Abstract
Cytochromes P450 (P450s) incur phosphorylation. Although the precise role of this post-translational modification is unclear, marking P450s for degradation is plausible. Indeed, we have found that after structural inactivation, CYP3A4, the major human liver P450, and its rat orthologs are phosphorylated during their ubiquitin-dependent proteasomal degradation. Peptide mapping coupled with mass spectrometric analyses of CYP3A4 phosphorylated in vitro by protein kinase C (PKC) previously identified two target sites, Thr264 and Ser420. We now document that liver cytosolic kinases additionally target Ser478 as a major site. To determine whether such phosphorylation is relevant to in vivo CYP3A4 degradation, wild type and CYP3A4 with single, double, or triple Ala mutations of these residues were heterologously expressed in Saccharomyces cerevisiae pep4Δ strains. We found that relative to CYP3A4wt, its S478A mutant was significantly stabilized in these yeast, and this was greatly to markedly enhanced for its S478A/T264A, S478A/S420A, and S478A/T264A/S420A double and triple mutants. Similar relative S478A/T264A/S420A mutant stabilization was also observed in HEK293T cells. To determine whether phosphorylation enhances CYP3A4 degradation by enhancing its ubiquitination, CYP3A4 ubiquitination was examined in an in vitro UBC7/gp78-reconstituted system with and without cAMP-dependent protein kinase A and PKC, two liver cytosolic kinases involved in CYP3A4 phosphorylation. cAMP-dependent protein kinase A/PKC-mediated phosphorylation of CYP3A4wt but not its S478A/T264A/S420A mutant enhanced its ubiquitination in this system. Together, these findings indicate that phosphorylation of CYP3A4 Ser478, Thr264, and Ser420 residues by cytosolic kinases is important both for its ubiquitination and proteasomal degradation and suggest a direct link between P450 phosphorylation, ubiquitination, and degradation.
Hepatic cytochromes P450 (P450s)3 are integral endoplasmic reticulum (ER)-anchored hemoproteins engaged in the oxidative biotransformation of various endo- and xenobiotics. Of these, human CYP3A4 is the most dominant liver enzyme, accounting for >30% of the hepatic microsomal P450 complement, and responsible for the oxidative metabolism of over 50% of clinically relevant drugs (1). In common with all the other ER-bound P450s, CYP3A4 is a monotopic protein with its N-terminal ≈33-residue domain embedded in the ER membrane with the bulk of its structure in the cytosol. Our in vivo studies of the heterologously expressed CYP3A4 in the yeast Saccharomyces cerevisiae as well as of its rat liver CYP3A2/3A23 orthologs in primary hepatocytes have revealed that human and rat liver CYPs 3A are turned over via ubiquitin (Ub)-dependent proteasomal degradation (UPD) (2–8). Thus, CYPs 3A represent excellent prototypic substrates of ER-associated degradation (ERAD), specifically of the ERAD-C pathway (6–11). Consistent with this CYP3A ERAD process, our studies of in vivo and/or in vitro reconstituted systems have led us to conclude that CYPs 3A are ubiquitinated by the UBC7/gp78 Ub-ligase complex and recruited by the p97-Npl4-Ufd1 complex before their degradation by the 26 S proteasome (4–8, 12). Because all these processes are energy-dependent, it is not surprising that in vitro reconstitution of CYP3A4 UPD requires ATP. However, inclusion of γ-S-[32P]ATP in an in vitro reconstituted CYP3A4 ubiquitination system catalyzed by rat liver cytosolic fraction II (FII) resulted in CYP3A4 protein phosphorylation, i.e. γ-[32P]phosphoryl transfer onto CYP3A4 target residues (13, 14). This phosphorylation was enhanced after cumene hydroperoxide (CuOOH)-mediated CYP3A4 inactivation. The physiological role, if any, of this CYP3A4 post-translational modification is unclear.
CYP3A4 is not the only P450 that is phosphorylated. Since the in vitro phosphorylation of a hepatic P450 (CYP2B4) by cAMP-dependent protein kinase A (PKA) was first described (15), various P450s, particularly those belonging to the subfamily 2, were documented to be phosphorylated in cell-free systems, hepatocyte incubations, and intact animals (16–32). Common features of such P450 phosphorylation were the presence of a cytosolically exposed PKA recognition sequence (RRXS) with the Ser residue as the exclusive kinase target, and the ensuing loss of prosthetic heme, conversion to the inactive P420 species, and consequent dramatic functional inactivation (15–20). Studies in intact rats also identified CYPs 3A and 2C6 as kinase targets (21). Although both these P450s lack the hallmark PKA recognition sequence, apparently they possess secondary PKA targeting sequences or are phosphorylated by other protein kinases such as PKC. Indeed, in vitro studies revealed that P450s were phosphorylated in an isoform-dependent manner by either PKA or PKC, except for CYP2B1, which was heavily phosphorylated by both (20). Over the years since this particular post-translational P450 modification was recognized, it has been assigned various functional roles (17, 29–33). Among these, as first proposed by Taniguchi et al. (16) and later explored both by Eliasson et al. (23–26) and us (13, 14), P450 phosphorylation served as a marker for its degradation. Accordingly, the phosphorylation of CYP2E1Ser129 and CYP3A1Ser393 by a microsomal cAMP-dependent protein kinase has been proposed to predispose these P450s but not the similarly phosphorylated CYP2B1 to proteolytic degradation by an integral ER Mg2+-ATP-activated serine protease (23–27). However, heterologous expression of CYP2E1S129A/S129G site-directed mutants in COS7 cells apparently had no effect on its relative stability thereby revealing that if CYP2E1 phosphorylation is important for its degradation (34, 35), then alternate Ser/Thr residues (i.e. in plausible secondary PKA recognition sites, Lys-Lys-Ser209-Lys and Lys-Lys-Ser449-Ala) may be recruited.
On the other hand, on the basis of rapid phosphorylation of CuOOH-inactivated CYP3A4 that precedes its ubiquitination and 26 S proteasomal degradation in an in vitro liver cytosolic FII-catalyzed system, we have proposed that CYP3A4 phosphorylation was essential for targeting it to proteins participating in its UPD/ERAD (13). Indeed, several examples of similar phosphorylation for targeting proteins to UPD exist, of which IκBα phosphorylation is the most notable and perhaps the best documented (36–47; see “Discussion”).
Our in vitro studies with specific kinase inhibitors as probes identified both PKC and PKA as the major FII kinases responsible for CYP3A4 phosphorylation (14). Indeed, in vitro model studies of CYP3A4 with PKC as the kinase, coupled with lysylendopeptidase C (Lys-C) digestion of the phosphorylated protein and liquid chromatography-tandem mass spectrometric (LC-MS/MS) analyses of the Lys-C digests, identified two PKC-phosphorylated CYP3A4 peptides 258ESRLEDpTQK266 and 414FLPERFpSK421 unambiguously phosphorylated at Thr264 and Ser420 (14). These same residues were also phosphorylated in corresponding studies with PKA.4 Furthermore, although both native and CuOOH-inactivated CYP3A4 were phosphorylated at Thr264, Ser420 phosphorylation was particularly enhanced after CuOOH-mediated CYP3A4 inactivation (14). Corresponding studies of CuOOH-inactivated CYP3A4 using rat liver cytosolic FII as the source of the kinase(s), revealed 32P phosphorylation of both these peptides as well as that of an additional CYP3A4 peptide 477LS(p)LGGLLQPEKPVVLK492. Unlike the unambiguous mass spectrometric identification of Thr264 and Ser420 as the phosphorylated CYP3A4 residues, the phosphorylation of Ser478, the only plausible phosphorylatable residue in this 32P-labeled peptide, was not similarly established. Nevertheless, the predominant phosphorylation of Thr264 in native CYP3A4 (14), but of two additional residues in the CuOOH-inactivated enzyme, is consistent with the inactivation-induced structural unraveling of this enzyme with exposure of otherwise concealed and/or kinase-inaccessible domains (48). Such unraveling of CYP3A4 protein stems from the irreversible modification of its active site by fragments generated from CuOOH-mediated oxidative destruction of its prosthetic heme (49). In this study, using mass spectrometric analyses of Lys-C digests of FII-phosphorylated CYP3A4, we have provided unambiguous evidence that in addition to Thr264 and Ser420, Ser478 is indeed phosphorylated. More importantly, through alanine-scanning mutagenesis of these three residues, we now document that although neither the structural conformation nor the catalytic function of this triple CYP3A4T264A/S420A/S478A mutant is altered, its degradation after heterologous expression in S. cerevisiae is significantly impaired. This is also true of CYP3A4T264A/S420A/S478A mutant degradation in human embryonic kidney (HEK293T) cells. Furthermore, using an in vitro reconstituted CYP3A4 ubiquitination system, catalyzed by human Ub-conjugating E2 enzyme UBC7 and integral ER protein gp78 as the E3 Ub ligase (12), we document that PKA/PKC-mediated phosphorylation of the wild type CYP3A4 (CYP3A4wt) considerably enhanced its UBC7/gp78-mediated ubiquitination. Together these findings reveal the critical importance of CYP3A4 phosphorylation at these residues for its UPD and suggest a direct link between phosphorylation and its ubiquitination and degradation.
EXPERIMENTAL PROCEDURES
Materials—Calpain inhibitor I was purchased from Calbiochem, and okadaic acid was obtained from Invitrogen. Sodium vanadate, β-glycerophosphate, sodium fluoride, and phenylmethylsulfonyl fluoride were obtained from Fisher, whereas aprotinin, α-macroglobulin, leupeptin, pepstatin A, 1-chloro-3-tosylamido-7-amino-2-heptanone-HCl, creatine phosphate, creatine phosphokinase, calyculin A, ATP, Ub, CuOOH, DTT, glutathione (GSH, reduced form), and ammonium bicarbonate were obtained from Sigma. MG262 (benzyloxycarbonyl-Leu-Leu-Leu-B(OH)2) was purchased from BostonBiochem (Boston, MA). Nickel-nitrilotriacetic acid superflow resin was obtained from Qiagen (Valencia, CA); TALON Dynabeads was from Invitrogen, and MALDI matrix α-cyano-4-hydroxycinnamic acid was from Waters. POROS 50R2 beads were purchased from Applied Biosystems Inc (Foster City, CA). γ-S-[32P]ATP (specific activity, 6000 Ci/mmol) was obtained from PerkinElmer Life Sciences.
Rat brain PKC (catalytic subunit) was obtained from Calbiochem-Novabiochem, whereas recombinant PKA (catalytic subunit) was from New England Biolabs (Ipswich, MA). Endoproteinase Lys-C (sequencing grade) was from Roche Applied Science. Ub-activating enzyme E1 was purchased from Biomol (Plymouth Meeting, PA). Human cytosolic C-terminal gp78 (E3) and murine UBC7 (E2) were expressed in Escherichia coli and purified as described (50), respectively, from plasmid pGEX-gp78C encoding the human cytosolic C-terminal gp78 domain (residues 309–643) and pGEX-MmUBC7 encoding murine UBC7. Both plasmids were gifts from Dr. A. M. Weissman. All other buffers and reagents were of the highest commercial grade.
Alanine-scanning Mutagenesis—To determine the role of the three phosphorylated CYP3A4 Thr264, Ser420, and Ser478 residues in CYP3A4 degradation, single, double, and triple mutations of these three amino acids to Ala were introduced into expression vectors by Quick-Change site-directed mutagenesis (Stratagene, La Jolla, CA). PCR amplification was performed using mutant sense and antisense primers with pYES2-ADH-3A4-CYC as a template to produce mutant CYP3A4 constructs for heterologous expression in yeast strains. Corresponding mutations were also introduced first into pGEM-3A4(His)6 vector, and then the mutant CYP3A4(His)6 construct was cloned into pCWori+ vector to generate the mutant constructs for expression in E. coli DH5αF strain. The primers used in mutagenesis and the mutant constructs are described (Table 1). The authenticity of each plasmid construct, the presence of the desired mutation, and the absence of extraneous mutations were verified by DNA sequencing.
