Abstract
The molecular mechanisms that regulate invertebrate visual pigment absorption are poorly understood. Through sequence analysis and functional investigation of vertebrate visual pigments, numerous amino acid substitutions important for this adaptive process have been identified. Here we describe a serine/alanine (S/A) substitution in long wavelength-absorbing Drosophila visual pigments that occurs at a site corresponding to Ala-292 in bovine rhodopsin. This S/A substitution accounts for a 10–17-nm absorption shift in visual pigments of this class. Additionally, we demonstrate that substitution of a cysteine at the same site, as occurs in the blue-absorbing Rh5 pigment, accounts for a 4-nm shift. Substitutions at this site are the first spectrally significant amino acid changes to be identified for invertebrate pigments sensitive to visible light and are the first evidence of a conserved tuning mechanism in vertebrate and invertebrate pigments of this class.
Organisms use color vision for survival behaviors such as foraging, mating, and predator avoidance (1–3). Color vision in invertebrates ranges from trichromatic systems capable of detecting UV, blue, and green (e.g. bees and flies) to the highly complex mantis shrimps (stomatopods) having 12 spectrally distinct classes of photoreceptor cells (4). Despite the diversity of invertebrate color vision systems and the large collection of naturally occurring visual pigments, important questions remain concerning the molecular mechanisms that regulate color sensitivity.
In both vertebrates and invertebrates, the visual pigment rhodopsin consists of a chromophore (e.g. 11-cis retinal) covalently bound to an opsin apoprotein via a protonated Schiff base. Upon light absorption, the chromophore isomerizes from cis to all-trans, inducing conformational changes in the opsin that produce activated metarhodopsin. Specific interactions between the retinal chromophore and residues in the opsin tune the λmax of the chromophore. Studies have shown that Glu-113 (bovine position) serves as the retinylidene Schiff base counter-ion in vertebrate visual pigments (5–7). Removing the negative charge of the counter-ion from the binding pocket deprotonates the chromophore and yields a UV-absorbing pigment (5–7). Using sequence alignments, phylogenetic analysis, analysis of the bovine rhodopsin crystal structure (PDB2 entry 1U19), and functional experiments, a large number of amino acids involved in the spectral tuning of vertebrate visual pigments have been identified (8).
In contrast, the counter-ion for invertebrate rhodopsin remains unknown, and only one spectrally relevant residue has been identified: an amino acid substitution in Drosophila pigments responsible for UV versus visible sensitivity (9). Interestingly, this amino acid substitution (Gly-90 in bovine rhodopsin) coincides with a substitution that mediates UV versus blue sensitivity in several bird species (10, 11). This discovery highlights the value of a cross-phyla comparison of visual pigments as a means to identify structural differences that may regulate color vision in invertebrates.
In the present study, we identify an amino acid substitution in Drosophila visual pigments that regulates the color sensitivity of blue- and green-absorbing rhodopsins. For these studies, we employed sequence analysis of invertebrate and vertebrate visual pigments and a functional examination of mutant invertebrate opsins. This amino acid substitution red-shifts the λmax of the Drosophila Rh1 pigment and reciprocally blue-shifts the λmax of Rh6 pigment. Interestingly, this site also affects the spectral tuning of vertebrate pigments and corresponds to Ala-292 in bovine rhodopsin (8, 12–16).
EXPERIMENTAL PROCEDURES
Electrophysiology and Microspectrophotometry—Spectral sensitivity was measured as described previously (9). Spectral sensitivity recordings were performed on transgenic animals expressing modified forms of rhodopsin either in a ninaE17 background or in a modified norpA; ninaE17 mutant background. The latter strain also contained an additional transgene driving the norpA cDNA in the R1–6 photoreceptor cells under the control of the Rh1 promoter. This background strain allows the activity of the modified pigment to be examined without interference from the R7 and R8 cells that are not affected by the ninaE mutation (17).
Microspectrophotometry was performed as described previously (17). A high intensity adapting light was used to photoconvert the visual pigment from its rhodopsin (R) to its metarhodopsin (M) state. The transmission spectrum of each state was measured, and a difference spectrum was calculated as described previously (17).
