Abstract
Foxp3 is a 47-kDa transcription factor central to regulatory T cell (Treg) function. The importance of Foxp3+ Tregs in controlling self-reactive T cells and preventing autoimmunity is well established. Our analysis of Foxp3 expression in natural Tregs led to identification of a shorter 41-kDa Foxp3 species in activated Tregs, indicating that Foxp3 may be processed by proteolytic cleavage upon cell activation. Searches of murine and human Foxp3 sequences for potential cleavage sites responsible for the generation of the short Foxp3 species revealed the presence of two RXXR proprotein convertase (PC) motifs, 48RDLR51 and 414RKKR417, located near the N- and C-terminal ends, respectively. We show, using retroviral expression of Foxp3 in CD4+ T cells, that Foxp3 is cleaved at both the N- and C-terminal RXXR sites and that mutagenesis of the RXXR motif prevents cleavage. The cleaved forms of Foxp3 are found in the chromatin fraction but not in nuclear or cytoplasmic extracts. CD4+ T cells expressing Foxp3 species engineered to mimic N-terminally, C-terminally, or N- and C-terminally cleaved Foxp3 forms are functionally distinct, as indicated by differences in expression of key Treg genes, such as interleukin-10 and cytotoxic T-lymphocyte antigen 4 (CTLA-4). In addition, CD4+ cells expressing C-cleaved Foxp3 are superior to those that express WT Foxp3 in preventing experimental colitis. Coexpression of Foxp3 with PC1 or PC7 results in cleavage of the Foxp3 C terminus. The mechanism by which Foxp3 is processed likely extends to other members of the FoxP subfamily, because Foxp1 and Foxp2 also have N-terminal RXXR proteolytic cleavage motifs at similar locations to Foxp3. Our results indicate that the generation of fully functionally competent Tregs is complex and dependent on the generation of multiple forms of Foxp3 that have differing effects on Treg cytokine production and suppressive function.
CD4+CD25+ regulatory T cells (Tregs)2 are central to the maintenance of immune tolerance and prevention of autoimmunity (1). Ectopic expression in CD4+CD25– of Foxp3, a 47-kDa DNA-binding protein, is sufficient to convert them to a Treg phenotype (2, 3). Mutations in the Foxp3 gene cause IPEX (immunodysregulation, polyendocrinopathy, enteropathy, X-linked) syndrome in humans (4–6) and the Scurfy phenotype in mice (2, 7). The frameshift mutation in Scurfy mice results in the loss of the C-terminal DNA-binding forkhead domain and lethal autoimmunity by 2–3 weeks after birth, whereas in humans, multiple mutations can lead to disease (8). Foxp3 has zinc finger and leucine zipper domains in the midportions of the molecule that are also present in other members of the FoxP subfamily.
Foxp3 forms a functional complex of approximately ∼600 kDa with histone acetyl-transferases, histone deacetylases, and AML1/Runx1 (9–11). Foxp3 may physically associate with the transcription factors RORα and RORγt (12, 13). Its interactions with NFAT and NF-κB are also reported (14, 15). Upon T cell receptor stimulation, Foxp3 binds to the promoters of several genes and can function as a transcriptional repressor or as an activator (15, 16). Consistent with a multifunctional role, Foxp3 binding results in the deacetylation of some promoters (IL-2 and interferon γ) and acetylation of others (glucocorticoid-induced tumor necrosis factor receptor, CD25, and CTLA-4) (16). The expression and function of Foxp3 itself is regulated by acetylation, such that use of histone deacetylase inhibitors enhances Treg production and suppressive functions (17). Expression of the N-terminal sequences of Foxp3 are sufficient to maintain Treg unresponsiveness, but the forkhead domain is necessary for their suppressive function (18).
During comparison of Foxp3 expression by resting versus activated mouse natural Tregs, we detected a 41-kDa Foxp3 species in the chromatin fraction of activated Tregs that was not detectable in resting Tregs. Analysis of mouse and human Foxp3 sequences for proteolytic sites that might be responsible for the generation of the 41-kDa species led to identification of two proprotein convertase (PC) cleavage motifs. The identified RXXR motifs are potential recognition/cleavage sites for the enzymes of the PC family. One of these motifs is at the N-terminal side (48RXXR51) of Foxp3, and the other is very close to the C-terminal end (414RXXR417) of the protein. Here we show Foxp3 is cleaved at both sites. CD4+ cells expressing the engineered forms of Foxp3 that mimic the N-cleaved, C-cleaved, or N&C-cleaved proteins are functionally distinct, as is evident by differences in the expression profiles of critical markers, their ability to suppress Teff cell proliferation, and differences in efficacy in an IBD model. Hence, the regulation of Treg activity is complex and includes proteolytic processing of Foxp3.
EXPERIMENTAL PROCEDURES
Mice—Female C57BL/6 mice (Jackson Laboratory) and female Rag1–/– (C57BL/6) mice (Taconic) were used in animal studies approved by the institutional animal care and use committee of the Children's Hospital of Philadelphia.
Antibodies—Anti-mouse Foxp3 mAbs (FJK-16s, NRRF-30; eBioScience), and SP1 and Furin antibodies (catalog numbers sc-59 and sc-20801; Santa Cruz Biotechnology) were used for Western blots. Polyclonal anti-mouse Foxp3 antibody was generated by immunizing rabbits with a synthetic 11-amino acid (aa) peptide, QRPNKCSNPCP, corresponding to 419–429 aa of mFoxp3. Foxp3-specific polyclonal antibody was purified from high titer rabbit antiserum by affinity chromatography (Sigma-Genosys).
cDNA Cloning and Mutagenesis—mFoxp3 cDNA was amplified from thymus with Foxp3-specific forward and reverse primers (Integrated DNA Technologies). The mutations were introduced with a QuikChange site II-directed mutagenesis kit (Stratagene; catalog number 200523), and after sequence confirmation, fragments were recloned to Minr-1 vector for retroviral expression.