TABLE 1.
Primers and mutant constructs used in this study
| Constructs | Primers | Templates | Expression system |
|---|---|---|---|
| Single pYES2-ADH-3A4-CYC mutants | |||
| T264A | Sense, GTCGCCTCGAAGATgCACAAAAGCACCGAG | pYES2-ADH-3A4-CYC | Yeast |
| Antisense, CTCGGTGCTTTTGTGcATCTTCGAGGCGAC | |||
| S420A | Sense, GTTCCTCCCTGAAAGATTCgcCAAGAAGAACAAGGACAAC | pYES2-ADH-3A4-CYC | Yeast |
| Antisense, GTTGTCCTTGTTCTTCTTGgcGAATCTTTCAGGGAGGAAC | |||
| S478A | Sense, AGATCCCCCTGAAATTAgcCTTAGGAGGACTTCTTCAAC | pYES2-ADH-3A4-CYC | Yeast |
| Antisense, GTTGAAGAAGTCCTCCTAAGgcTAATTTCAGGGGGATCT | |||
| Double pYES2-ADH-3A4-CYC mutants | |||
| T264A/S420A | Sense, GTTCCTCCCTGAAAGATTCgcCAAGAAGAACAAGGACAAC | pYES2-ADH-3A4T264A-CYC | Yeast |
| Antisense, GTTGTCCTTGTTCTTCTTGgcGAATCTTTCAGGGAGGAAC | |||
| T264A/S478A | Sense, AGATCCCCCTGAAATTAgcCTTAGGAGGACTTCTTCAAC | pYES2-ADH-3A4T264A-CYC | Yeast |
| Antisense, GTTGAAGAAGTCCTCCTAAGgcTAATTTCAGGGGGATCT | |||
| S420A/S478A | Sense, AGATCCCCCTGAAATTAgcCTTAGGAGGACTTCTTCAAC | pYES2-ADH-3A4TS420A-CYC | Yeast |
| Antisense, GTTGAAGAAGTCCTCCTAAGgcTAATTTCAGGGGGATCT | |||
| Triple pYES2-ADH-3A4-CYC mutants | |||
| T264A/S420A/S478A | Sense, AGATCCCCCTGAAATTAgcCTTAGGAGGACTTCTTCAAC | pYES2-ADH-3A4T264AS420A-CYC | Yeast |
| Antisense, GTTGAAGAAGTCCTCCTAAGgcTAATTTCAGGGGGATCT | |||
| Single, double, or triple pCW3A4His6 mutants | |||
| T264A | Sense, GTCGCCTCGAAGATgCACAAAAGCACCGAG | pGEM-3A4-His6 | DH5αF |
| Antisense, CTCGGTGCTTTTGTGcATCTTCGAGGCGAC | |||
| S420A | Sense, GTTCCTCCCTGAAAGATTCgcCAAGAAGAACAAGGACAAC | pGEM-3A4-His6 | DH5αF |
| Antisense, GTTGTCCTTGTTCTTCTTGgcGAATCTTTCAGGGAGGAAC | |||
| S478A | Sense, AGATCCCCCTGAAATTAgcCTTAGGAGGACTTCTTCAAC | pGEM-3A4-His6 | DH5αF |
| Antisense, GTTGAAGAAGTCCTCCTAAGgcTAATTTCAGGGGGATCT | |||
| T264A/S420A | Sense, GTTCCTCCCTGAAAGATTCgcCAAGAAGAACAAGGACAAC | pGEM-3A4-T264A-His6 | DH5αF |
| Antisense, GTTGTCCTTGTTCTTCTTGgcGAATCTTTCAGGGAGGAAC | |||
| T264A/S478A | Sense, AGATCCCCCTGAAATTAgcCTTAGGAGGACTTCTTCAAC | pGEM-3A4-T264A-His6 | DH5αF |
| Antisense, GTTGAAGAAGTCCTCCTAAGgcTAATTTCAGGGGGATCT | |||
| S420A/S478A | Sense, AGATCCCCCTGAAATTAgcCTTAGGAGGACTTCTTCAAC | pGEM-3A4-S420A-His6 | DH5αF |
| Antisense, GTTGAAGAAGTCCTCCTAAGgcTAATTTCAGGGGGATCT | |||
| T264A/S420A/S478A | Sense, AGATCCCCCTGAAATTAgcCTTAGGAGGACTTCTTCAAC | pGEM-3A4-T264AS420A-His6 | DH5αF |
| Antisense, GTTGAAGAAGTCCTCCTAAGgcTAATTTCAGGGGGATCT |
E. coli Expression of CYP3A4(His)6 and Its Ala Mutants—Wild type, C-terminally His6-tagged CYP3A4 (CYP3A4wt) was generated as described (14), or its corresponding single, double, or triple Ala mutant incorporated into a pCWori+ vector was expressed in DH5αF cells grown in TB media at 37 °C and induced with isopropyl β-d-thiogalactopyranoside; the temperature was lowered to 30 °C and then allowed to grow as described previously (14). Recombinant CYP3A4wt and its mutants were purified by Ni2+-affinity chromatography with hydroxylapatite chromatography for detergent exchange, as described previously (14). Stock aliquots were stored at –80 °C until use.
Effect of Ala-scanning Mutagenesis on CYP3A4 Structure and Function—To determine whether mutation of Thr264, Ser420, and/or Ser478 to Ala affected CYP3A4 folding and thus its tertiary structure, the CO-induced spectral analyses of purified recombinant wild type and single, double, and triple mutants were carried out as described (51). The corresponding protein concentration of the purified recombinant enzymes was determined by bicinchoninic acid assay. Their relative testosterone 6β-hydroxylase was also assayed as a functional CYP3A4 marker to verify their relative functional competence after site-directed mutagenesis (51).
CYP3A4 Inactivation by CuOOH—Recombinant CYP3A4(His)6 expressed in E. coli was purified by Ni2+-affinity chromatography coupled with detergent exchange on hydroxylapatite and stored in potassium phosphate buffer (KPB; 50 mm, pH 7.6), containing 20% glycerol, EDTA (0.1 mm), and DTT (0.5 mm), exactly as described previously (14). CYP3A4(His)6 (1.1 nmol) was inactivated by incubation with CuOOH (1.5 mm), in the presence of GSH (1 mm) and EDTA (1 mm) in KPB, in a final volume of 40 μl at 37 °C for 15 min in a shaking water bath. The reaction was terminated by the addition of DTT to a final concentration of 5 mm. The mixture was placed on ice, until immediate use in phosphorylation reactions. In preliminary studies, the inactivation of CYP3A4 or its triple mutant by CuOOH was confirmed by >95% loss of the typical P450-reduced CO spectra relative to a non-CuOOH-treated control incubated in parallel (14).
Phosphorylation of CuOOH-inactivated CYP3A4 by Rat Liver FII—CuOOH-inactivated CYP3A4 (1 nmol, 30 μl) was next incubated with FII (4 mg/ml; prepared as detailed in the supplementary material), Hepes buffer (50 mm, pH 7.5), 15% glycerol, MgCl2 (10 mm), and freshly prepared protease inhibitors as follows: α2-macroglobulin (1 μg/μl), aprotinin (2.4 μg/μl), leupeptin (0.5 μg/μl), calpain inhibitor I (176 μm), soybean inhibitor I (10 μg/ml), E-64 (50 μm), pepstatin (1.4 μg/μl), creatine phosphate (10 mm), creatine phosphokinase (10 units/ml), EGTA (1 mm), okadaic acid (1.5 mm), calyculin A (1 mm), ATP (0.5 mm), and γ-[32P]ATP (10 μCi/μl, when 32P-labeled CYP3A4 was generated) in a final volume of 250 μl, at 30 °C for 30 min in a water bath shaker at 250 rpm.
Isolation of Phosphorylated CuOOH-inactivated CYP3A4—After incubation, a binding buffer consisting of sodium phosphate buffer (50 mm, pH 8.0), 300 mm NaCl, and 0.01% Tween 20 containing 50 mm NaF, 1 mm Na3VO4, 25 mm β-glycerophosphate was added to the above P450 phosphorylation mixture followed by 40 μl of TALON Dynabeads (Invitrogen; 40 mg/ml suspension; binding ratio, 1 mg of beads, 10 μgofa 30-kDa His-tagged protein), and the mixture was shaken for 20 min at room temperature. The beads were then sequentially washed free of contaminating proteins and sedimented magnetically four times with the same binding buffer. Bound CYP3A4(His)6 was eluted from the Dynabeads with a buffer consisting of 50 mm phosphate-buffered saline, 300 mm NaCl, 250 mm imidazole, 0.01% Tween 20, 1% Triton X-100, and 2% β-mercaptoethanol. The eluates were then mixed with an equal volume of SDS-PAGE loading buffer (Tris (62.5 mm, pH 6.8), 25% glycerol, 10% SDS containing 50 mm DTT, and 5% β-mercaptoethanol), boiled at 95 °C for 5 min, and subjected to SDS-PAGE on 9% Tris-HCl gels.
The relative protein phosphorylation of CuOOH-inactivated CYP3A4WT and its T264A/S420A/S478A triple mutant was similarly determined after inclusion of [γ-32P]ATP in the phosphorylation reaction. The corresponding native P450 proteins incubated in parallel in the absence of CuOOH were included as plausible controls for this phosphorylation reaction. SDS-PAGE was carried out on 4–20% Tris-HCl gels, following which the gels were dried and then exposed to PhosphorImaging screens, and visualized using a Typhoon scanner.
Mapping CYP3A4 Phosphorylation Sites by Mass Spectrometry—Sample preparation for mapping phosphorylation sites in CYP3A4(His)6 was carried out exactly as described above except that [γ-32P]ATP was omitted from the phosphorylation reaction. At the end of the reaction, buffer containing phosphatase inhibitors (NaF, sodium vanadate, and β-glycerophosphate) was added as described above, and the phosphorylated CYP3A4(His)6 was “fished” out of the mixture with magnetic TALON Dynabeads. At this point, several experimental strategies to map phosphorylation sites on the isolated CYP3A4 protein were explored. In the first strategy, the purified protein was directly digested on the cobalt magnetic TALON beads with Lys-C (Lys-C:CYP3A4, 1:20, w/w, in 25 mm ammonium bicarbonate, pH 8) at 37 °C for about 12–16 h with constant rotation. The digestion was terminated by the addition of 1.5 volumes of 7% formic acid, 0.2% trifluoroacetic acid, and the enzymatic peptides were analyzed in the modular mass spectrometric tool without any pre-fractionation as described (52). In another strategy, the isolated CYP3A4 protein was further separated by SDS-PAGE; the gel was stained with Coomassie Brilliant Blue (Pierce); the gel band (≈55 kDa) corresponding to CYP3A4 was excised, and the protein was analyzed by the standard in-gel digestion/peptide extraction/mass spectrometric procedure, except that Lys-C was used as the protease (52). In yet another strategy, the CYP3A4 phosphopeptides were enriched after digestion of the protein with Lys-C protease, using a phosphopeptide enrichment kit from Pierce, essentially as recommended by the manufacturer. Phosphopeptides were then purified using a step-elution reversed phase procedure (OligoR3:POROS 50R2 (1:2) loaded on top of Zip Tip), washed with 0.1% trifluoroacetic acid, and eluted with 50% MeOH, 50% acetonitrile, containing 0.1% trifluoroacetic acid as described (52–54). The eluates were directly loaded on the sample plate and subjected to analyses by the above-mentioned modular mass spectrometer (52).