Nomogram Curve Fitting—Rhodopsin and metarhodopsin theoretical absorption spectra were calculated from sensitivity and difference spectra as described previously (9, 17) using the spectral shape of the rhodopsin α-band absorption described by the following lognormal function
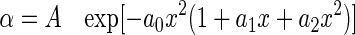 |
(Eq. 1) |
where
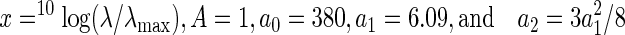 |
(Eq. 2) |
Homology Modeling and Dynamics Methods—Three protein models were generated using molecular dynamics methods similar to Ramos et al. (18). The structure of wild-type Drosophila Rh1 (Swiss-Prot accession number P06002) (19) was generated using PHYRE (20, 21) and differs from the original sequence by the deletion of residues 1–6 and 242–254. The structure was minimized 5000 steps using NAMD (22) and aligned to bovine rhodopsin (PDB entry 1U19) (23) using STRAP (24). The retinal molecule from bovine rhodopsin was placed into Rh1 at lysine 319, creating a lysine-bound retinal (LYR-319). The retinal molecule was modified to 3-hydroxyretinal using VMD (25). Topology and parameter NAMD input for LYR-319 was modified from previous studies (26–31). In the mutant models, alanine 315 was mutated to serine (Rh1A315S) or cysteine (Rh1A315C) and minimized 5000 steps with NAMD. Subsequently, all three models underwent the same procedure. Internal water molecules were placed using DOWSER (32) followed by a 6000-step minimization. The proteins were embedded in a membrane of 1-palmitoyl-2-oleoylphosphatidylchlorine with VMD. Full solvation with TIP3 water molecules, neutralization, and the addition of sodium and chloride ions were performed with VMD. The resulting models were 93 × 94 × 97 Å containing 12,525 water molecules. Full particle mesh Ewald calculations for electrostatics were used for all simulations. To relax the system, the following were used: a 20-ps run with protein backbone and LYR-319 fixed; a 50-ps run with protein backbone and LYR-319 residue harmonically constrained; a 50-ps run with LYR-319 residue harmonically constrained; and then a 2-ns run with no constraints. For comparison of the wild-type Drosophila Rh1 model with the published squid rhodopsin crystal structure (PDB entry 2z73), we used the STRAP program as noted above (24, 33).
RESULTS
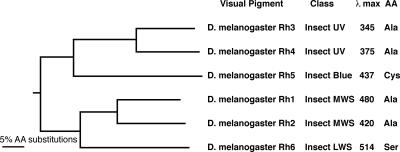
To identify spectral tuning sites in the invertebrate pigment population, we performed an alignment of the six well characterized Drosophila opsins and generated a phylogenetic tree to evaluate their relatedness. A comprehensive phylogenetic analysis of invertebrate pigments was published recently (34). The Drosophila visual pigments represent almost all of the major classes of visual pigments within the invertebrate lineage including ultraviolet, blue, middle wavelength-sensitive (MWS), and long wavelength-sensitive (LWS) pigments, Fig. 1. The phylogenetic relationship between pigments of different spectral types is conserved in most other species (e.g. insect UV and blue pigments are derived from the same lineage).
FIGURE 1.
Phylogenetic relationship between Drosophila melanogaster opsins. The tree was constructed from a ClustalW alignment with bovine rhodopsin as an outgroup. Both neighbor joining and unweighted pair group methods were used. The trees were bootstrapped 1000 replications, and each node was supported 100% by both methods. The class of each pigment is indicated, referred to in Ref. 34. λmax corresponds to the maximal sensitivity of flies expressing each visual pigment in the R1–6 photoreceptor cells (17, 45, 46). The amino acid (AA) residue present at the site corresponding to bovine rhodopsin Ala-292 is indicated.
We then examined the sequences of the Drosophila pigments at sites known to influence the spectral tuning of vertebrate visual pigments (reviewed in Ref. 8). We isolated potentially spectrally relevant residues by identifying amino acids that were conserved in a given spectral family of opsins but that differed in the other opsin families. Such a relationship between amino acid substitution and absorption maxima variation has been extensively substantiated in the vertebrate visual pigment lineages.