Retroviral Expression—Retroviruses were generated by cotransfection of Foxp3 mutants (in Minr-1 vector) with pCLeco (Invitrogen) helper plasmid into the 293T-based Phoenix ecotropic packaging cell line, using Lipofectamine 2000 reagent (Invitrogen; catalog number 52887). Virus containing supernatant was collected and used to infect purified CD4+ or CD4+CD25– T cells. Prior to infection with the retrovirus, CD4+ T cells were isolated by magnetic sorting, activated with PMA (3 ng/ml), ionomycin (1 mm), and IL-2 (5 units/ml) for 24 h, washed, and transduced using the 48- and 72-h viral supernatants obtained from transfected Phoenix cells (16). Transduced cells were expanded for 1–3 days, unless stated, and used in the suppression assays or for subcellular fractionation (16).
Treg Suppression Assays—5 × 104 carboxyfluorescein succinimidyl ester-labeled CD4+CD25– (Teff) cells, isolated using magnetic beads (Miltenyi), were stimulated with CD3 mAb in the presence of irradiated syngeneic T-cell depleted splenocytes and varying ratios of activated CD4+ cells transduced with different Foxp3 constructs (17). Suppression of proliferation was determined from the CFSE profile of dividing Teff cells at 72 h.
Nuclear and Cytoplasmic Extraction—Subcellular fractions were prepared as described (19). For analysis of histones, cellular fractions extracted with 0.2 m H2SO4 were analyzed on acetic-acid urea gels (20).
Western Blots—The proteins were separated by 14 or 20% SDS-PAGE, blotted, and probed with the primary antibody followed by secondary antibody horseradish peroxidase and Luminol reagent (Santa Cruz Biotechnology; catalog number sc-2048).
DNA and Protein Estimation—DNA in the subcellular fractions was estimated using the diphenylamine-based color reaction assay described by Zaccharias Dische in 1930 and subsequently modified by Burton (21). Protein estimation was done using a Bio-Rad DC protein assay kit (catalog number 5000112).
Adoptive Transfer Model of IBD—For the adoptive transfer model of colitis, CD4+CD25– T cells were isolated from spleen and mesenteric lymph nodes were isolated using magnetic beads (Miltenyi Biotec) to >95% purity (by flow cytometry), and 1 × 106 cells were injected intraperitoneal into C57BL/6 background Rag1–/– mice together with Thy1.1+CD4+ cells (5 × 105) transfected with WT or mutated Foxp3 (22). The mice were monitored biweekly for clinical evidence of colitis, including weight loss, fecal blood, and stool consistency.
Quantitative PCR—Total RNA was prepared by lysing 1 × 106 cells with Qiashredder (Qiagen; catalog number 79654) and purification of RNA using RNeasy mini kit (Qiagen; catalog number 74104). cDNA was synthesized with TaqMan reverse transcription reagents (Applied Biosystems), primer/probe sets were obtained from Applied Biosystems, and quantitative PCR was performed using an ABI Prism 7000 Analyzer.
Pathology—Gut samples were paraffin-embedded and stained by hematoxylin and eosin, or snap frozen and stained by immunoperoxidase using anti-Foxp3 or an isotype control mAb (23).
RESULTS
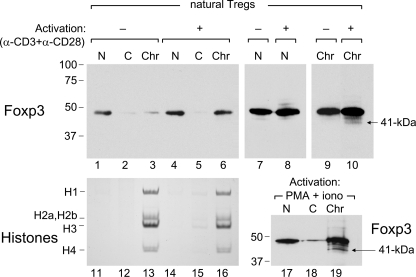
Detection of the 41-kDa Foxp3 Species—Subcellular localization of Foxp3 is regulated by signal transduction (16). To determine the effect of Treg activation on Foxp3 expression and its cellular distribution, naturally occurring thymic-derived CD4+CD25+ Tregs from peripheral lymph nodes and spleen were isolated and activated with CD3 and CD28 mAbs, followed by subcellular fractionation and Western blot analysis. Treg cell activation resulted in increased levels of Foxp3, detected as a single 47-kDa species in nuclear and cytoplasmic extracts and especially in the chromatin fraction (Fig. 1, lane 6). In resting Tregs, the ratio of chromatin-bound Foxp3 to Foxp3 in nuclear extracts was very low, but with TCR stimulation almost equal amounts of Foxp3 were detected in these fractions (Fig. 1, lanes 1, 3, 4, and 6). Increased exposure revealed the presence of additional Foxp3 species, migrating both more quickly and more slowly than the major 47-kDa Foxp3 species (Fig. 1, lanes 7–10). Although most of these additional species were common to resting and activated Tregs, a 41-kDa Foxp3 species was found exclusively in the chromatin fraction of activated Tregs (Fig. 1, lane 10). The chromatin fraction used in this study had the characteristic histone profile of chromatin (24), indicating that subcellular fractionation was efficiently performed (Fig. 1, lanes 11–16). Activation of Tregs by PMA and ionomycin also showed the 41-kDa Foxp3 species to be exclusively in the chromatin fraction (Fig. 1, lane 19).
FIGURE 1.
41-kDa Foxp3 species is detected in the chromatin fraction of activated natural Tregs. Foxp3 expression was analyzed by Western blotting in nuclear (N) and cytoplasmic extracts (C) and chromatin (Chr) fraction using FJK-16s mAb. Lanes 1–3, fractions from resting Tregs; lanes 4–6, fractions from CD3/CD28 mAb-activated Tregs; lanes 7 and 8, nuclear extracts, similar to lanes 1 and 4 but overexposed; lanes 9 and 10, chromatin fractions similar to lanes 3 and 6 but overexposed. These lanes (lanes 9 and 10) were from the same experiment with the same exposure and placed together using Adobe Photoshop without any other manipulation. Lanes 11–16, parallel samples to the ones shown in lanes 1–6 were analyzed for histone content by acetic-acid urea-PAGE. Lanes 17–19, N and C extracts and Chr fraction were prepared from Tregs after activation with PMA/ionomycin. The arrows next to lanes 10 and 19 mark the 41-kDa Foxp3 species.