Phosphosite Identification by MALDI-MS and MALDI-MSn Analyses—For peptide extraction, digests were mixed with POROS 50R2 beads (2.5 μl) and incubated at 4 °C for about 8–10 h with strong shaking. The peptide-bound beads were packed into a ZipTip C18 column, washed with 0.1% trifluoroacetic acid (20 μl), and eluted with two sequential 5-μl aliquots of 30% acetonitrile in 0.1% trifluoroacetic acid, followed by four sequential elutions with 5-μl aliquots of 60% acetonitrile in 0.1% trifluoroacetic acid. Aliquots (5 μl) of each peptide eluate were directly spotted onto the MALDI target plate and covered with a saturated α-cyano-4-hydroxycinnamic acid matrix solution (2.5 μl), as described (52–55). The peptides were then subjected to MALDI-MS/MSn analyses on an orthogonal time-of-flight prTOF mass spectrometer (PerkinElmer Life Sciences) in tandem with a vMALDI-Ion Trap mass spectrometer (Thermo Electron) with an interchangeable MALDI target plate. MS/MS data with neutral loss of 98 Da and MS/MS/MS data for identity of phosphopeptide were analyzed exactly as detailed (52).
Yeast Expression of CYP3A4wt and Its Single, Double, or Triple T264A, S420A, and/or S478A Mutants—Expression vector pYes2-ADH-3A4-CYC (modified from pYES2/CT, URA-marked, under the control of ADH1 instead of GAL1 promoter) harboring CYP3A4wt or one of its mutants (Table 1) was heterologously expressed in pep4Δ yeast strain (RHY106-4; ade-2-101 met2 his3Δ200 lys2-801 ura3-52 pep4Δ; kindly provided by Prof. Randy Hampton, University of California, San Diego; 56, 57), exactly as described previously (5). This strain is defective in Pep4p master protease required for maturational processing of proteases critical for vacuolar protein degradation, but it contains a fully active UBC and HRD gene complement and thus a functional UPD as its sole P450 degradation machinery (HRD indicates genes involved in HMG-CoA reductase degradation). Details of the yeast cell transformation protocol and culture conditions have been described previously (4, 5, 8).
As discussed in detail previously (5, 8), of the existing methods for monitoring protein degradation in yeast, most are suitable for relatively short turnover but not long turnover proteins such as the P450s (56–58). Unlike cultured hepatocytes wherein protein synthesis and degradation occur continuously and concurrently, protein degradation in yeast occurs largely in the stationary phase. At this point the de novo protein synthesis is drastically reduced, making pulse-chase degradation analyses of proteins such as P450s extremely inefficient on one hand and erratic on the other. The latter is apparently because of the fact that a fraction of the newly labeled pool gets diverted to a stable pool (58). Similarly, the cycloheximide chase approach (56, 57) is also unsuitable for the longer lived P450s. Cycloheximide can perturb cellular physiology via translational shut-off of protein synthesis during the relatively longer time courses required to monitor P450 degradation thereby altering this process (58, 59). A regulatable promoter such as GAL1–10 would be ideal, but then in yeast, P450s, including CYP3A4, are very poorly expressed from plasmids under such promoters (4, 5, 60). For these combined reasons, P450 degradation was as described previously (4, 5), monitored by the stationary chase analyses. Briefly, yeast strains transformed with CYP3A4wt or CYP3A4 mutant expression plasmid or the corresponding empty vector were grown at 30 °C in SD medium with appropriate supplements, as specifically indicated. Cells were harvested at an early culture stage during the logarithmic growth phase of the culture (A of ≈0.8 at 600 nm), at a mid-stage after “stationary chase” (generally 10–12 h after reaching an A of 0.5 at 600 nm), or at a late stage, i.e. 12 h after reaching an A600 = 0.8.
Microsomal Preparation—Yeast microsomal fractions were prepared from spheroplasts and enriched by removal of the other cellular contaminants by a differential sucrose gradient ultracentrifugation exactly as described previously (5). The microsomal pellet was overlaid with KPB, pH 7.4, containing 1 mm DTT, 0.1 mm EDTA, 20% (v/v) glycerol, and protease inhibitors and stored at –80 °C until used.
CYP3A4wt/CYP3A4 Mutant Immunoblotting Analyses—Microsomal protein (50 μg) from early, mid-stage, and late stage cultures was used in these analyses. The protein content was normalized after methanol/H2SO4 precipitation and acetone washes of yeast microsomes to eliminate interference in the protein assay of variable amounts of adventitious chromophoric material (4, 5). Microsomal CYP3A4wt/CYP3A4 mutant protein content was assayed by Western immunoblotting analyses after transfer onto a nitrocellulose membrane using goat anti-CYP3A4 IgG as the primary antibody (1:5,000, v/v) and rabbit anti-goat horseradish peroxidase-coupled secondary antibody (1:30,000, v/v), with a Pierce Pico ECL detection system. The immunoblots were densitometrically quantitated using ImageJ software as described previously (4, 5). The relative CYP3A4wt/CYP3A4 mutant content at the mid- or late stages of culture was expressed as the percentage of the corresponding CYP3A4wt/CYP3A4 mutant content (100%) at the early stage. Values depicted represent the mean ± S.D. of at the least 3–5 individual experiments.
Degradation Analyses of CYP3A4wt and Its T264A/S420A-S478A Mutant in HEK293T Cells—This human cell line is commonly used in studies of mammalian protein degradation and therefore chosen for verification of our yeast findings (50, 61–64). The mammalian expression plasmids were first generated by PCR amplification of pYES2-ADH-3A4-CYC and pYES2-ADH-3A4T264A/S420A/S478A-CYC templates using 3A4-forward (5′-AAGGATCCATGGCTCTCATCCCAGAC-3′) and 3A4-reverse (5′-AACTCGAGTCAGGCTCCACTTACGGT-3′) primers to generate CYP3A4wt and T264A/S420A/S478A mutant constructs containing 5′-BamHI and 3′-XhoI restriction sites. The lentiviral vector (pHRCMV-GFP-Wsin8) derived from human immunodeficiency virus 1, known to effectively infect human cells, was used for HEK293T cell transfection. pHRCMV-GFP-Wsin8 was digested with BamHI and XhoI restriction enzymes to excise the green fluorescent protein sequence and create a BamHI-XhoI pHRCMV-Wsin8 vector. CYP3A4wt and T264A/S420A/S478A mutant constructs containing 5′-BamHI and 3′-XhoI restriction sites were digested with XhoI to facilitate directional cloning into the BamHI-XhoI pHRCMV-Wsin8 vector to generate the corresponding pHRCMV-3A4wt-Wsin8 and pHRCMV-T264A-S420A-S478A-Wsin8 expression plasmids. A corresponding plasmid without the CYP3A4 sequence served as the empty vector control.
HEK293T cells were grown to 90% confluency in 6-well collagen-coated plates in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 1% penicillin/streptomycin (culture medium). Before transfection this medium was replaced with reduced serum minimal essential medium, Opti-MEM®. Cells were transfected individually with pHRCMV-3A4WT-Wsin18, pHRCMV-T264A-S420A-S478A-Wsin18, or pHRCMV-vector control-Wsin18 using Lipofectamine 2000 (4 μg of DNA, 16.7 μl of Lipofectamine). The transfection was terminated after 6 h by replacing the medium with the culture medium. In preliminary studies, to determine the time course of CYP3A4 expression, the medium was changed 24, 30, 36, 48, 60, and 72 h after transfection. The cells were washed with phosphate-buffered saline and harvested at each of these time points in lysis buffer consisting of Tris-HCl (20 mm, pH 7.5) 1% Triton, NaCl (150 mm), 10% glycerol, EDTA (1 mm), EGTA (1 mm), NaF (100 mm), tetrabasic sodium pyrophosphate (10 mm), β-glycerophosphate (17.5 mm), N-ethylmaleimide (5 mm), Na3VO4 (1 mm), and protease inhibitors phenylmethylsulfonyl fluoride (1 mm), leupeptin (20 μm), aprotinin (1.5 μm), E-64 (50 μm), pepstatin (10 μm), antipain (10 μm), 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (1 mm), and bestatin (60 μm). The cells were lysed using an OMNI™-TH homogenizer and sonicated for 40 s. Lysates were clarified by sedimentation at maximum speed in a table-top microcentrifuge at 4 °C for 15 min. Lysate supernatants (30 μg) were subjected to Western CYP3A4 immunoblotting analyses (see above) and densitometric quantitation using ImageJ software as described above.
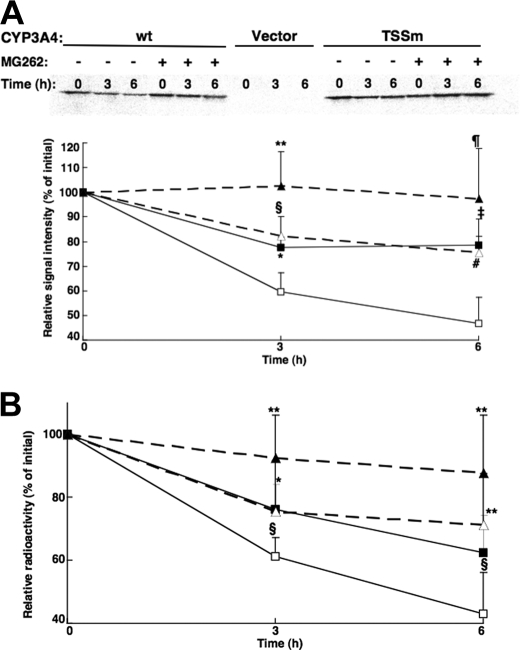
Pulse-Chase Analyses—HEK293T cells were cultured and transfected as described above. Thirty h after transfection, the medium was replaced with Met/Cys-free Dulbecco's modified Eagle's medium for 1 h and pulsed with 20 μCi/ml of Easy-Tag 35S-EXPRESS for 1 h. Each 35S-labeled well was then washed twice with ice-cold phosphate-buffered saline containing excess Met (10 mm) and Cys (1.4 mm). The 35S-labeled protein was chased with 10 mm Met, 1.4 mm Cys medium with or without the proteasomal inhibitor MG262 (10 μm). Cells were harvested in the lysis buffer at the time of chase (0-time) and at 3 or 6 h thereafter. Lysates were prepared as described above.
Aliquots (600 μg of protein; 200 μl) were immunoprecipitated with goat anti-P450 3A4 IgG (1 mg), essentially as described below. Immunoprecipitated CYP3A4 was eluted with 50 μl of an SDS-PAGE loading buffer (described above) by boiling at 95 °C for 5 min. Aliquots of these eluates (30 μl) were subjected to SDS-PAGE on 4–20% Tris-HCl gels and subsequent PhosphorImager analyses. Other aliquots (15 μl) were diluted with KPB buffer to 100 μl and mixed with 6 ml of scintillation mixture for radioactivity determination.