As the result of this analysis, we identified an amino acid substitution within the Drosophila opsins that corresponds to a spectral tuning site identified in the vertebrate pigments at amino acid position 292 (bovine opsin numbering). Mutagenesis experiments at this site in a variety of pigments have shown that A292S or S292A mutants demonstrate substantial blue and red spectral shifts, respectively (8, 12–16). As shown in Fig. 1, the LWS Drosophila Rh6 has a serine at this position, whereas the UV-sensitive Rh3 and Rh4 and MWS Rh1 and Rh2 have an alanine, and the blue-sensitive Rh5 has a cysteine at this position. This relationship is conserved in other invertebrate pigments. The mollusk rhodopsins contain valine, the arthropod LWS pigments contain serine or tyrosine, the arthropod MWS pigments contain alanine, and the insect blue and UV pigments contain alanine at this position, except for Drosophila Rh5 noted above.
To investigate the role that this amino acid substitution plays in tuning the spectral properties of invertebrate pigments, we constructed mutant forms of the blue-absorbing (Rh1) and green-absorbing (Rh6) Drosophila opsins, as described previously (9). In these mutant forms, we replaced the residue in one pigment with the corresponding residue found in the other (i.e. Rh1 A315S and Rh6 S313A). In addition, we generated an Rh1 mutant that contains the cysteine found in the same position of Rh5. To determine the effect that these amino acid changes have on the absorption properties of the visual pigments, we introduced the genes encoding these modified pigments (as well as their non-modified counterparts) into the germline of transgenic Drosophila containing the ninaE mutation. ninaE is a deletion in the endogenous Rh1 gene that is expressed in the R1–6 photoreceptor cells (19, 35). By placing the transgene under the control of the Rh1 promoter, we exchanged the function of the endogenous Rh1 pigment with the pigment of interest.
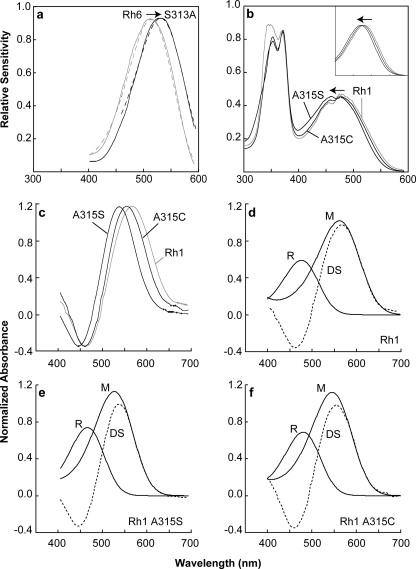
As shown in Fig. 2A, electrophysiological analysis of the transgenic animals expressing Rh6 S313A showed a dramatic red shift in spectral sensitivity when compared with the animals expressing the unmodified form of the Rh6 pigment (from a λmax = 514 nm to a λmax = 531 nm). In comparison, transgenic animals expressing Rh1 A315S had a complimentary blue shift in sensitivity (from a λmax = 480 nm to a λmax = 470 nm) when compared with wild-type animals that express the endogenous Rh1 pigment, Fig. 2b. Furthermore, transgenic animals expressing Rh1 A315C also had a blue shift in sensitivity (from a λmax = 480 nm to a λmax = 476 nm) (Fig. 2b). Thus, the Ser versus Ala substitution appears to be responsible for dramatic shifts in the absorption of Rh6 (∼17-nm shift) and Rh1 (∼10-nm shift), whereas the Cys versus Ala substitution seems to be responsible for a smaller but significant shift in absorption for Rh1 (∼4-nm shift). These results confirm the importance of this site for spectral tuning of invertebrate pigments and are consistent with the results reported for substitutions at this site in the vertebrate pigments.
FIGURE 2.