Only the N-terminal RXXR Motif Is Present in Other FoxP Subfamily Members—The detection of a shorter Foxp3 species only in activated Tregs suggested Foxp3 may be endoproteolytically processed upon activation to yield the 41-kDa species. Our search for proteolytic sites in both mouse and human Foxp3 revealed two RXXR motifs (48RDLR51 and 414RKKR417) that represent potential cleavage sites for PC enzymes (Fig. 2). The presence of a serine residue immediately after the C-terminal RKKR (RKKRS) in mouse and human Foxp3 indicated Foxp3 may be cleaved at this site, because serine is most commonly found next to the RXXR motifs recognized by PCs (25, 26). The N-terminal RDLR is also followed by serine (RDLRS) in mouse and by glycine (RDLRG) in the human, another acceptable amino acid for cleavage by PCs (27). The search for RXXR motifs in other FoxP subfamily members showed that Foxp3 is unique in having the C-terminal RXXR motif (Fig. 2). However, N-terminal RXXR motifs are also found at similar locations in mouse and human Foxp1 and Foxp2. These N-terminal RXXR motifs in Foxp1–3, located within the first 51 aa, are in a region that does not include the zinc finger, leucine zipper, or forkhead domains common to FoxP subfamily members. Within the first 51 aa, Foxp1 and Foxp2 are only 6 and 14% identical to the N-terminal end of Foxp3. The presence of the N-terminal RXXR motifs in Foxp1–3, in a region of limited homology, suggests these members are processed by a common mechanism (Fig. 2).
FIGURE 2.
Schematic diagram showing the RXXR proprotein convertase recognition/cleavage sites in mouse Foxp1, Foxp2, and Foxp3. RXXR motifs are shown in gray boxes, and the numbers above the RXXRs indicate the precise locations of cleavage. Corresponding human residues are shown beneath the mouse residues, as marked. The percentages indicate the identical aa residues for the first 51 aa between Foxp1, Foxp2, and Foxp3 (highlighted). The second Foxp3 diagram beneath the full-length 429-aa Foxp3 diagram shows the sizes of the fragments that would be generated after processing of Foxp3 at the N- and C-terminal RXXR sites. QKRR sequences in Foxp1 and Foxp4 (corresponding to the RKKR sequence of Foxp3) are followed by proline residues (shown in red), an amino acid not favored by most PCs at this position. ZF, zinc finger; LZ, leucine zipper, FHD, forkhead domain.
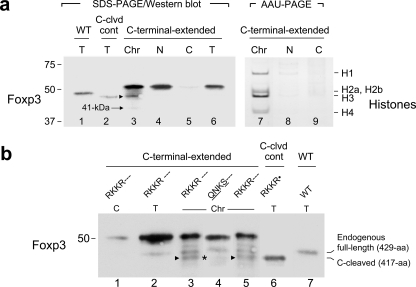
Foxp3 Is Cleaved at the N-terminal RXXR Motif—To determine whether Foxp3 is cleaved at the RXXR sites, we mutated the N-terminal 48RDLR51 sequence to 48HDLH51, effectively abolishing the RXXR motif without altering the positive charge. This mutation was introduced to full-length Foxp3 (429 aa), as well as to a construct that encodes a short 417-aa Foxp3 mimicking Foxp3 cleaved at the C-terminal cleavage site immediately following the RKKR (C-cleaved Foxp3; RKKR followed by a stop codon, missing the last 12 aa). We also engineered a size control Foxp3 that is the same size (366 aa) as a Foxp3 cleaved at both the N and C termini. These constructs were retrovirally expressed in CD4+CD25– T cells, separated into subcellular fractions, and analyzed by Western blotting; a schematic diagram of these constructs is shown in Fig. 3a. Western blot of the chromatin fractions shows the 366-aa size control and the shorter Foxp3 species generated from C-cleaved Foxp3 species comigrate, in agreement with the possibility that the 41-kDa species may be the result of N-terminal cleavage of Foxp3. Moreover, the expression of the Foxp3 mutants that have lost their N-terminal RXXR motif did not yield any detectable 41-kDa species (Fig. 3b, lanes 4 and 5), indicating the N-terminal cleavage event in Foxp3 is dependent on the presence of an intact RXXR motif. We then analyzed the nuclear and cytoplasmic extracts, and similar to natural Tregs, the 41-kDa species was detected in the chromatin fraction but not in the nuclear or cytoplasmic extracts (Fig. 3c), suggesting a distinct function for this Foxp3 species.
FIGURE 3.
N-terminal Foxp3 cleavage depends on an intact RXXR motif. a, schematic of the constructs used in retroviral expression. The vertical arrows indicate the expected cleavage sites. Mutant residues are underlined, and • indicates engineered ends). Row 1, empty vector (control); row 2, size control for N&C-cleaved Foxp3 (366 aa), addition of the translational initiator methionine was compensated by removal of R417; row 3, C-cleaved Foxp3; row 4, C-cleaved Foxp3 with RDLR mutated to HDLH; row 5, full-length (429 aa) Foxp3 with RDLR mutated to HDLH; row 6, full-length WT Foxp3, a control. b, demonstration of N-terminal cleavage of Foxp3 and its dependence on an intact RXXR motif. N-terminal cleavage and its absence in the RXXR mutants was determined by analysis of chromatin-bound Foxp3 obtained from CD4+ cells expressing the constructs 1–6 shown in a. The 41-kDa Foxp3 species is marked with a short arrow. c, Western blot analysis of Foxp3 expression in N, C, and Chr fractions and localization of the 41-kDa Foxp3 species exclusively to the chromatin fraction. The numbers on top indicate the construct numbers described in a. Arrows mark the shorter Foxp3 species, and the asterisk indicates its absence in construct 4. Following treatment with the Foxp3 mAb FJK-16s, the membrane was treated with SP1 antibody to determine the efficiency of subcellular fractionation. N, nuclear extract; C, cytoplasmic extract; Chr, chromatin fraction. d, differential recognition of Foxp3 species by the Foxp3 mAbs NRRF-30 and FJK-16s. An arrow marks the N-terminal 41-kDa species not recognized by the N-terminal specific NRRF-30.
An N-terminal Specific Foxp3 mAb Does Not Recognize the 41-kDa Foxp3—Analysis using mAbs also showed that the 41-kDa species is generated by N-terminal cleavage of Foxp3 because NRRF-30 mAb, specific to the Foxp3 N-terminal end, did not recognize the 41-kDa species, in contrast to the binding of another mAb, FJK-16s, specific to a more central Foxp3 epitope (Fig. 3d). Thus, a subpopulation of chromatin-bound Foxp3 in activated Tregs is N-cleaved. N-terminal cleavage of Foxp3 may explain the discrepancy in results from different groups and help resolve the controversy over the specificity of N-terminal specific human PCH101 mAb versus other Foxp3-specific mAbs (28).