Ubiquitination of CuOOH-inactivated CYP3A4wt and Its Triple Mutant—CYP3A4wt or mutant (250 pmol) was CuOOH inactivated as described above and then incubated at 30 °C for 5–90 min in a reconstituted ubiquitination system consisting of E1 (0.68 μm), human UBC7 (4.25 μm), gp78C (the cytosolic 63-kDa E3 ligase domain; 1 μm), ATP (5 mm), creatine phosphokinase (20 units), creatinine phosphate (20 mm), MgCl2 (12 mm), 32P-Ub (115 μm; prepared as described (12)), Hepes buffer (50 mm, pH 7.4, containing 20% glycerol), EGTA (0.5 mm), EDTA (0.5 mm), PKC (0.004 units), PKA (0.004 units) in a final volume of 70 μl. Control reactions were incubated in parallel for 90 min in the absence of ATP, CYP3A4wt, CYP3A4 triple mutant, or PKA/PKC. Each reaction was terminated with 30 μl of SDS-PAGE 5× loading buffer (250 mm Tris, pH 6.8, 20% SDS, 0.02% Coomassie Blue, 30% glycerol, DTT (584 mm), and 25% β-mercaptoethanol) and boiled at 95 °C for 5 min before being subjected to SDS-PAGE on 4–20% Tris-HCl gels. The gels were then dried and exposed to PhosphorImaging screens and visualized using a Typhoon scanner.
CYP3A4 Immunoprecipitation Analyses—To establish that CYP3A4 was indeed ubiquitinated, CYP3A4wt or its mutant was immunoprecipitated from 90-min reactions identical to those detailed above, including control incubations in the absence of ATP, CYP3A4wt, CYP3A4 triple mutant, or PKA/PKC. At the end of each reaction, N-ethylmaleimide (5 mm, final concentration) was added followed by 10% SDS to a final concentration of 2%, and the samples were boiled for 5 min. CYP3A4 was immunoprecipitated with goat anti-CYP3A4 IgG (3 mg) bound to protein A-Sepharose CL-4B (200 μl) after the mixture was rotated end-to-end at 4 °C overnight essentially as described (3, 6). The immunoprecipitated CYP3A4 was solubilized with 100 μl of SDS-PAGE loading buffer containing 5% SDS, 20% glycerol, DTT (50 mm), and 5% β-mercaptoethanol in 50 mm Tris buffer, pH 6.8, boiled for 5 min, and then equivalent aliquots (48 μl) were subjected to SDS-PAGE on 4–20% Tris-HCl gels. The gels were then dried and exposed to PhosphorImaging screens and visualized using a Typhoon scanner.
Statistical Analyses—Analyses were performed by Student's t test using Microsoft Excel. A value of p < 0.05 was considered statistically significant.
RESULTS
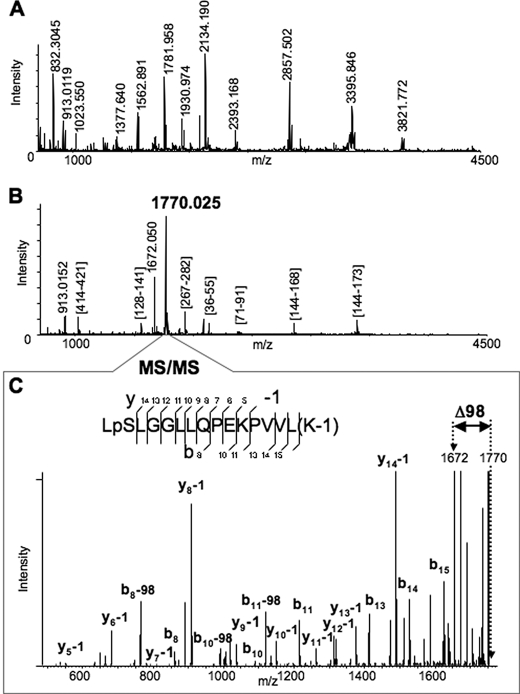
Rat Liver Cytosolic FII-mediated CYP3A4 Phosphorylation, Mass Spectrometric Analyses—Following incubation with rat liver cytosolic FII, CYP3A4 protein isolated with TALON Dynabeads was subjected to mapping of phosphorylation sites by mass spectrometry. The typical MALDI-MS spectrum of Lys-C-digested CYP3A4 is shown in Fig. 1A. After ∼80 ion peaks with the signal-to-noise ratio above 1.5 were detected in the spectrum, the detected species were further subjected to MS/MS analysis (52). All fragmentation spectra were examined for the presence of a fragment distanced by 98 Da from the parent peptide, which is a very characteristic signature of phosphorylation arising because of the losses of elements of phosphoric acid (65). Such analysis consistently revealed the ion species at m/z 1770.025 that exhibited the neutral loss of 98 Da. Furthermore, the same ion peak at m/z 1770.025 was also the most abundant species in the MALDI-MS spectrum of the peptides obtained after phosphopeptide enrichment of the Lys-C digest (Fig. 1B). Although the majority of the peaks in this spectrum was mapped as Lys-C fragments of the digested protein, the identity of the ion peak at m/z 1770.025 could not be readily assigned. Interpretation of the MS/MS spectrum of the ion species at m/z 1770.25 revealed the identity of the peptide (residues 477–492) from CYP3A4 with two modifications (Fig. 1C). One modification is phosphorylation of Ser478 residue. The other is modification of Lys492 by ∼1 Da, which can be due to the possible conversion of the C-terminal carboxyl group to amide or oxidative conversion of lysine to aminoadipic semialdehyde (66, 67). The spectrum also revealed a prominent peptide peak with loss of 98 Da from the parent peptide, which corresponds to the fragment at m/z 1672.050 also observed in the spectrum in Fig. 1B. Thus, our findings reveal that CYP3A4 Ser478 is phosphorylated by liver cytosolic kinases within LSLLGGLLQPEKPVVLK (where underline indicates Ser478) situated in its substrate-recognition sequence 6 domain. Aliquots of the CYP3A4 digests were also subjected to LC-MS/MS that once again revealed Ser478 to be the major phosphosite along with the previously identified Thr264 and Ser420 residues (14) (data not shown). CYP3A4 protein phosphorylation at Ser478, Thr264, and Ser420 thus may have an important biological role.
FIGURE 1.
Identification of the Ser478 phosphorylation site of CYP3A4 by MALDI-MSn analyses. A, MALDI-MS spectrum of Lys-C digested CYP3A4. Some of the enzymatic peptides are labeled in the spectrum. B, MALDI-MS spectrum of Lys-C digested CYP3A4 after the phosphopeptide enrichment performed as described in the text. All the detected peaks were subjected to MS/MS analysis to identify the peptides. The majority of the observed peaks was assigned to Lys-C enzymatic peptides of CYP3A4, except the two most abundant peaks, which were observed at m/z 1770.025 and 1672.050. C, MS/MS spectrum of the peptide at m/z 1770.025. The assignment of the observed fragments revealed the identity of the peptide (477–492) from CYP3A4 with two modifications. One modification is phosphorylation of Ser478 residue. Another is modification of Lys492 by 1 Da (see possible explanations in the text). The spectrum also revealed a prominent peak of loss of 98 Da from the parent peptide, which corresponds to the fragment with m/z 1672.050 also observed in the previous spectrum (B).
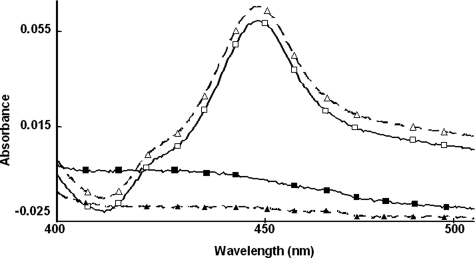
Retention of Typical CYP3A4 Structure-Function Characteristics after Alanine-scanning Mutagenesis—To determine the relative importance of these phosphorylation sites to CYP3A4 degradation, Thr264, Ser420, and/or Ser478 were mutated to Ala singly or in combination via site-directed mutagenesis of an E. coli CYP3A4 expression plasmid, and the corresponding recombinant CYP3A4 proteins were purified to homogeneity. UV-visible spectral analyses of each recombinant CYP3A4 Ala mutant revealed comparable CO-reduced binding spectrum to that of the wild type, as shown for the extreme case of the triple mutant (Fig. 2). The recombinant CYP3A4 proteins also exhibited comparable holo-P450 content (nmol/nmol protein) (Table 2), thereby indicating no gross structural alterations of the single, double, or triple mutants. Functional reconstitution of the purified recombinant proteins also revealed comparable activity as determined by the CYP3A4-selective functional marker, testosterone 6β-hydroxylase (Table 2). Together, these findings verify that mutation of Thr264, Ser420, and/or Ser478 to Ala, singly or in combination, had little effect on CYP3A4 structure and/or function.
FIGURE 2.
CO-binding spectra of reduced CYP3A4wt and its T264A/S420A/S478A (TSSm) mutant before and after CuOOH inactivation. The relative spectra of equivalent protein concentrations of CYP3A4wt (□) and its TSSm mutant (Δ) reveal a comparable maximum absorption at 450 nm, the typical P450 structural signature. The spectra of corresponding CuOOH-inactivated proteins (▪, ▴) reveal that this spectral signature is abolished after P450 inactivation. Spectral analyses of all purified CYP3A4 mutants revealed comparable spectra and spectral content to that of CYP3A4wt (Table 2).
TABLE 2.
Effects of Ala-scanning mutagenesis of Thr264, Ser420, and/or Ser478 on CYP3A4 structure and function
For experimental details see under “Experimental Procedures.” Values are mean ± S.D. of three separate determinations. Testosterone 6β-hydroxylase activity was determined with purified recombinant CYP3A4wt and mutant CYP3A4 proteins, functionally reconstituted with P450 reductase and cytochrome b5 as described (50).
| CYP3A4 | Total CYP3A4 protein content | Holo-P450 content | Holo-P450/total P450 protein content | Testosterone 6β-hydroxylase activity |
|---|---|---|---|---|
| nmol/ml | nmol/ml | nmol/ml | pmol 6β-hydroxytestosterone formed/pmol P450/min | |
| Wild type | 4.17 ± 0.15 | 3.49 ± 0.15 | 0.81 ± 0.17 | 15.0 ± 0.6 |
| T264A | 5.98 ± 0.35 | 4.25 ± 0.44 | 0.71 ± 0.21 | 11.9 ± 0.3 |
| S420A | 10.5 ± 0.75 | 8.71 ± 0.57 | 0.84 ± 0.21 | 15.6 ± 0.4 |
| S478A | 7.22 ± 0.65 | 5.38 ± 0.40 | 0.75 ± 0.17 | 14.4 ± 0.6 |
| T264A/S420A | 5.57 ± 0.20 | 4.54 ± 0.26 | 0.82 ± 0.15 | 12.5 ± 0.4 |
| T264A/S478A | 14.7 ± 0.38 | 11.1 ± 0.36 | 0.76 ± 0.13 | 12.6 ± 0.3 |
| S420A/S478A | 3.46 ± 0.15 | 2.53 ± 0.29 | 0.73 ± 0.19 | 14.2 ± 0.4 |
| T264A/S420A/S478A | 5.63 ± 0.30 | 4.34 ± 0.11 | 0.77 ± 0.13 | 17.5 ± 0.7 |
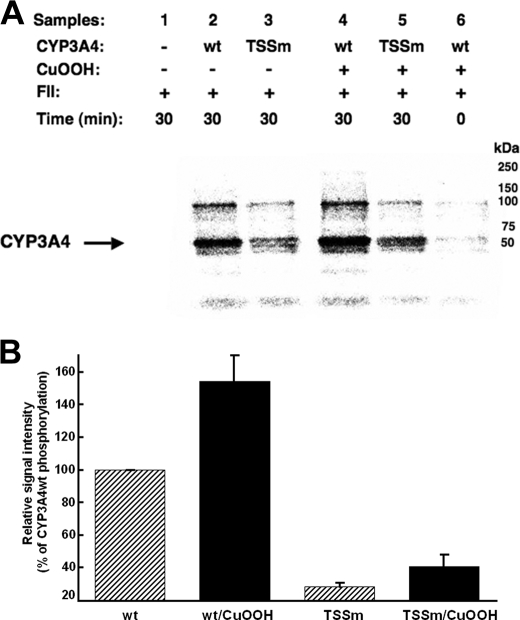
Relative Phosphorylation of CYP3A4wt and Its T264A/S420A/S478A Mutant—CYP3A4wt and its triple mutant were first CuOOH-inactivated (Fig. 2) and then incubated in parallel with rat liver cytosol in the presence of [γ-32P]ATP and an ATP-generating system, which confirmed the previously observed phosphorylation of CYP3A4 protein (Fig. 3) (14). As expected, the triple mutant was phosphorylated to a considerably lesser extent than the wild type protein by rat liver cytosolic FII. Although the extent of the phosphorylation was greatly attenuated, it was not completely abolished in the triple mutant, thereby revealing the plausible existence of additional phosphorylatable residues other than Thr264, Ser420, and Ser478. However, it is noteworthy that the “native” P450 controls incubated in parallel were also phosphorylated, albeit to a lesser extent than the corresponding CuOOH-inactivated enzymes, consistent with our previous findings (14).