Rh1 and Rh6 spectral sensitivities and rhodopsin/metarhodopsin difference spectra. a and b, measured spectral sensitivities of flies expressing Rh1, Rh6, or one of the modified rhodopsin pigments in the R1–6 photoreceptor cells (solid lines). Each sensitivity spectrum obtained in this study was also fit to a rhodopsin absorption nomogram (dotted lines). a, mean spectral sensitivities of flies expressing Rh6 S313A (black line) when compared with flies expressing the unmodified Rh6 pigment (gray line). The nomogram curve fit for Rh6 S313A has a λmax red-shifted from the unmodified pigment 17 nm (dotted lines). For all sensitivity data, the λmax, correlation coefficient, and number of flies examined are indicated in Table 1. b, mean spectral sensitivities of flies expressing the rhodopsin mutants, Rh1 A315S, and Rh1 A315C (black lines) when compared with the unmodified Rh1 pigment (gray line). The large peak in the UV region occurs because of the action of a sensitizing pigment that absorbs in the UV and activates the Rh1 rhodopsin through energy transfer. The peaks in the visible region are due to direct absorption by the visual pigment itself. The nomogram curve fits for Rh1 A315C and Rh1 A315S λmax are blue-shifted 4 and 10 nm, respectively (inset). The fine structure noted in the sensitivity spectra in the region of 475 nm is an artifact and results from spectral spikes in xenon lamp output in this region. c–f, difference spectra were measured by in vivo MSP of flies expressing Rh1 or one of the modified rhodopsin pigments. c, mean difference spectra of Rh1 (gray line) when compared with Rh1 A315S and Rh1 A315C (black lines). d–f, calculated R and M absorption spectra based on nomogram curve fitting to the measured difference spectra (DS). In all calculations, the λmax for R spectra was set to the λmax measured physiologically. For MSP data, the λmax, correlation coefficient, and number of flies examined are indicated in Table 1.
Photoactivation of rhodopsin occurs upon absorption of a photon by the retinal chromophore. This induces the isomerization of the 11–12-cis double bond to the trans configuration, which then induces a series of conformational changes in the protein that leads to its activation and the formation of metarhodopsin. To determine the absorption profile of the metarhodopsin (M) forms of the modified Rh1 pigments, we used microspectrophotometry (MSP) to examine dissected retina from transgenic flies and wild-type flies (Fig. 2c). The M-form of Rh1 A315S has a λmax = 524 nm, whereas the M-form of Rh1 A315C has a λmax = 548 nm, dramatic shifts of 36 and 12 nm, respectively, from wild-type Rh1 (Fig. 2, d–f). We were unable to record a difference spectrum from transgenic animals expressing the Rh6 S313A construct, consistent with previous studies of unmodified Rh6 (17). These results indicate that this amino acid substitution is important for regulating the tuning of both R-form and M-form absorption.
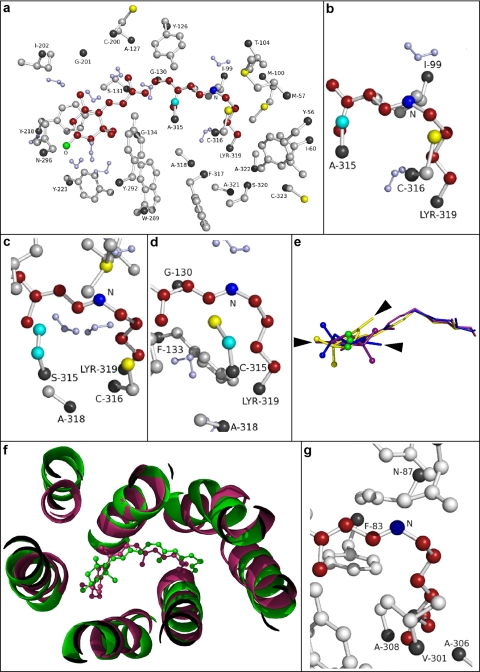
To evaluate the structural effects of amino acid substitutions, we generated structural models of Rh1 wild type, Rh1A315S, and Rh1A315C using molecular dynamics methods. As expected, the overall topology of the structural models is very similar to that of bovine rhodopsin, upon which they are based. The root mean square deviation, the average distance between the atoms of superimposed proteins, for each pairwise structural model comparison was as follows: Rh1 wild type – Rh1A315C = 5.6 Å, Rh1 wild type – Rh1A315S = 4.2 Å, and Rh1A315S – Rh1A315C = 4.9 Å. Overall, this demonstrates a very close fit between the structures as a whole, although Rh1 wild type and Rh1A315S show a higher degree of overlap with each other than they do with the Rh1A315C mutant. The amino side chains within 3.2 Å of the retinal chromophore in the model of Rh1 wild type are shown in Fig. 3a. Fig. 3, b–d, show residue 315 in the wild-type and mutant pigments in very close proximity to the Schiff base nitrogen. The distances were 4.8 Å to Ala in Rh1 wild type, 4.2 Å to Ser in Rh1A315S, and 4.3 Å to Cys in Rh1A315C. We examined the topology of the retinal chromophore and found that although the geometry of the polyene chain in each model overlaps to a substantial degree, the β-ionone ring is rotated in the models of the mutant pigments by 140 (Rh1A315S) or 190 (Rh1A315C) degrees when compared with Rh1 wild type (Fig. 3e). Higher resolution images of the models can be found in supplemental Figs. 1–3. We also compared the model of wild-type Rh1 with the recently published squid rhodopsin crystal structure (33) and found a high degree of similarity between the two structures (Fig. 3f). There is a substantial overlap between the helical regions of the proteins and close approximation between the two chromophore structures. Root mean square deviation for the comparison between the two structures was 2.7 Å. We examined the region of the squid rhodopsin chromophore binding site and found that, similar to the three Drosophila models, the residue corresponding to Ala-315 in Rh1 (squid Val-301) is in close proximity to the Schiff base nitrogen (Fig. 3g). The distances from the Schiff base nitrogen to the carbon atoms of the two methyl groups in the Val-301 side chain were 4.5 and 5.5 Å. A higher resolution image of the squid rhodopsin structure can be found in supplemental Fig. 4.