Foxp3 Is Cleaved at the C-terminal RXXR Motif—To determine whether C-terminal cleavage takes place, we made additional constructs to resolve the uncleaved and C-cleaved (missing the last 12 aa) Foxp3s, because the size difference between the two forms was too small to resolve satisfactorily by SDS-PAGE. Two new constructs were made that encode C-terminally extended Foxp3, so that the C-cleaved and uncleaved forms could be resolved. These constructs were retrovirally expressed in CD4+ T cells, and one of them, with an additional 19 aa at its C-terminal end (sequences past RKKR extended from 12 to 31 aa) was found to be suppressive in the in vitro Treg assay (data not shown) and could be C-cleaved (Fig. 4). Thus, Foxp3 gets cleaved very close to its C-terminal end, in addition to being cleaved at the N-terminal 48RDLR↓S52 (Fig. 4a, arrowhead marks the C-cleaved Foxp3 and arrow marks the 41-kDa species resulting from N-terminal cleavage). In control experiments, the samples were analyzed for their DNA and protein content, and the protein/DNA ratio of the chromatin fraction was determined to be 2.8 ± 0.2, in agreement with the protein/DNA ratios reported for chromatin (29, 30) (Table 1). In addition, we confirmed that the histone profile of the chromatin fraction matches the histone profile of chromatin (24) (Fig. 4a, lane 7). To find out whether the C-terminal proteolytic cleavage is dependent on an intact RXXR motif, the RKKR in the C-terminally extended Foxp3 was mutated to QNKS, effectively abolishing the motif. The constructs were expressed in CD4+ T cells, and chromatin proteins were separated on large gels and analyzed by Western blotting. Analysis of C-terminally extended Foxp3 (RKKR---) expression showed the presence of uncleaved C-terminally extended Foxp3, endogenous Foxp3 of the CD4+ cells, and C-cleaved Foxp3 (comigrating with the 417-aa size control protein) (Fig. 4b, C-cleaved Foxp3 marked with arrowheads, lanes 3 and 5; and size control, lane 6). In contrast, the corresponding sample from the QNKS-- mutant showed the presence of uncleaved C-terminally extended mutant Foxp3, the endogenous Foxp3 of the CD4+ cells, but lacked any detectable C-cleaved (417 aa) species, indicating that the QNKS-- mutant does not get cleaved at the C terminus in the absence of an intact RXXR motif (empty area marked with an asterisk, Fig. 4b, lane 4). Thus, Foxp3 can be proteolytically cleaved at both the N-terminal and C-terminal sites (48RDLR↓S52 and 414RKKR↓S418), and both of these cleavages require intact RXXR motifs.
FIGURE 4.
C-terminal Foxp3 cleavage depends on an intact RXXR motif. a, demonstration of C-terminal cleavage of Foxp3. Lane 1, size control for endogenous full-length Foxp3 (429 aa); lane 2, size control for C-cleaved Foxp3, expressed from a construct mimicking C-cleaved Foxp3 (417 aa); lanes 3–6, Foxp3 species in subcellular fractions (Chr, N, and C) and total extracts (T) from CD4+ cells expressing C-terminally extended Foxp3; the arrowhead marks the C-cleaved Foxp3, and the arrow marks the 41-kDa species; lanes 7–9, histone profile of samples shown in lanes 3–5, determined by AAU-PAGE. AAU-PAGE, acetic acid-urea-PAGE; C-clvd cont, C-cleaved control. The other abbreviations as described in Fig. 3c. b, dependence of C-terminal cleavage of Foxp3 on an intact RXXR motif. Lanes 1–3, C, T, and Chr fractions from cells expressing C-terminally extended Foxp3 (RKKR---); lane 4, Chr fraction from cells expressing the mutant C-terminally extended Foxp3 (RKKR replaced with QNKS; designated as QNKS---); lane 5, same as lane 3; lane 6, size control for C-cleaved Foxp3 (417 aa) (RKKR·); lane 7, WT Foxp3, control for endogenous full-length Foxp3 (429 aa). The arrowheads mark the C-cleaved 417-aa Foxp3, and the asterisk marks its absence in the QNKS-- mutant. The three hyphens in RKKR-- and QNKS-- indicate the additional C-terminal 19 aa in these C-terminally extended Foxp3s.
TABLE 1.
DNA and protein distribution in the subcellular fractions of CD4+ T cells retrovirally expressing C-terminal extended Foxp3
DNA and protein measurements are described under “Experimental Procedures.” N, nuclear extract; C, cytoplasmic extract; Chr, chromatin fraction; T, total extract.
| DNA | Protein | Protein/DNA | |
|---|---|---|---|
| % | % | mg/ml | |
| Chr | 73 ± 2 | 36 ± 3 | 2.8 ± 0.2 |
| N | 1 ± 1 | 12 ± 1 | >200 |
| C | 26 ± 1 | 52 ± 4 | 11.5 ± 1 |
| T | 100 | 100 | 5.1 ± 0.1 |
Detection of the Cleaved 12-aa Foxp3 Peptide—To determine the presence of the 12-aa peptide cleaved from the C-terminal end of Foxp3, an anti-peptide polyclonal antibody was generated in rabbits directed to the last 11 aa of the mouse Foxp3 sequence (Fig. 5, a and b). This polyclonal antibody was then used in the Western blot analysis of spleen and heart extracts and recognized a ∼1.3-kDa peptide in the spleen extracts (theoretical molecular mass of cleaved C-terminal peptide, 1.34 kDa). Nonlymphoid tissues, such as heart, did not show any trace of this peptide (Fig. 5c).
FIGURE 5.
Detection of the cleaved 12-aa C-terminal Foxp3 peptide by Western blotting. a, schematic diagram showing the locations of the N- and C-terminal RXXR motifs in Foxp3. The vertical arrows indicate the cleavage sites (48RDLR↓S52 and 414RKKR↓S418), and the diagonal arrow marks the C-terminal 12-aa tail released from Foxp3 following cleavage. b, the C-terminal sequences of mouse and human Foxp3s. The last 35 aa of the forkhead domain is shaded in gray, and the RKKR is sequence marked with a bold line. The cleavage site is indicated with arrowheads. A peptide having the sequence of the last 11 aa of Foxp3 was synthesized (boxed) and was then used to generate a polyclonal anti-peptide antibody in rabbits to detect the cleaved 12-aa C-terminal Foxp3 peptide. c, Western blot detection of the 12-aa C-cleaved peptide. Spleen and heart extracts were analyzed on large gels by 20% SDS-PAGE followed by Western blotting. Detection was with the anti-peptide antibody, and the 11-mer synthetic peptide (molecular mass, 1.24 kDa) was included in the Western blot as size control, in addition to other commercial size markers.