FIGURE 3.
In vitro phosphorylation of CYP3A4wt and its T264A/S420A/S478A mutant (TSSm) by FII in the presence of γ-S-[32P]ATP. A, relative extent of phosphorylation of each protein was examined by PhosphorImager scanning after Dynabead TALON extraction and SDS-PAGE analyses of comparably sized aliquots as detailed (see under “Experimental Procedures”). These data are representative of incubations carried out in at least three separate experiments. B, P450 phosphorylation (region between 50 and 250 kDa) was quantitated from the PhosphorImager scanned gels using ImageJ software. Values depicted represent the mean ± S.D. of three individual experiments. The phosphorylation of CuOOH-inactivated CYP3A4wt protein was increased statistically significantly over the corresponding native CYP3A4wt at p < 0.01. Similar statistically significant differences at p < 0.01 were also observed between the phosphorylation of native CYP3A4wt and that of the native TSSm protein, as well as between that of CuOOH-inactivated CYP3A4wt and CuOOH-inactivated TSSm protein. The difference between the phosphorylation of the native TSSm protein and its corresponding CuOOH-inactivated species was statistically significant at p < 0.05. wt, wild type.
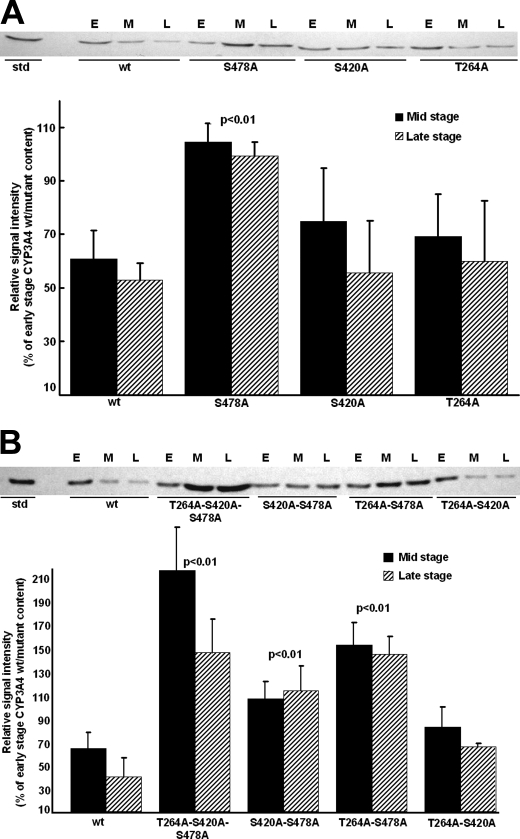
Relative Role of Thr264, Ser420, and Ser478 Phosphorylation in CYP3A4 UPD—To determine the relative importance of each of these phosphorylatable residues to CYP3A4 ERAD, the in vivo degradation of single, double, and triple CYP3A4 mutants was examined in UBC/HRD/pep4Δ S. cerevisiae strains that have Pep4p-dependent vacuolar/autophagic lysosomal degradation deleted leaving UPD as the sole functional proteolytic pathway. CYP3A4wt and its T264A, S420A, and S478A single, double, and triple mutants were heterologously expressed in yeast, and their relative stability was monitored as described previously (4, 5, 8). Western immunoblotting analyses revealed comparable losses of the initial (early stage) content of either T264A or S420A and the wild type at mid and late stages, thereby excluding a major role for either Thr264 or Ser420 phosphorylation by itself in CYP3A4 ERAD (Fig. 4A). In contrast, a statistically significant S478A mutant stabilization relative to the wild type was observed at these times, indicating that Ser478 phosphorylation may be relevant to CYP3A4 degradation (Fig. 4A). To probe whether CYP3A4 degradation requires phosphorylation at more than one site, corresponding double (T264A/S420A, T264A/S478A, or S420A/S478A) and triple (T264A/S420A/S478A) CYP3A4 mutants were similarly probed to determine whether they more profoundly impaired CYP3A4 ERAD than the single mutant (Fig. 4B). Intriguingly, both the double (T264A/S478A and S420A/S478A) mutants containing S478A mutation showed a statistically significant doubling of their stability at both the end points examined, whereas the triple (T264A/S420A/S478A) mutant showed a dramatic 4-fold stabilization of the protein (Fig. 4B), thereby revealing that mutation of all these phosphorylatable CYP3A4 residues considerably influences its degradation. In contrast to the significant stabilization of the degradation of either CYP3A4 S478A double mutant, that of the CYP3A4 T264A/S420A double mutant was not similarly stabilized (Fig. 4B). These findings thus indicate that of the three identified phosphorylated CYP3A4 residues Ser478 was by far the most critical to CYP3A4 degradation.
FIGURE 4.
Relative degradation of CYP3A4wt and T264A, S420A, and/or S478A mutants in S. cerevisiae. A, Western immunoblotting analyses of microsomal proteins (50 μg) at early (E), mid (M), and late (L) stages of culture of CYP3A4wt and single mutants of T264A, S420A, and S478A. B, corresponding analyses of CYP3A4wt and double and triple mutants of T264A, S420A, and S478A. Immunoblots of microsomes from individual yeast cultures (n = 3–4) were subjected to Western immunoblotting analyses and densitometric quantitation as detailed (see under “Experimental Procedures”). Values (mean ± S.D.) are expressed as percent of early stage CYP3A4wt or mutant content. Statistically significant differences by the Student's t test in CYP3A4 mutant protein stabilization from CYP3A4wt at corresponding stages of culture are indicated by the p value. wt, wild type.
Degradation Analyses of CYP3A4wt and Its T264A/S420A/S478A Mutant in HEK293T Cells—To verify that the differential degradation of these CYP3A4 proteins in yeast was relevant to mammalian cells, their degradation was monitored in HEK293T cells after transient expression of a lentiviral vector encoding CYP3A4wt and the triple mutant proteins. In preliminary experiments, the time course (24–72 h) for maximal expression of these CYP3A4 proteins was found to be between 30 and 36 h, with CYP3A4 degradation predominating thereafter. CYP3A4 degradation was monitored by [35S]Met/Cys pulse-chase analyses (Fig. 5, A and B) as detailed under “Experimental Procedures.” After initial 35S labeling, CYP3A4 immunoprecipitation analyses revealed that both CYP3A4wt and its triple mutant were degraded over the 6-h time course, irrespective of whether the data were monitored by PhosphorImager screening and ImageJ analyses (Fig. 5A) or scintillation counting (Fig. 5B) of the corresponding CYP3A4 immunoprecipitates. However, the degradation of the triple mutant was significantly slower than that of CYP3A4wt. These findings from pulse-chase analyses of relative CYP3A4wt and its triple mutant degradation in HEK293T cells (Fig. 5), despite the shorter experimental time course, are thus essentially similar to those observed in the yeast (Fig. 4B). Furthermore, consistent with a UPD process, the degradation of either protein in HEK293T cells was inhibited by the proteasomal inhibitor MG262 (10 μm).
FIGURE 5.
Degradation of CYP3A4wt and its T264A/S420A/S478A mutant (TSSm) by pulse-chase analyses in HEK293T cells. Pulse-chase analyses of HEK293T cells were carried out as detailed after 30 h of transfection of the CYP3A4wt and TSSm expression plasmids and the corresponding vector control, in the presence or absence of the proteasome inhibitor, MG262 (10 μm). At 0, 3, and 6 h after the chase, cells were harvested and lysates subjected to CYP3A4 immunoprecipitation as detailed (see under “Experimental Procedures”). Aliquots of CYP3A4 immunoprecipitates were subjected to SDS-PAGE analyses and PhosphorImager scanning. A representative scan is shown in A. The corresponding 35S-labeled CYP3A4 over the time course was monitored using ImageJ software. Values depicted represent the mean ± S.D. of three individual experiments. Open symbols represent CYP3A4wt (□) and its TSSm mutant (Δ) from cells treated without MG262. Closed symbols (▪, ▴) represent the corresponding values from cells treated with MG262. Statistically significant differences at p < 0.05 were observed between CYP3Awt ± MG262 (*), TSSm ± MG262 (**), and CYP3Awt versus TSSm (§) at 3 h, and between TSSm ± MG262 (¶) at 6 h. Statistically significant differences at p < 0.01 were observed between CYP3Awt ± MG262 (‡) and CYP3Awt versus TSSm (#) at 6 h. B, aliquots of 35S-CYP3A4 immunoprecipitates were also monitored by scintillation counting. Values depicted represent the mean ± S.D. from the three individual experiments in A. Statistically significant differences at p < 0.05 were observed between CYP3Awt ± MG262 (*), TSSm ± MG262 (**), and CYP3Awt versus TSSm (§) at 3 and 6 h.