FIGURE 3.
Molecular modeling of Drosophila Rh1 wild-type, Rh1A315S, and Rh1A315C mutant pigments. a, model of Drosophila Rh1 wild-type pigment showing all residues within 3.2 Å of the lysine-bound Schiff base chromophore. b–d, close-up views of the Schiff base nitrogen and residue 315 in Rh1 wild type (b), Rh1A315S (c), and Rh1A315C (d). The 3-hydroxy-11-cis-retinal chromophore is shown in red with the Schiff base nitrogen in dark blue and the 3-hydroxyl oxygen in green. The α-carbons are shown in black. Hydrogen atoms are omitted except on water molecules, which are shown in light purple. Wild-type and mutated residues (315) are colored cyan. Sequence numbering is based on Swiss-Prot accession number P06002 (19). e, alignment of the 3-hydroxy-retinal chromophore from Rh1 wild type (purple), Rh1A315S (yellow), and Rh1A315C (blue). The 3-hydroxyl oxygen atoms are shown in green. The arrowheads illustrate the rotation of the chromophore ring in the mutant models Rh1A315S (two o'clock) and Rh1A315C (three o'clock) relative to Rh1 wild type (nine o'clock). f, alignment of the Drosophila Rh1 model (green) and squid rhodopsin crystal structure (PDB entry 2z73; purple). The panel is a ribbon diagram showing the orientation of the helices in the region of the chromophore. g, close-up view of the Schiff base nitrogen and residue Val-301 (corresponding to Rh1 Ala-315) in the region of the chromophore. Sequence numbering is based on Swiss-Prot Accession Number P31356 (47). Higher resolution images of the Drosophila Rh1 model structures and squid rhodopsin crystal structure are shown in supplemental Figs. 1–4.
DISCUSSION
The principal result from this study is that the Ala versus (Ser or Cys) substitution present at residue 315 in the Drosophila visual pigments (corresponding to bovine rhodopsin Ala-292) serves to shift the absorption of the LWS Rh6 opsin and blue-sensitive Rh5 opsin to shorter wavelengths. These results are consistent with previous studies on vertebrate visual pigments that demonstrated a 9–10-nm blue shift in A292S mutant forms of bovine rhodopsin and other pigments (8, 12–14, 16). The results are also consistent with studies of the mouse green cone pigment, dolphin LWS pigment, and coelacanth rhodopsin having S292A substitutions that red-shifted pigment absorption by 8–28 nm (12, 13, 15). In addition, substitutions of Ile, Ser, Ala, or Thr at this site are also thought to play a role in rhodopsin spectral differences in both cottoid and deep sea fish species and have arisen multiple times within fish lineages (36–39).
The residue 315 site in Drosophila Rh1 is within ∼4Åofthe Schiff base nitrogen of the chromophore in the structures we have modeled, suggesting that amino acid substitutions here can directly interact with the chromophore. Substitution of a serine in place of an alanine at this position would introduce a highly polar side chain and a strong hydrogen bond acceptor that would be ideally positioned to interact with the chromophore.