Differential Effects of Foxp3 Processing on CTLA-4 and IL-10 mRNA Levels—We found that the shorter 41-kDa Foxp3 species is generated only upon the activation of Tregs, indicating that different Foxp3 forms may have distinct functions. To study this further, we used quantitative PCR to determine mRNA levels of the CTLA-4 inhibitory receptor and the anti-inflammatory and immunosuppressive cytokine, IL-10, in CD4+ cells retrovirally transduced to express the engineered Foxp3 forms (mimicking either N-cleaved, C-cleaved, or N&C-cleaved (double-cleaved) Foxp3. We found that resting cells expressing WT Foxp3 had the highest level of CTLA-4 mRNA. In contrast, up to 8-fold more IL-10 mRNA was detected in activated cells expressing C-cleaved or N&C-cleaved Foxp3, compared with those expressing WT Foxp3 (Fig. 6). Interferon γ mRNA expression was found to be somewhat lower in cells expressing WT Foxp3 (data not shown). Hence, alternative processing of Foxp3 has distinct effects on the expression of several Treg-associated genes.
FIGURE 6.
CTLA-4 and IL-10 mRNA levels in resting and activated CD4+ T cells expressing the different Foxp3 forms. a, Western blot showing the migration properties and equal expression of WT, N-cleaved, C-cleaved, and N&C-cleaved Foxp3 by transduced bulk CD4+ cells. Total cell extracts were used in the Western blot analysis. Equal expression of different Foxp3 forms was also confirmed by flow cytometry (data not shown). b, determination of CTLA-4 mRNA expression in samples shown in a. Statistical analysis (analysis of variance) showed p < 0.05 for WT versus N, C, N&C, and vector groups using resting cells. c, determination of IL-10 mRNA in the same samples. Statistical analysis (analysis of variance) showed p < 0.001 for WT versus C and NC groups using activated cells. The results are representative of three independent experiments. The bars show the means ± S.D. of triplicate samples. RNA was extracted from resting cells (Rest.) or from cells activated (Activ.) for 4 h with plate-bound CD3/CD28 mAbs (coated with 1 μg of CD3 and 0.5 μgof CD28), followed by cDNA preparation and quantitative PCR analysis. WT, wild type Foxp3; N, N-cleaved Foxp3; C, C-cleaved Foxp3; NC, N- and C-cleaved Foxp3; V, empty Minr-1 vector (control).
CD4 Cells Expressing C-cleaved and N&C-cleaved Foxp3 Strongly Suppress T Cell Proliferation—To test how proteolytic processing affects the suppression capability of CD4+ T cells that express different forms of Foxp3, transduced cells were tested in vitro using standard Treg suppression assays. We found that cells expressing N-cleaved Foxp3 suppressed effector T cell proliferation comparably with WT Foxp3. However, expression of double-cleaved Foxp3 resulted in suppression of effector T cell proliferation about 2.5 times more strongly than WT Foxp3 at the ratio of one Treg to two T effector cells (Fig. 7). It should be noted that WT Foxp3 expressed in the suppression assays is not present in the cells as a homogenous population, but rather as a mixture of uncleaved and cleaved (N-, C-, and double-cleaved) forms and therefore does not represent a pure population. We have precluded the use of C-terminal cleavage-resistant mutants in our experiments because the C-terminal RKKR residues are also involved in DNA binding. Thus, the use of such uncleavable Foxp3 mutants would make functional interpretation of cleavage resistance difficult. In agreement with this reasoning, we found an almost total loss of suppressive activity using a Foxp3 mutant in which the RKKR is replaced with PNNW· (PNNW, followed by a stop codon; mutant residues underlined), highlighting the importance of the basic four-amino acid sequence RKKR in the function of Foxp3 (Fig. 7).
FIGURE 7.
Suppression of Teff proliferation by CD4+ cells expressing different Foxp3 forms. The different Foxp3 forms expressed by the CD4+ cells used in the study are schematically shown in the inset. Teff proliferation in samples corresponding to untransduced CD4+ T cells (or transduced with empty vector) is identical to the PNNW· mutant, therefore, are not shown. The variation in retroviral infections between samples was minimal (<10%), and similar suppression profiles were obtained with sorted and unsorted cells. The data shown is representative of three experiments, with the indicated samples.
CD4 Cells Expressing C-cleaved Foxp3 Prevent IBD—Our hypothesis that one form of Foxp3 may be more efficacious in a particular in vivo model than other forms was tested in vivo using an adoptive transfer model of colitis (31). Rag1–/– (C57BL/6) mice were injected with a limiting number of 5 × 105 transduced Thy1.1+ CD4+ cells expressing either WT Foxp3 or C-cleaved Foxp3 (RKKR·), or empty vector (control), plus 1 × 106 CD4+CD25– Teff cells. Additional mice received CD4+CD25– Teff cells alone. 5 × 105 transduced CD4+ cells were transferred to mice based on prior experiments demonstrating that this number of cells resulted in only partial response. The weights of the animals in each group were monitored weekly for 45 days. Mice that received cells expressing empty vector or no Tregs progressively lost weight and succumbed to disease, whereas mice receiving cells expressing WT Foxp3 showed minor weight loss but survived (p < 0.01 versus Minr-1 or no Treg groups) (Fig. 8a). In contrast to the use of WT Foxp3, mice receiving C-cleaved Foxp3 (RKKR·) continued to gain weight (p < 0.05 versus WT Foxp3) (Fig. 8a). Histologic analysis showed mild mononuclear cell infiltration and edema in gut sections from mice receiving WT Foxp3, whereas mice receiving C-cleaved Foxp3 had essentially normal histology (Fig. 8b). Both groups showed considerable numbers of Foxp3+ mononuclear cells by immunoperoxidase staining, suggesting differences in weight loss were not caused by differences in cell recruitment to inflamed gut tissues (Fig. 8b). The study was terminated at day 45, and splenocytes from each group were analyzed for Thy1.1 and Foxp3 expression. Flow cytometry showed most splenic Thy1.1+ cells expressed Foxp3, consistent with the survival of transferred Foxp3+ cells throughout the study (data not shown).