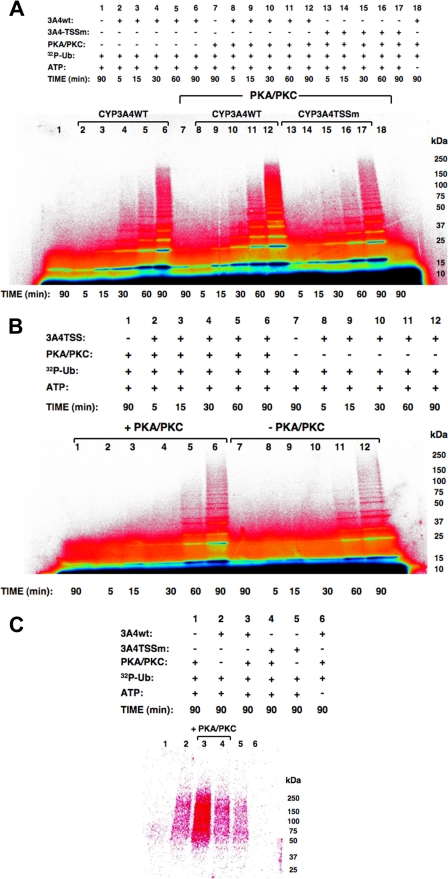
Relative Role of Thr264, Ser420, and Ser478 Phosphorylation in CYP3A4 Ubiquitination—Conceivably, the observed CYP3A4 stabilization from degradation after Ala mutation of the three phosphorylatable residues was because CYP3A4 protein phosphorylation was required for targeting it to an ERAD participant such as an E3 Ub ligase for ubiquitination. To explore this particular possibility, we examined the relative potential of the purified CuOOH-inactivated recombinant CYP3A4wt and its triple mutant to be ubiquitinated in an in vitro mammalian UBC7/gp78-catalyzed system (12; see below). A time-dependent ubiquitination of CuOOH-inactivated CYP3A4wt reaching a maximum at 90 min was observed (Fig. 6A, lanes 2–6), and this ubiquitination pattern was enhanced when PKA/PKC was included in the reaction mixture (Fig. 6A, lanes 8–12). Note the strikingly increased detection of high molecular mass (HMM) ubiquitinated CYP3A4 species in the 75–250-kDa range on inclusion of PKA/PKC (Fig. 6A, lane 6 versus lane 12). No corresponding HMM pattern was observed when CYP3A4 was omitted from this reaction (Fig. 6A, lane 7), thereby revealing that this enhanced ubiquitination profile was not simply due to PKA/PKC-mediated enhancement of the UBC7-gp78 polyUb chain formation, detected as diffuse Ub-trimers/tetramers in the 18–36-kDa range (Fig. 6A, lane 1). Such an HMM or Ub-tri/tetramer profile was also not observed when ATP was excluded from the ubiquitination mixtures5 (Fig. 6A, lane 18). By contrast, inclusion of PKA/PKC in the CYP3A4 triple mutant ubiquitination system resulted in no visibly increased HMM profile at 90 min of incubation (Fig. 6A, lane 17 versus lane 12). Indeed its ubiquitination profile was even slightly fainter than that detected with CYP3A4wt in the absence of PKA/PKC (Fig. 6A, lane 17 versus lane 6). However, direct comparison of CYP3A4 mutant ubiquitination profiles in the presence and absence of PKA/PKC (Fig. 6B, lanes 6 versus lane 12) reveals that its basal ubiquitination is slightly enhanced, possibly because of PKA/PKC-mediated phosphorylation of a residue other than Thr264, Ser420, or Ser478. This finding is also consistent with the considerably lower extent but not abolition of its phosphorylation relative to that of CYP3A4wt (Fig. 3).
FIGURE 6.
Relative role of Thr264, Ser420, and Ser478 phosphorylation in CYP3A4 ubiquitination. A, PhosphorImager analyses of time-dependent ubiquitination of CuOOH-inactivated CYP3A4wt and its triple mutant (TSSm) in the presence or absence of PKA/PKC in the reaction. B, corresponding time-dependent ubiquitination of CuOOH-inactivated CYP3A4 triple mutant in the presence or absence of PKA/PKC in the reaction. Note that in B, the reaction mixture aliquots used were half the size of those in A. C, PhosphorImager analyses of CYP3A4 immunoprecipitates from ubiquitination reactions and corresponding controls carried out at 90 min containing either CuOOH-inactivated CYP3A4wt or TSSm and in the presence or absence of PKA/PKC in the reaction. Data from a typical experiment are shown. Each experiment was conducted in its entirety at the least three separate times. The color wheel intensity code is as follows: black > dark blue > light blue > green > yellow > orange > red > magenta > white.
CYP3A4 immunoprecipitation analyses of aliquots from identical 90-min reactions exhibiting maximal CYP3A4 ubiquitination (Fig. 6A) indeed confirmed that CYP3A4wt was ubiquitinated to a greater extent than its triple mutant in the absence of PKA/PKC (Fig. 6C, lanes 2 versus 5), and this difference was greatly accentuated in PKA/PKC presence (Fig. 6C, lanes 3 versus 4). As expected, no protein 32P ubiquitination was detected in immunoprecipitates from reactions that either lacked CYP3A4 or ATP. Together these findings suggest that phosphorylation indeed enhances UBC7/gp78-mediated CYP3A4 ubiquitination, and thus could promote its degradation via the UPD pathway.
DISCUSSION
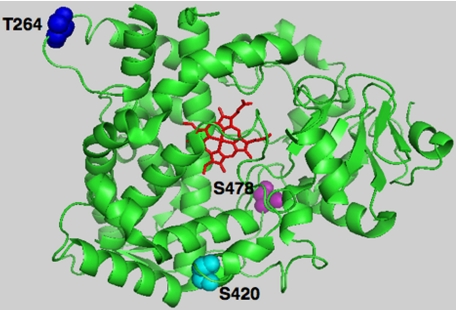
P450s are known to incur post-translational protein phosphorylation, and the target sites identified lie within cytosolically accessible domains harboring kinase consensus recognition motifs (13–32). We have previously reported that cytosolically exposed Thr264 (14) is one such residue that is phosphorylated by PKC, PKA, and/or rat liver cytosolic kinases in both native and suicidally inactivated CYP3A4, whereas other normally concealed and deeply buried residues such as Ser420 become accessible only after CYP3A4 inactivation (14). In this study, we have identified yet one more phosphorylated residue, Ser478, which is located within CYP3A4 substrate recognition sequence 6 (48), and thus in a bona fide active site region normally protected from cytosolic kinases (Fig. 7). The biological relevance of the observed phosphorylation sites was examined through heterologous expression of each single, double, or triple CYP3A4 mutant in yeast following site-directed mutagenesis of these CYP3A4 phosphorylatable residues (Thr264, Ser420, and Ser478) to Ala. These findings documented that mutation of Ser478 by itself significantly abrogated its UPD and markedly stabilized the protein relative to the native enzyme. This effect was greatly magnified when Ser478 mutation was coupled with that of either Thr264 or Ser420. It was further exacerbated when all three residues were jointly mutated, thereby indicating that multisite protein phosphorylation at these particular CYP3A4 residues is indeed biologically relevant to its UPD.
FIGURE 7.
PyMol depiction of CYP3A4 phosphorylatable Thr264, Ser420, and Ser478 residues. The CYP3A4 crystal structure (48) was used as the template, depicting the prosthetic heme in red, phosphorylatable residues Thr264 in blue, Ser420 in cyan, and Ser478 in magenta.
That such biological relevance of P450 phosphorylation to yeast UPD extends to that of the higher eukaryotes was documented by P450 degradation analyses in HEK293T cells. Transient CYP3A4 transfection of these cells coupled with pulse-chase analyses not only verified that CYP3A4wt and its T264A/S420A/S478A mutant were both proteasomally degraded, but also that the degradation of the triple mutant was significantly slower (Fig. 5). It is to be noted however that such T264A/S420A/S478A mutant stabilization is more pronounced in the pep4Δ -yeast strain. This is not only because of the relatively longer degradation time course employed but also possibly because of the fact that on genetic deletion of the yeast vacuolar (autophagic lysosomal) degradation pathway, UPD remains its major if not the sole P450 degradation pathway. By contrast, HEK293T cells in our studies had both pathways functionally intact and thus could provide alternative pathways for P450 degradation. However, the virtual abolition of the phosphorylation-impaired CYP3A4 T264A/S420A/S478A mutant turnover by MG262-induced proteasomal inhibition (Fig. 5A) reveals that phosphorylation is indeed relevant to global CYP3A4 proteolytic turnover in these cells. Collectively, these findings once again underscore the remarkable similarities in the yeast and mammalian P450 degradation pathways and thus the usefulness and validity of the yeast model for P450 degradation studies.
The issue that this in vivo documentation of the relevance of protein phosphorylation to yeast and HEK293T cell P450 degradation ostensibly utilized native CYP3A4 proteins, whereas our in vitro CYP3A4 phosphorylation studies were conducted with CuOOH-inactivated P450s, requires a clarification. Both native and suicidally inactivated CYP3A incur UPD in a classical ERAD process, and this may in part be determined by intrinsic molecular degrons in the CYP3A4 structure. However, the relative rate of this degradation is considerably increased after CYP3A inactivation (2, 3, 6). Indeed, CYP3A suicide inactivation by 3,5-dicarbethoxy-2,6-dimethyl-4-ethyl-1,4-dihydropyridine, the agent employed in our previous studies (2, 3, 6, 68), and that by CuOOH or H2O2 is mechanistically very similar, involving oxidative fragmentation of the heme to reactive products that irreversibly bind to the P450 active site (49, 69, 70). This structural insult damages the P450 protein, marking it for ubiquitination and thus rapid clearance via UPD (2, 3, 6, 7, 68).
Similar CYP3A heme modification is also physiologically detected (71, 72). It apparently stems from H2O2 generated during futile oxidative cycling (73–76) with consequently similar structural damage that could trigger their normal turnover. This may also be true of other P450s, such as CYP2E1, that are also degraded via UPD (77–81). Plausible evidence for this possibility is the marked stabilization of these P450 proteins on inhibition or abortion of this futile oxidative uncoupling by the following: (i) their substrates (EtOH, acetone, and clotrimazole; Refs. 26, 80, 81); (ii) troleandomycin, a quasi-irreversible inactivator, that by complexing CYP3A heme renders it catalytically inactive (82); or (iii) conditional deletion (83) or chemical inhibition (84, 85) of P450 reductase and consequent impairment of the electron flux through the P450 heme.
Given its inherent mechanistic similarities to the physiological CYP3A inactivation, CuOOH inactivation is thus merely a means to accelerate this process, enabling the scrutiny of the phosphorylation/ubiquitination reactions within the limits of viable incubation periods for purified P450 proteins. At a practical level, it also obviates the need for inclusion of P450 reductase and/or cytochrome b5 in the in vitro systems thereby simplifying the LC-MS/MS characterization of post-translationally modified CYP3A4 peptides.
We find it noteworthy that both in yeast and HEK293T cells, CYP3A4 UPD occurs without any deliberate coexpression of P450 reductase. Thus, in these eukaryotic cells, the basal level of futile oxidative cycling of CYP3A4 could be low, consistent with their relatively low endogenous content of P450 reductase. The fact that under these conditions, CYP3A4 UPD is still a Thr264, Ser420, and Ser478 phosphorylation-dependent process (Fig. 5) implies the possible existence of cellular mechanisms such as alternate P450 reducing systems and/or P450 unfoldases, involved in exposing some of these otherwise concealed CYP3A4 residues to cytosolic kinases.
Intriguingly, however, the above findings of multisite phosphorylation of CYP3A4 (Thr264, Ser420, Ser478, and possibly other residues), and its significant attenuation by inhibitors of both PKA and/or PKC (14), reveal that multiple kinases are most likely involved in this process. However, it remains to be determined whether this process is hierarchical with initial phosphorylation of one residue by one particular kinase serving to prime the recognition of another residue by a different kinase (86, 87), or whether multiple kinases concomitantly target CYP3A4 as a substrate.
However, the specific biological purpose of this multisite CYP3A4 phosphorylation remained unclear. Well established precedents for a causal association between protein phosphorylation and UPD exist in the turnover of several short lived cell regulatory proteins (36–47). Of these, the degradation of the prototypic NF-κB inhibitor, IκBα may be the most instructive (42–45). Phosphorylation of IκBα Ser32/Ser36 is apparently required for its ubiquitination at Lys21/Lys22, which targets the ubiquitinated protein to the cellular AAA ATPase p97 (Cdc48p in yeast), that in turn ferries it to the 26 S proteasome (42–45). Studies with IκBα S32A/S36A and K21R/K22R mutants indicated that both Lys21/Lys22 and Ser32/Ser36 are required for phosphorylation, ubiquitination, p97 binding, and subsequent UPD (42–45). Thus it was conceivable that phosphorylation of CYP3A4 Thr264, Ser420, and Ser478 residues could similarly target the protein for its ubiquitination by an ERAD-associated Ub ligase.
