., Smith, D. F., Blackwell, J. M., Stuart, K. D., Barrell, B., and Myler, P. J. (2005) Science 309 436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Libusova, L., Sulimenko, T., Sulimenko, V., Hozak, P., and Draber, P. (2004) Exp. Cell Res. 295 375–386 [DOI] [PubMed] [Google Scholar]
- 62.Carrillo, E., Crusat, M., Nieto, J., Chicharro, C., Thomas Mdel, C., Martinez, E., Valladares, B., Canavate, C., Requena, J. M., Lopez, M. C., Alvar, J., and Moreno, J. (2008) Vaccine 26 1902–1911 [DOI] [PubMed] [Google Scholar]
- 63.Li, Z., and Wang, C. C. (2008) Eukaryot Cell 7 1941–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abraham, R. T. (2004) DNA Repair (Amst) 3 883–887 [DOI] [PubMed] [Google Scholar]
- 65.Nagle, D. L., Karim, M. A., Woolf, E. A., Holmgren, L., Bork, P., Misumi, D. J., McGrail, S. H., Dussault, B. J., Jr., Perou, C. M., Boissy, R. E., Duyk, G. M., Spritz, R. A., and Moore, K. J. (1996) Nat. Genet. 14 307–311 [DOI] [PubMed] [Google Scholar]
- 66.Inoue, Y., and Shingyoji, C. (2007) Cell Motil Cytoskeleton 64 690–704 [DOI] [PubMed] [Google Scholar]
- 67.Yagi, T. (2000) Cell Struct. Funct. 25 263–267 [DOI] [PubMed] [Google Scholar]
- 68.Bonini, N. M., and Nelson, D. L. (1988) J. Cell Biol. 106 1615–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bonini, N. M., Evans, T. C., Miglietta, L. A., and Nelson, D. L. (1991) Adv Second Messenger Phosphoprotein Res 23 227–272 [PubMed] [Google Scholar]
- 70.Naitoh, Y., and Kaneko, H. (1972) Science 176 523–524 [DOI] [PubMed] [Google Scholar]
- 71.Hyams, J. S., and Borisy, G. G. (1978) J. Cell Sci. 33 235–253 [DOI] [PubMed] [Google Scholar]
- 72.Brokaw, C. J. (1979) J. Cell Biol. 82 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johnson, R. E., and Brokaw, C. J. (1979) Biophys J 25 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Okuno, M., and Brokaw, C. J. (1979) J. Cell Sci. 38 105–123 [DOI] [PubMed] [Google Scholar]
- 75.Bessen, M., Fay, R. B., and Witman, G. B. (1980) J. Cell Biol. 86 446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cosson, M. P., Tang, W. J., and Gibbons, I. R. (1983) J. Cell Sci. 60 231–249 [DOI] [PubMed] [Google Scholar]
- 77.Kamiya, R., and Witman, G. B. (1984) J. Cell Biol. 98 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Omoto, C. K., and Brokaw, C. J. (1985) Cell Motil 5 53–60 [DOI] [PubMed] [Google Scholar]
- 79.Bannai, H., Yoshimura, M., Takahashi, K., and Shingyoji, C. (2000) J. Cell Sci. 113 (Pt 5), 831–839 [DOI] [PubMed] [Google Scholar]
- 80.Smith, E. F. (2002) Mol. Biol. Cell 13 3303–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Holwill, M. E., and McGregor, J. L. (1976) J Exp Biol 65 229–242 [DOI] [PubMed] [Google Scholar]
- 82.Gadelha, C., Wickstead, B., and Gull, K. (2007) Cell Motil Cytoskeleton 64 629–643 [DOI] [PubMed] [Google Scholar]
- 83.Quill, T. A., Sugden, S. A., Rossi, K. L., Doolittle, L. K., Hammer, R. E., and Garbers, D. L. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 14869–14874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smith, E. F., and Yang, P. (2004) Cell Motil Cytoskeleton 57 8–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brokaw, C. J., and Kamiya, R. (1987) Cell Motil Cytoskeleton 8 68–75 [DOI] [PubMed] [Google Scholar]
- 86.Notari, L., Pepe, I. M., Cugnoli, C., and Morelli, A. (2001) Biochim Biophys Acta 1504 438–443 [DOI] [PubMed] [Google Scholar]
- 87.Notari, L., Morelli, A., and Pepe, I. M. (2003) Photochem Photobiol Sci 2 1299–1302 [DOI] [PubMed] [Google Scholar]
- 88.Kinukawa, M., and Vacquier, V. D. (2007) J Biochem. 142 501–506 [DOI] [PubMed] [Google Scholar]
- 89.Cao, W., Gerton, G. L., and Moss, S. B. (2006) Mol Cell Proteomics 5 801–810 [DOI] [PubMed] [Google Scholar]
- 90.Wrightsman, R. A., Miller, M. J., Saborio, J. L., and Manning, J. E. (1995) Infect Immun 63 122–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Saravia, N. G., Hazbon, M. H., Osorio, Y., Valderrama, L., Walker, J., Santrich, C., Cortazar, T., Lebowitz, J. H., and Travi, B. L. (2005) Vaccine 23 984–995 [DOI] [PubMed] [Google Scholar]
- 92.Basu, R., Bhaumik, S., Basu, J. M., Naskar, K., De, T., and Roy, S. (2005) J. Immunol. 174 7160–7171 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.