Quantum mechanical and molecular mechanical calculations examining the electrostatic, electronic polarization, and geometry effects of the A292S mutation in bovine rhodopsin indicate that the serine side chain hydroxyl may form hydrogen bonds with the carbonyl oxygen of Met-288 and an adjacent water molecule and may be oriented toward the hydroxyl of Ser-186, resulting in an increase in chromophore bond length alternation and a blue shift in absorption (40). Consistent with this view is the observation of a potential hydrogen bond network between the Schiff base nitrogen, Asn-87 and Tyr-111 within the squid rhodopsin crystal structure (33). Thus, introduction of a polar serine or cysteine at position 315 in Drosophila Rh1, at the position corresponding to squid Val-301, may lead to a blue spectral shift by increasing chromophore bond length by altering hydrogen bonding in this region.
With regard to the smaller blue shift observed in the Rh1A315C mutation, the sulfhydryl group of cysteine has a weaker dipole moment and would be expected to be a weaker hydrogen bond acceptor than the hydroxyl group of serine (12). Our finding that the spectral shift associated with Rh1 A315C (–4 nm) is smaller than that of Rh1 A315S (–10 nm) is consistent with this expectation and the observed –1-nm shift in the bovine rhodopsin A292C mutant (12). Similar results were obtained in our analysis of metarhodopsin, which demonstrated –36- and –12-nm shifts in the Rh1A315S and Rh1315C mutants respectively (Fig. 2, d–f, and Table 1). Although the magnitude of the spectral shifts differ in the rhodopsin and metarhodopsin forms of the pigment, the observation of shifts in both states indicates that in both the native and the activated forms of the pigment, amino acid 315 is positioned close to the chromophore.
TABLE 1.
λmax and rhodopsin absorption nomogram curve data for each wild type and modified visual pigment examined in the study
NA = not available (see “Results”). Correlation coefficient = coefficient of the fit of the nomogram curve to the absorption or sensitivity data. R = rhodopsin, M = metarhodopsin, SS = spectral sensitivity data.
|
Visual pigment
|
λmax
|
Correlation coefficient
|
Number of flies analyzed
|
Figure panels
|
|||
|---|---|---|---|---|---|---|---|
| R | M | SS | MSP | SS | MSP | ||
| Rh1 | 480 | 560 | 0.981 | 0.997 | 3 | 7 | Fig. 2b, inset, 2c, and 2d |
| Rh1 A315S | 470 | 524 | 0.976 | 0.999 | 15 | 10 | Fig. 2b, inset, 2c, and 2e |
| Rh1 A315C | 476 | 548 | 0.979 | 0.992 | 8 | 11 | Fig. 2b, inset, 2c, and 2f |
| Rh6 | 514 | NA | 0.997 | NA | 10 | NA | Fig. 2a |
| Rh6 S313A | 531 | NA | 0.996 | NA | 10 | NA | Fig. 2a |
Interestingly, this amino acid position (292 in human rhodopsin) is associated with human congenital stationary night blindness. A patient with this disorder was found to carry a missense mutation A292E (41). Although the mutant forms a functional pigment and activates transducin in a light-dependent manner, in the absence of chromophore, the mutant opsin constitutively activates transducin. The λmax of the A292E human mutant rhodopsin is unchanged despite the introduction of a negatively charged side chain at this site (41, 42). Although this differs from the –10-nm shift found in the bovine rhodopsin mutant A292D (43), it may be that there is a difference in the overall context of the chromophore binding pocket that prevents the substitution in human rhodopsin from having the expected effect, as suggested by the case of the lack of spectral shift for the S292A mutant of the human blue cone pigment (44).
In summary, the present studies demonstrate that as is the case in vertebrates, amino acid substitutions in invertebrate visual pigments at the site corresponding to Ala-292 in bovine rhodopsin are responsible for specific spectral changes of 10–17 nm. Not only do amino acid substitutions at this site alter the absorption of the native form of the visual pigment, but they are also responsible for as much as 36-nm spectral shift in the activated form of the visual pigment, metarhodopsin. These findings represent the first naturally occurring amino acid substitution in the insect blue, MWS, and LWS visual pigments that has been shown to play a role in spectral tuning and suggests that although some aspects of vertebrate and invertebrate visual pigment tuning are different, the tuning mechanism at Ala-292 is conserved.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant R01-EY10759 from the NEI (to S. G. B.). This work was also supported by Grant IOB-0449367 from the National Science Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains four supplemental figures.