FIGURE 8.
Percent weight loss after adoptive transfer of colitis in mice and treatment of disease by CD4+ cells expressing different Foxp3 forms. a, serial analysis of weight loss showing the benefit of therapy with C-cleaved Foxp3. b, comparison of duodenal samples collected at day 45. Injection of cells expressing WT Foxp3 was associated with mild mononuclear cell recruitment and villous edema, whereas animals receiving cells expressing C-cleaved Foxp3 had essentially normal histology (hematoxylin and eosin-stained paraffin sections; original magnifications, ×200). Immunoperoxidase staining showed infiltration by Foxp3+ mononuclear cells in both cases (hematoxylin-counterstained cryostat sections; original magnifications, ×400). The inset shows a lack of staining using isotype-matched control mAb.
PC1 and PC7 Process the C-terminal End of Foxp3—As a first step toward identifying which PCs may be involved in processing Foxp3, the mRNA levels of the seven PCs that recognize RXXR motifs were assessed in resting versus activated CD4+CD25– (non-Treg) and CD4+CD25+ (Treg) cells. The mRNA levels of four of the PCs were too low to be detected, but Furin, PC1, and PC7 mRNAs were detectable and upon activation of cells increased in Tregs and decreased in non-Tregs (Fig. 9a). To determine whether these three PCs are able to cleave Foxp3, their cDNAs were cloned into the retroviral Minr-1 vector and coexpressed with C-terminally extended Foxp3 in CD4+CD25– T cells. C-terminally extended Foxp3 was used to enable detection of C-terminal cleavage, and CD4+CD25– cells were used to minimize background from endogenous PC activity, because endogenous PC levels were found to drop in these cells upon activation (Fig. 9a). Analysis of subcellular fractions obtained from transduced cells showed that none of the three PCs cleaved the N-terminal end of Foxp3, whereas the C-terminal end was cleaved by both PC1 and PC7, and the cleaved form was detectable only in the chromatin fraction (Fig. 9b).
FIGURE 9.
Determination of Furin, PC1, PC7, and Foxp3 mRNA expression by quantitative PCR in resting versus activated murine CD4+CD25– and CD4+CD25+ cells and demonstration of C-terminal cleavage of Foxp3 by PC1 and PC7. a, cells harvested from spleen and lymph nodes were purified using magnetic beads, cultured with or without CD3 mAb for 3 days prior to RNA isolation; the values are relative to 18 S ribosomal RNA, and p < 0.01 for resting versus activated levels of Furin, PC1, and PC7. R, resting; A, activated. The bars show the means ± S.D. of triplicate samples. b, C-terminally extended Foxp3 was coexpressed in retrovirally transduced CD4+CD25– cells with empty vector (lane 1), Furin (lane 2), PC1 (lane 3), and PC7 (lane 4). Lane 5 is a control (empty vector). Exp. 1 and Exp. 2 indicate results from two different experiments. The subcellular fractions from the transduced T cells were analyzed on Western blots using Foxp3 mAb and Furin polyclonal Abs.
DISCUSSION
In this study we demonstrate that activation of Tregs leads to proteolytic cleavage of Foxp3 and that the RXXR motifs located close to the N- and C-terminal ends, 48RDLR51 and 414RKKR417, are sites for proteolytic cleavage. We show Foxp3 can be cleaved at either the N- or C-terminal site, or at both sites, and the processed protein is exclusively found in the chromatin fraction. We find CD4+ cells expressing the different engineered forms of Foxp3 show differences in their expression of the CTLA-4 receptor and IL-10. CTLA-4 expression is highest in resting cells that express WT Foxp3. In contrast, cells expressing the C-cleaved or the double-cleaved forms of Foxp3 have a high level of IL-10 expression. Samples corresponding to WT Foxp3 were found to have very low levels of IL-10, similar to samples corresponding to empty vector. The induction of IL-10 expression in cells expressing C-cleaved Foxp3 is particularly meaningful because IL-10 is reported to be important in preventing IBD in the adoptive transfer model of colitis (32), and mice with CD4+ cells deficient in IL-10 expression develop spontaneous disease (33). Tregs that express IL-10 are found to be localized in large and small intestines (34). Exogenous addition of IL-10 to CD4+ cell cultures during their activation differentiates them into a phenotype that produces high levels of this cytokine, and these IL-10-producing cells are found to be capable of preventing colitis in SCID mice when coadministered with pathogenic splenic T cells (32). It is likely that expression of C-cleaved Foxp3 in transduced CD4+ T cells converts them into a Tr1-like (contact-independent type 1-like) regulatory T cell phenotype (35). Tr1 cells are a subset of CD4+ T cells that have only been partially characterized and are defined by their expression of high levels of IL-10. Tr-1-like subsets of Tregs have also recently been reported (36).
The finding that Foxp3, a nuclear, DNA-binding protein, is processed by enzymes of the PC family is surprising because PCs are known to process secreted proteins and peptide hormones (25, 37, 38). Our data indicate that both PC1 and PC7 are capable of processing Foxp3 at its C-terminal end when coexpressed. PC7 mRNA levels in active Tregs are ∼15-fold more than PC1, and in agreement with our observations, high level PC7 expression is reported in lymphoid organs (39, 40). Hence, PC7 may be the principal enzyme to process the C-terminal end of Foxp3 in natural Tregs, but additional work is needed to prove this point. The N-terminal end of Foxp3 is not processed by Furin, PC1, or PC7, indicating involvement of another PC in N-terminal processing. Substrate specificities of PCs are affected by amino acid residues at positions P2–P4 (R4X3X2R1↓X1′X2′). In addition, the surrounding residues at both sides of the RXXR motif (extending to P8 and P4′) contribute to recognition specificity (26, 41, 42). Cleavage preference of individual PCs is rather complex, and the lack of information makes the design of specific inhibitors very difficult (42). Foxp3 C-terminal RXXR motif is totally comprised of basic amino acids. In contrast, the N-terminal RDLR has an aspartic acid at P3 and leucine at P2, an indication that N-terminal processing of Foxp3 likely requires a PC with a different substrate specificity.