We therefore examined the role of CYP3A4 phosphorylation in its ubiquitination using an in vitro functionally reconstituted system that contained human Ub-activating enzyme E1, human Ub-conjugating E2 enzyme UBC7, and mouse gp78 E3 Ub-ligase in the presence or absence of PKA and PKC as the kinases. Our choice of these particular enzymes was dictated by the following: (i) our findings of PKA and PKC as the major catalysts of CYP3A4 phosphorylation using specific kinase inhibitors as probes (14); (ii) our P450 ERAD studies in S. cerevisiae (4, 5) that identified Ubc7p as the specific E2 involved in CYP3A4 ERAD along with its ER-anchor Cue1p; and (iii) our recent in vitro findings with functionally reconstituted CYP3A4 ubiquitination systems using ERAD-associated mammalian/human E2 Ub-conjugating enzymes and E3 Ub-ligases that not only confirmed UBC7, the human homolog of Ubc7p as the E2, but also identified its cognate integral ER-RING-finger gp78 protein as a dominant CYP3A4 E3 Ub-ligase (12). Our findings described above clearly indicated that CYP3A4 phosphorylation by PKA/PKC indeed enhanced CYP3A4 ubiquitination by the UBC7/gp78 system. By contrast, no comparably enhanced ubiquitination of the CYP3A4 triple mutant was observed in the presence of PKA/PKC, consistent with its intrinsic lack of relevant phosphorylatable residues and consequently its inability to be phosphorylated. The slightly enhanced ubiquitination of this mutant after PKA/PKC phosphorylation over that of its non-phosphorylated species (Fig. 6, B and C) possibly reflects the existence of phosphorylatable CYP3A4 residue(s) other than Thr264, Ser420, and Ser478, which could potentially serve as minor targets thereby accounting for the observed minor extent of CYP3A4 triple (T264A/S420A/S478A) mutant phosphorylation (Fig. 3).
The precise molecular mechanism of how protein phosphorylation enhances CYP3A4 ubiquitination is presently unclear. At pH ≈7.5, the incorporation of a phosphate moiety into a polar Ser, Thr, or Tyr residue imparts two negative charges onto the protein, thereby converting a hydrophobic protein into a polar and extremely hydrophilic one, and thus keeping the protein soluble and less aggregation-prone. Aberrant proteins are believed to be cytotoxic because through misfolding or unfolding they expose hydrophobic domains that can promote oligomerization and/or free cysteines that can cross-link, resulting in rampant protein aggregation (88). CYPs 3A are notoriously aggregation-prone, and this feature is greatly enhanced after structural damage and unraveling of the structural fold of the protein by insults such as CuOOH inactivation, which could expose otherwise concealed hydrophobic domains as well as free sulfhydryl groups. Conceivably, CYP3A phosphorylation could help solubilize these structurally damaged aggregation-prone proteins and thus accelerate their triage from the intracellular environment by targeting them for ubiquitination.
On the other hand, the incorporation of a phosphate into the protein structure may also alter CYP3A4 conformation through exposure of “specific recognition domains.” In this regard, the strength and specificity of interactions with gp78 may in fact be enhanced, if not induced, by the multisite phosphorylation of CYP3A4, which by altering its conformation to a much greater extent than that by a single site phosphorylation could promote its substrate recognition by gp78 E3 Ub-ligase and thus enhance its ubiquitination. Furthermore, in vivo, such multisite phosphorylation-induced conformational alterations could also potentially enhance CYP3A4-protein interactions with other relevant proteins along its degradation trail, such as the Hsp70/90 molecular chaperones for sorting, p97 for ER extraction, and/or delivery to the 26 S proteasome for subsequent destruction, consistent with a classical ERAD process. Unequivocally, such combined solubilizing effects of protein phosphorylation, ubiquitination, and/or molecular chaperone binding would be essential for preventing the aggregation of senescent and/or structurally damaged P450 proteins, thereby averting their cellular toxicity.
In summary, our findings indicate that after structural and functional inactivation, CYP3A4 is mainly phosphorylated on three residues (Thr264, Ser420, and Ser478; see Fig. 7). The latter two are buried deep within the CYP3A4 active site cavity and thus normally inaccessible to cytosolic kinases. Their phosphorylation results in enhanced CYP3A4 ubiquitination by UBC7/gp78 E2-E3 system and is apparently required for their UPD. Mutation of these residues, and particularly of Ser478, significantly abrogates CYP3A4 ubiquitination and proteasomal degradation, thereby implicating a direct causal link between P450 phosphorylation and its proteasomal degradation. The precise molecular mechanism involved in this process remains to be elucidated. However, it is noteworthy that unlike previous studies, wherein protein phosphorylation was presumed to be the initial insult that triggers P450 degradation, our findings suggest that initial structural unraveling of the protein is necessary for targeting Ser478 to phosphorylation. Such structural unraveling can occur physiologically during futile oxidative cycling of CYP3A4, a process that is blocked in the presence of relevant CYP3A4 substrates that consequently “induce” this P450 via protein stabilization.
Supplementary Material
Acknowledgments
We thank Dr. Michael Pabarcus for provision of reagents used in initial ubiquitination studies and Prof. Randy Hampton, University of California, San Diego, for the pep4Δ yeast strain. We are indebted to Dr. A. M. Weissman (NCI, National Institutes of Health, Frederick, MD) for the gift of the GEX plasmids. We also thank Dr. Michelle Trester-Zeidlitz (University of California, San Francisco) for valuable discussions on phosphopeptide enrichment for MS analyses.
This work was supported, in whole or in part, by National Institutes of Health Grants GM44037 and DK26506 (to M. A. C.) and R21 RR023120 (to A. N. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental text.
Footnotes
The abbreviations used are: P450, CYPs, cytochrome P450; CuOOH, cumene hydroperoxide; ER, endoplasmic reticulum; ERAD, ER-associated degradation; FII, fraction II; HMM, high molecular mass; LC-MS/MS, liquid chromatography-tandem mass spectrometry; Lys-C, lysylendopeptidase C; PKA, cAMP-dependent protein kinase A; PKC, protein kinase C; Ub, ubiquitin; UBC, genes for Ub-conjugating enzymes; UPD, Ub-dependent proteasomal degradation; MALDI, matrix-assisted laser desorption ionization; MS, mass spectrometry; E1, ubiquitin-activating enzyme; E2, ubiquitin-conjugating enzyme; E3, ubiquitin-protein isopeptide ligase; DTT, dithiothreitol.
X. Wang, K. F. Medzihradzsky, and M. A. Correia, unpublished observations.
No “native” CYP3A4 proteins were specifically included in the ubiquitination reactions as parallel controls. This is because purified CYPs 3A tend to denature during the prolonged cumulative in vitro incubation periods at ≥30 °C required to control first for CuOOH inactivation and then the phosphorylation/ubiquitination reactions. Failing to retain their native character, the denatured proteins are also phosphorylated (Fig. 3) and/or ubiquitinated in vitro (not shown), albeit more slowly. Thus, in contrast to their in vivo counterparts (2, 3, and 6), they do not qualify as truly native controls in vitro.
References
- 1.Guengerich, F. P. (2005) Cytochrome P450: Structure, Mechanism and Biochemistry, (Ortiz de Montellano, P., ed) pp. 377–530, Kluwer-Academic/Plenum Press, New York
- 2.Correia, M. A., Davoll, S. H., Wrighton, S. A., and Thomas, P. E. (1992) Arch. Biochem. Biophys. 297 228–238 [DOI] [PubMed] [Google Scholar]
- 3.Wang, H. F., Figueiredo Pereira, M. E., and Correia, M. A. (1999) Arch. Biochem. Biophys. 365 45–53 [DOI] [PubMed] [Google Scholar]
- 4.Murray, B. P., and Correia, M. A. (2001) Arch. Biochem. Biophys. 393 106–116 [DOI] [PubMed] [Google Scholar]
- 5.Liao, M., Faouzi, S., Karyakin, A., and Correia, M. A. (2006) Mol. Pharmacol. 69 1897–1904 [DOI] [PubMed] [Google Scholar]
- 6.Faouzi, S., Medzihradszky, K. F., Hefner, C., Maher, J. J., and Correia, M. A. (2007) Biochemistry 46 7793–7803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Correia, M. A., Sadeghi, S., and Mundo-Paredes, E. (2005) Annu. Rev. Pharmacol. Toxicol. 45 439–464 [DOI] [PubMed] [Google Scholar]
- 8.Correia, M. A., and Liao, M. (2007) Expert Opin. Drug Metab. Toxicol. 3 33–49 [DOI] [PubMed] [Google Scholar]
- 9.Vashist, S., and Ng, D. T. (2004) J. Cell Biol. 165 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taxis, C., Hitt, R., Park, S. H., Deak, P. M., Kostova, Z., and Wolf, D. H. (2003) J. Biol. Chem. 278 35903–35913 [DOI] [PubMed] [Google Scholar]
- 11.Ahner, A., and Brodsky, J. L. (2004) Trends Cell Biol. 14 474–478 [DOI] [PubMed] [Google Scholar]
- 12.Pabarcus, M., Hoe, N., Sadeghi, S., Patterson, C., Wiertz, E., and Correia, M. A. (December 10, 2008) Arch. Biochem. Biophys. 10.1016/j.abb. 2008.12.001 [DOI] [PMC free article] [PubMed]
- 13.Korsmeyer, K. K., Davoll, S., Figueiredo-Pereira, M. E., and Correia, M. A. (1999) Arch. Biochem. Biophys. 365 31–44 [DOI] [PubMed] [Google Scholar]
- 14.Wang, X., Medzihradszky, K. F., Maltby, D., and Correia, M. A. (2001) Biochemistry 40 11318–11326 [DOI] [PubMed] [Google Scholar]
- 15.Pyerin, W., Wolf, C. R., Kinzel, V., Kubler, D., and Oesch, F. (1983) Carcinogenesis 4 573–576 [DOI] [PubMed] [Google Scholar]
- 16.Taniguchi, H., Pyerin, W., and Stier, A. (1985) Biochem. Pharmacol. 34 1835–1837 [DOI] [PubMed] [Google Scholar]