The fact that only a minor portion of Foxp3 is found processed may reflect the level of cellular demand for the processed form. The rate of Foxp3 processing may also depend on the availability of active PCs (members of the PC family of enzymes themselves require processing to become active) or depend on additional factors for proper presentation of Foxp3 to the PC. More 41-kDa Foxp3 species is generated in natural Tregs following activation with PMA plus ionomycin, compared with activation with CD3/CD28 stimulation, indicating that the type and duration of activation contribute to how much 41-kDa species is detected. PMA plus ionomycin treatment also results in the appearance of several additional Foxp3 species ranging between 41 and 47 kDa, and these species remain to be identified.
Furin processes many substrates, including pro-transforming growth factor-β, to their active form (43). Mice with furin-deficient Tregs have recently been shown to be unable to keep T cell expansion and activation under control, and these mice develop IBD at approximately 6 months, highlighting the importance of PCs in the maintenance of peripheral tolerance (44). Our studies here suggest that PCs may control Treg function via additional mechanisms, namely by direct cleavage and regulation of Foxp3 activity.
The identification of the molecules that interact with the N- and C-terminal ends of Foxp3 will likely help explain how Foxp3 achieves diverse effects in altering the expression of different genes. It is possible that N- or C-terminal cleavages of Foxp3 lead to major topological changes and alter its DNA binding properties. In support of such conformational changes, we consistently find that the N-terminal cleavage in an C-cleaved form of Foxp3 takes place more efficiently compared with WT Foxp3, indicating the release of the C-terminal tail results in a structural change, making the N-terminal site more accessible for processing by the PC(s) (Fig. 3b, lane 3 versus lane 6). Proteolytic cleavages at the N- and C-terminal ends may also enable the shorter forms of Foxp3 to associate with different factors to achieve diverse roles. Devising methods that alter the ability of the PCs in processing Foxp3 should make possible generation of Tregs with functions tailored for specific therapeutic purposes.
Acknowledgments
We thank Hong Sai and Rongxiang Han for assistance with in vivo studies and immunohistology.
This work was supported, in whole or in part, by National Institutes of Health Grant AI54720 (to W. W. H.). This work was also supported by a Foerderer-Murray Award (to E. Ö.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Treg, regulatory T cell; PC, proprotein convertase; IL, interleukin; N-cleaved, N-terminally cleaved; C-cleaved, C-terminally cleaved; N&C-cleaved, N- and C-terminally cleaved; mAb, monoclonal antibody; WT, wild type; aa, amino acid(s); PMA, phorbol 12-myristate 13-acetate; IBD, inflammatory bowel disease; CTLA-4, cytotoxic T-lymphocyte antigen 4.
References
- 1.Sakaguchi, S., Sakaguchi, N., Asano, M., Itoh, M., and Toda, M. (1995) J. Immunol. 155 1151–1164 [PubMed] [Google Scholar]
- 2.Fontenot, J. D., Gavin, M. A., and Rudensky, A. Y. (2003) Nat. Immunol. 4 330–336 [DOI] [PubMed] [Google Scholar]
- 3.Allan, S. E., Alstad, A. N., Merindol, N., Crellin, N. K., Amendola, M., Bacchetta, R., Naldini, L., Roncarolo, M. G., Soudeyns, H., and Levings, M. K. (2008) Mol. Ther. 16 194–202 [DOI] [PubMed] [Google Scholar]
- 4.Bennett, C. L., Christie, J., Ramsdell, F., Brunkow, M. E., Ferguson, P. J., Whitesell, L., Kelly, T. E., Saulsbury, F. T., Chance, P. F., and Ochs, H. D. (2001) Nat. Genet. 27 20–21 [DOI] [PubMed] [Google Scholar]
- 5.Wildin, R. S., Smyk-Pearson, S., and Filipovich, A. H. (2002) J. Med. Genet. 39 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gambineri, E., Torgerson, T. R., and Ochs, H. D. (2003) Curr. Opin. Rheumatol. 15 430–435 [DOI] [PubMed] [Google Scholar]
- 7.Brunkow, M. E., Jeffery, E. W., Hjerrild, K. A., Paeper, B., Clark, L. B., Yasayko, S. A., Wilkinson, J. E., Galas, D., Ziegler, S. F., and Ramsdell, F. (2001) Nat. Genet. 27 68–73 [DOI] [PubMed] [Google Scholar]
- 8.Wildin, R. S., Ramsdell, F., Peake, J., Faravelli, F., Casanova, J. L., Buist, N., Levy-Lahad, E., Mazzella, M., Goulet, O., Perroni, L., Bricarelli, F. D., Byrne, G., McEuen, M., Proll, S., Appleby, M., and Brunkow, M. E. (2001) Nat. Genet. 27 18–20 [DOI] [PubMed] [Google Scholar]
- 9.Li, B., Samanta, A., Song, X., Iacono, K. T., Brennan, P., Chatila, T. A., Roncador, G., Banham, A. H., Riley, J. L., Wang, Q., Shen, Y., Saouaf, S. J., and Greene, M. I. (2007) Int. Immunol. 19 825–835 [DOI] [PubMed] [Google Scholar]
- 10.Li, B., Samanta, A., Song, X., Iacono, K. T., Bembas, K., Tao, R., Basu, S., Riley, J. L., Hancock, W. W., Shen, Y., Saouaf, S. J., and Greene, M. I. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 4571–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ono, M., Yaguchi, H., Ohkura, N., Kitabayashi, I., Nagamura, Y., Nomura, T., Miyachi, Y., Tsukada, T., and Sakaguchi, S. (2007) Nature 446 685–689 [DOI] [PubMed] [Google Scholar]
- 12.Du, J., Huang, C., Zhou, B., and Ziegler, S. F. (2008) J. Immunol. 180 4785–4792 [DOI] [PubMed] [Google Scholar]
- 13.Zhou, L., Lopes, J. E., Chong, M. M., Ivanov, II, Min, R., Victora, G. D., Shen, Y., Du, J., Rubtsov, Y. P., Rudensky, A. Y., Ziegler, S. F., and Littman, D. R. (2008) Nature 453 236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bettelli, E., Dastrange, M., and Oukka, M. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 5138–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu, Y., Borde, M., Heissmeyer, V., Feuerer, M., Lapan, A. D., Stroud, J. C., Bates, D. L., Guo, L., Han, A., Ziegler, S. F., Mathis, D., Benoist, C., Chen, L., and Rao, A. (2006) Cell 126 375–387 [DOI] [PubMed] [Google Scholar]
- 16.Chen, C., Rowell, E. A., Thomas, R. M., Hancock, W. W., and Wells, A. D. (2006) J. Biol. Chem. 281 36828–36834 [DOI] [PubMed] [Google Scholar]
- 17.Tao, R., de Zoeten, E. F., Özkaynak, E., Chen, C., Wang, L., Porrett, P. M., Li, B., Turka, L. A., Olson, E. N., Greene, M. I., Wells, A. D., and Hancock, W. W. (2007) Nat. Med. 13 1299–1307 [DOI] [PubMed] [Google Scholar]
- 18.Lee, S. M., Gao, B., and Fang, D. (2008) Blood 111 3599–3606 [DOI] [PubMed] [Google Scholar]
- 19.Schreiber, E., Matthias, P., Muller, M. M., and Schaffner, W. (1989) Nucleic Acids Res. 17 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panyim, S., and Chalkley, R. (1969) Arch. Biochem. Biophys. 130 337–346 [DOI] [PubMed] [Google Scholar]
- 21.Burton, K. (1956) Biochem. J. 62 315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ostanin, D. V., Pavlick, K. P., Bharwani, S., D'Souza, D., Furr, K. L., Brown, C. M., and Grisham, M. B. (2006) Am. J. Physiol. 290 G109–G119 [DOI] [PubMed] [Google Scholar]
- 23.Lee, I., Wang, L., Wells, A. D., Dorf, M. E., Özkaynak, E., and Hancock, W. W. (2005) J. Exp. Med. 201 1037–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garrard, W. T., Pearson, W. R., Wake, S. K., and Bonner, J. (1974) Biochem. Biophys. Res. Commun. 58 50–57 [DOI] [PubMed] [Google Scholar]
- 25.Özkaynak, E., Schnegelsberg, P. N., Jin, D. F., Clifford, G. M., Warren, F. D., Drier, E. A., and Oppermann, H. (1992) J. Biol. Chem. 267 25220–25227 [PubMed] [Google Scholar]
- 26.Duckert, P., Brunak, S., and Blom, N. (2004) Protein Eng. Des. Sel. 17 107–112 [DOI] [PubMed] [Google Scholar]
- 27.Constam, D. B., and Robertson, E. J. (1999) J. Cell Biol. 144 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran, D. Q., Ramsey, H., and Shevach, E. M. (2007) Blood 110 2983–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russev, G., Venkov, C., and Tsanev, R. (1974) Eur. J. Biochem. 43 253–256 [DOI] [PubMed] [Google Scholar]
- 30.von der Decken, A., and Andersson, G. M. (1977) J. Nutr. 107 949–958 [DOI] [PubMed] [Google Scholar]
- 31.Mottet, C., Uhlig, H. H., and Powrie, F. (2003) J. Immunol. 170 3939–3943 [DOI] [PubMed] [Google Scholar]
- 32.Groux, H., O'Garra, A., Bigler, M., Rouleau, M., Antonenko, S., de Vries, J. E., and Roncarolo, M. G. (1997) Nature 389 737–742 [DOI] [PubMed] [Google Scholar]
- 33.Roers, A., Siewe, L., Strittmatter, E., Deckert, M., Schluter, D., Stenzel, W., Gruber, A. D., Krieg, T., Rajewsky, K., and Muller, W. (2004) J. Exp. Med. 200 1289–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maynard, C. L., Harrington, L. E., Janowski, K. M., Oliver, J. R., Zindl, C. L., Rudensky, A. Y., and Weaver, C. T. (2007) Nat. Immunol. 8 931–941 [DOI] [PubMed] [Google Scholar]
- 35.Dieckmann, D., Bruett, C. H., Ploettner, H., Lutz, M. B., and Schuler, G. (2002) J. Exp. Med. 196 247–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stassen, M., Fondel, S., Bopp, T., Richter, C., Muller, C., Kubach, J., Becker, C., Knop, J., Enk, A. H., Schmitt, S., Schmitt, E., and Jonuleit, H. (2004) Eur. J. Immunol. 34 1303–1311 [DOI] [PubMed] [Google Scholar]
- 37.Zhou, A., Webb, G., Zhu, X., and Steiner, D. F. (1999) J. Biol. Chem. 274 20745–20748 [DOI] [PubMed] [Google Scholar]
- 38.Scamuffa, N., Calvo, F., Chretien, M., Seidah, N. G., and Khatib, A. M. (2006) FASEB J. 20 1954–1963 [DOI] [PubMed] [Google Scholar]
- 39.Munzer, J. S., Basak, A., Zhong, M., Mamarbachi, A., Hamelin, J., Savaria, D., Lazure, C., Hendy, G. N., Benjannet, S., Chretien, M., and Seidah, N. G. (1997) J. Biol. Chem. 272 19672–19681 [DOI] [PubMed] [Google Scholar]
- 40.Seidah, N. G., Hamelin, J., Mamarbachi, M., Dong, W., Tardos, H., Mbikay, M., Chretien, M., and Day, R. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 3388–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou, Y., and Lindberg, I. (1993) J. Biol. Chem. 268 5615–5623 [PubMed] [Google Scholar]
- 42.Remacle, A. G., Shiryaev, S. A., Oh, E. S., Cieplak, P., Srinivasan, A., Wei, G., Liddington, R. C., Ratnikov, B. I., Parent, A., Desjardins, R., Day, R., Smith, J. W., Lebl, M., and Strongin, A. Y. (2008) J. Biol. Chem. 283 20897–20906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas, G. (2002) Nat. Rev. Mol. Cell Biol. 3 753–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pesu, M., Watford, W. T., Wei, L., Xu, L., Fuss, I., Strober, W., Andersson, J., Shevach, E. M., Quezado, M., Bouladoux, N., Roebroek, A., Belkaid, Y., Creemers, J., and O'Shea, J. J. (2008) Nature 455 246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]