- 17.Vilgrain, I., Defaye, G., and Chambaz, E. M. (1984) Biochem. Biophys. Res. Commun. 125 554–561 [DOI] [PubMed] [Google Scholar]
- 18.Muller, R., Schmidt, W. E., and Stier, A. (1985) FEBS Lett. 187 21–24 [DOI] [PubMed] [Google Scholar]
- 19.Pyerin, W., Taniguchi, H., Horn, F., Oesch, F., Amelizad, Z., Friedberg, T., and Wolf, C. R. (1987) Biochem. Biophys. Res. Commun. 142 885–892 [DOI] [PubMed] [Google Scholar]
- 20.Pyerin, W., and Taniguchi, H. (1989) EMBO J. 8 3003–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch, J. A., and Waxman, D. J. (1989) Biochemistry 28 3145–3152 [DOI] [PubMed] [Google Scholar]
- 22.Jansson, I., Curti, M., Epstein, P. M., Peterson, J. A., and Schenkman, J. B. (1990) Arch. Biochem. Biophys. 283 285–292 [DOI] [PubMed] [Google Scholar]
- 23.Eliasson, E., Johansson, I., and Ingelman-Sundberg, M. (1988) Biochem. Biophys. Res. Commun. 150 436–443 [DOI] [PubMed] [Google Scholar]
- 24.Eliasson, E., Johansson, I., and Ingelman-Sundberg, M. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 3225–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eliasson, E., Mkrtchian, S., and Ingelman-Sundberg, M. (1992) J. Biol. Chem. 267 15765–15769 [PubMed] [Google Scholar]
- 26.Eliasson, E., Mkrtchian, S., Halpert, J. R., and Ingelman-Sundberg, M. (1994) J. Biol. Chem. 269 18378–18383 [PubMed] [Google Scholar]
- 27.Zhukov, A., Werlinder, V., and Ingelman-Sundberg, M. (1993) Biochem. Biophys. Res. Commun. 197 221–228 [DOI] [PubMed] [Google Scholar]
- 28.Menez, J. F., Machu, T. K., Song, B. J., Browning, M. D., and Deitrich, R. A. (1993) Alcohol Alcohol. 28 445–451 [PubMed] [Google Scholar]
- 29.Lohr, J. B., and Kuhn-Velten, W. N. (1997) Biochem. Biophys. Res. Commun. 231 403–408 [DOI] [PubMed] [Google Scholar]
- 30.Oesch-Bartlomowicz, B., and Oesch, F. (2003) Arch. Biochem. Biophys. 409 228–234 [DOI] [PubMed] [Google Scholar]
- 31.Oesch-Bartlomowicz, B., and Oesch, F. (2005) Biochem. Biophys. Res. Commun. 338 446–449 [DOI] [PubMed] [Google Scholar]
- 32.Aguiar, M., Masse, R., and Gibbs, B. F. (2005) Drug Metab. Rev. 37 379–404 [DOI] [PubMed] [Google Scholar]
- 33.Sepuri, N. B., Yadav, S., Anandatheerthavarada, H. K., and Avadhani, N. G. (2007) FEBS J. 274 4615–4630 [DOI] [PubMed] [Google Scholar]
- 34.Freeman, J. E., and Wolf, C. R. (1994) Biochemistry 33 13963–13966 [DOI] [PubMed] [Google Scholar]
- 35.Oesch-Bartlomowicz, B., Padma, P. R., Becker, R., Richter, B., Hengstler, J. G., Freeman, J. E., Wolf, C. R., and Oesch, F. (1998) Exp. Cell Res. 242 294–302 [DOI] [PubMed] [Google Scholar]
- 36.Willems, A. R., Lanker, S., Patton, E. E., Craig, K. L., Nason, T. F., Mathias, N., Kobayashi, R., Wittenberg, C., and Tyers, M. (1996) Cell 86 453–463 [DOI] [PubMed] [Google Scholar]
- 37.Kaplan, K. B., Hyman, A. A., and Sorger, P. K. (1997) Cell 91 491–500 [DOI] [PubMed] [Google Scholar]
- 38.Butler, M. P., Hanly, J. A., and Moynagh, P. N. (2007) J. Biol. Chem. 282 29729–29737 [DOI] [PubMed] [Google Scholar]
- 39.Feldman, R. M., Correll, C. C., Kaplan, K. B., and Deshaies, R. J. (1997) Cell 91 221–230 [DOI] [PubMed] [Google Scholar]
- 40.Verma, R., Annan, R. S., Huddleston, M. J., Carr, S. A., Reynard, G., and Deshaies, R. J. (1997) Science 278 455–460 [DOI] [PubMed] [Google Scholar]
- 41.Miyamoto, S., Maki, M., Schmitt, M. J., Hatanaka, M., and Verma, I. M. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 12740–12744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown, K., Gerstberger, S., Carlson, L., Franzoso, G., and Siebenlist, U. (1995) Science 267 1485–1488 [DOI] [PubMed] [Google Scholar]
- 43.Chen, Z., Hagler, J., Palombella, V. J., Melandri, F., Scherer, D., Ballard, D., and Maniatis, T. (1995) Genes Dev. 9 1586–1597 [DOI] [PubMed] [Google Scholar]
- 44.Chen, Z. J., Parent, L., and Maniatis, T. (1996) Cell 84 853–862 [DOI] [PubMed] [Google Scholar]
- 45.Dai, R. M., Chen, E., Longo, D. L., Gorbea, C. M., and Li, C. C. (1998) J. Biol. Chem. 273 3562–3573 [DOI] [PubMed] [Google Scholar]
- 46.Miranda, M., Wu, C. C., Sorkina, T., Korstjens, D. R., and Sorkin, A. (2005) J. Biol. Chem. 280 35617–35624 [DOI] [PubMed] [Google Scholar]
- 47.Shimura, H., Schwartz, D., Gygi, S. P., and Kosik, K. S. (2004) J. Biol. Chem. 279 4869–4876 [DOI] [PubMed] [Google Scholar]
- 48.Yano, J. K., Wester, M. R., Schoch, G. A., Griffin, K. J., Stout, C. D., and Johnson, E. F. (2004) J. Biol. Chem. 279 38091–38094 [DOI] [PubMed] [Google Scholar]
- 49.He, K., Bornheim, L. M., Falick, A. M., Maltby, D., Yin, H., and Correia, M. A. (1998) Biochemistry 37 17448–17457 [DOI] [PubMed] [Google Scholar]
- 50.Fang, S., Ferrone, M., Yang, C., Jensen, J. P., Tiwari, S., and Weissman, A. M. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, H., Dick, R., Yin, H., Licad-Coles, E., Kroetz, D. L., Szklarz, G., Harlow, G., Halpert, J. R., and Correia, M. A. (1998) Biochemistry 37 12536–12545 [DOI] [PubMed] [Google Scholar]
- 52.Blethrow, J. D., Tang, C., Deng, C., and Krutchinsky, A. N. (2007) PLoS ONE 2 e358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi, B. K., Cho, Y. M., Bae, S. H., Zoubaulis, C. C., and Paik, Y. K. (2003) Proteomics 3 1955–1961 [DOI] [PubMed] [Google Scholar]
- 54.Larsen, M. R., Thingholm, T. E., Jensen, O. N., Roepstorff, P., and Jørgensen, T. J. (2005) Mol. Cell. Proteomics 4 873–886 [DOI] [PubMed] [Google Scholar]
- 55.Trester-Zedlitz, M., Burlingame, A., Kobilka, B., and von Zastrow, M. (2005) Biochemistry 44 6133–6143 [DOI] [PubMed] [Google Scholar]
- 56.Hampton, R. Y., Gardner, R. G., and Rine, J. (1996) Mol. Biol. Cell 7 2029–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilhovsky, S., Gardner, R., and Hampton, R. (2000) Mol. Biol. Cell 11 1697–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kornitzer, D. (2002) Methods Enzymol. 351 639–647 [DOI] [PubMed] [Google Scholar]
- 59.Volland, C., Urban-Grimal, D., Geraud, G., and Haguenauer-Tsapis, R. (1994) J. Biol. Chem. 269 9833–9841 [PubMed] [Google Scholar]
- 60.Callen, D. F., and Philpot, R. M. (1977) Mutat. Res. 45 309–324 [DOI] [PubMed] [Google Scholar]
- 61.Doolman, R., Leichner, G. S., Avner, R., and Roitelman, J. (2004) J. Biol. Chem. 279 38184–38193 [DOI] [PubMed] [Google Scholar]
- 62.Ballar, P., Shen, Y., Yang, H., and Fang, S. (2006) J. Biol. Chem. 281 35359–35368 [DOI] [PubMed] [Google Scholar]
- 63.Kikkert, M., Doolman, R., Dai, M., Avner, R., Hassink, G., van Voorden, S., Thanedar, S., Roitelman, J., Chau, V., and Wiertz, E. (2004) J. Biol. Chem. 279 3525–3534 [DOI] [PubMed] [Google Scholar]
- 64.Chen, B., Mariano, J., Tsai, Y. C., Chan, A. H., Cohen, M., and Weissman, A. M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Annan, R. S., and Carr, S. A. (1996) Anal. Chem. 68 3413–3421 [DOI] [PubMed] [Google Scholar]
- 66.Amici, A., Levine, R. L., Tsai, L., and Stadtman, E. R. (1989) J. Biol. Chem. 264 3341–3346 [PubMed] [Google Scholar]
- 67.Requena, J. R., Chao, C. C., Levine, R. L., and Stadtman, E. R. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Correia, M. A., Decker, C., Sugiyama, K., Caldera, P., Bornheim, L., Wrighton, S. A., Rettie, A. E., and Trager, W. F. (1987) Arch. Biochem. Biophys. 258 436–451 [DOI] [PubMed] [Google Scholar]
- 69.Schaefer, W. H., Harris, T. M., and Guengerich, F. P. (1985) Biochemistry 24 3254–3263 [DOI] [PubMed] [Google Scholar]
- 70.Guengerich, F. P. (1986) Biochem. Biophys. Res. Commun. 138 193–198 [DOI] [PubMed] [Google Scholar]
- 71.Correia, M. A., Sugiyama, K., and Yao, K. Q. (1989) Drug Metab. Rev. 20 615–628 [DOI] [PubMed] [Google Scholar]
- 72.Riddick, D. S., and Marks, G. S. (1990) Biochem. Pharmacol. 40 1915–1921 [DOI] [PubMed] [Google Scholar]
- 73.Oprian, D. D., Gorsky, L. D., and Coon, M. J. (1983) J. Biol. Chem. 258 8684–8691 [PubMed] [Google Scholar]
- 74.Guengerich, F. P., and Johnson, W. W. (1997) Biochemistry 36 14741–14750 [DOI] [PubMed] [Google Scholar]
- 75.Zangar, R. C., Davydov, D. R., and Verma, S. (2004) Toxicol. Appl. Pharmacol. 199 316–331 [DOI] [PubMed] [Google Scholar]
- 76.Denisov, I. G., Grinkova, Y. V., McLean, M. A., and Sligar, S. G. (2007) J. Biol. Chem. 282 26865–26873 [DOI] [PubMed] [Google Scholar]
- 77.Gorsky, L. D., Koop, D. R., and Coon, M. J. (1984) J. Biol. Chem. 259 6812–6817 [PubMed] [Google Scholar]
- 78.Ekstrom, G., and Ingelman-Sundberg, M. (1989) Biochem. Pharmacol. 38 1313–1319 [DOI] [PubMed] [Google Scholar]
- 79.Dai, Y., Rashba-Step, J., and Cederbaum, A. I. (1993) Biochemistry 32 6928–6937 [DOI] [PubMed] [Google Scholar]
- 80.Roberts, B. J., Song, B. J., Soh, Y., Park, S. S., and Shoaf, S. E. (1995) J. Biol. Chem. 270 29632–29635 [DOI] [PubMed] [Google Scholar]
- 81.Bardag-Gorce, F., Li, J., French, B. A., and French, S. W. (2002) Free Radic. Biol. Med. 32 17–21 [DOI] [PubMed] [Google Scholar]
- 82.Watkins, P. B., Wrighton, S. A., Schuetz, E. G., Maurel, P., and Guzelian, P. S. (1986) J. Biol. Chem. 261 6264–6271 [PubMed] [Google Scholar]
- 83.Henderson, C. J., Otto, D. M., Carrie, D., Magnuson, M. A., McLaren, A. W., Rosewell, I., and Wolf, C. R. (2003) J. Biol. Chem. 278 13480–13486 [DOI] [PubMed] [Google Scholar]
- 84.Zhukov, A., and Ingelman-Sundberg, M. (1999) Biochem. J. 340 453–458 [PMC free article] [PubMed] [Google Scholar]
- 85.Goasduff, T., and Cederbaum, A. I. (1999) Arch. Biochem. Biophys. 370 258–270 [DOI] [PubMed] [Google Scholar]
- 86.Woods, Y. L., Cohen, P., Becker, W., Jakes, R., Goedert, M., Wang, X., and Proud, C. G. (2001) Biochem. J. 355 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cohen, P. (2000) Trends Biochem. Sci. 25 596–601 [DOI] [PubMed] [Google Scholar]
- 88.Nishikawa, S., Brodsky, J. L., and Nakatsukasa, K. (2005) J. Biochem. (Tokyo) 137 551–555 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.