Footnotes
The abbreviations used are: PDB, Protein Data Bank; R, rhodopsin; M, metarhodopsin; MWS, middle wavelength-sensitive; LWS, long wavelength-sensitive; MSP, microspectrophotometry.
References
- 1.Maximov, V. V. (2000) Philos. Trans. R. Soc. Lond. B Biol. Sci. 355 1239–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dyer, A. G., and Chittka, L. (2004) J. Exp. Biol. 207 1683–1688 [DOI] [PubMed] [Google Scholar]
- 3.Detto, T. (2007) Proc. Biol. Sci. R. Soc. Lond. 274 2785–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marshall, J., Cronin, T. W., and Kleinlogel, S. (2007) Arthropod Struct. Dev. 36 420–448 [DOI] [PubMed] [Google Scholar]
- 5.Zhukovsky, E. A., and Oprian, D. D. (1989) Science 246 928–930 [DOI] [PubMed] [Google Scholar]
- 6.Sakmar, T. P., Franke, R. R., and Khorana, H. G. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 8309–8313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nathans, J. (1990) Biochemistry 29 9746–9752 [DOI] [PubMed] [Google Scholar]
- 8.Takahashi, Y., and Ebrey, T. G. (2003) Biochemistry 42 6025–6034 [DOI] [PubMed] [Google Scholar]
- 9.Salcedo, E., Zheng, L., Phistry, M., Bagg, E. E., and Britt, S. G. (2003) J. Neurosci. 23 10873–10878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokoyama, S., Radlwimmer, F. B., and Blow, N. S. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 7366–7371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilkie, S. E., Robinson, P. R., Cronin, T. W., Poopalasundaram, S., Bowmaker, J. K., and Hunt, D. M. (2000) Biochemistry 39 7895–7901 [DOI] [PubMed] [Google Scholar]
- 12.Sun, H., Macke, J. P., and Nathans, J. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 8860–8865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fasick, J. I., and Robsinson, P. R. (1998) Biochemistry 37 433–438 [DOI] [PubMed] [Google Scholar]
- 14.Lin, S. W., Kochendoerfer, G. G., Carroll, K. S., Wang, D., Mathies, R. A., and Sakmar, T. P. (1998) J. Biol. Chem. 273 24583–24591 [DOI] [PubMed] [Google Scholar]
- 15.Yokoyama, S., Zhang, H., Radlwimmer, F. B., and Blow, N. S. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 6279–6284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janz, J. M., and Farrens, D. L. (2001) Biochemistry 40 7219–7227 [DOI] [PubMed] [Google Scholar]
- 17.Salcedo, E., Huber, A., Henrich, S., Chadwell, L. V., Chou, W. H., Paulsen, R., and Britt, S. G. (1999) J. Neurosci. 19 10716–10726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramos, L. S., Chen, M. H., Knox, B. E., and Birge, R. R. (2007) Biochemistry 46 5330–5340 [DOI] [PubMed] [Google Scholar]
- 19.O'Tousa, J. E., Baehr, W., Martin, R. L., Hirsh, J., Pak, W. L., and Applebury, M. L. (1985) Cell 40 839–850 [DOI] [PubMed] [Google Scholar]
- 20.Bennett-Lovsey, R. M., Herbert, A. D., Sternberg, M. J., and Kelley, L. A. (2008) Proteins 70 611–625 [DOI] [PubMed] [Google Scholar]
- 21.Kelley, L. A., MacCallum, R. M., and Sternberg, M. J. (2000) J. Mol. Biol. 299 499–520 [DOI] [PubMed] [Google Scholar]
- 22.Phillips, J. C., Braun, R., Wang, W., Gumbart, J., Tajkhorshid, E., Villa, E., Chipot, C., Skeel, R. D., Kale, L., and Schulten, K. (2005) J. Comput. Chem. 26 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okada, T., Sugihara, M., Bondar, A. N., Elstner, M., Entel, P., and Buss, V. (2004) J. Mol. Biol. 342 571–583 [DOI] [PubMed] [Google Scholar]
- 24.Gille, C., and Frommel, C. (2001) Bioinformatics (Oxf.) 17 377–378 [DOI] [PubMed] [Google Scholar]
- 25.Humphrey, W., Dalke, A., and Schulten, K. (1996) J. Mol. Graph. 14 33–38 [DOI] [PubMed] [Google Scholar]
- 26.Hayashi, S., and Ohmine, I. (2000) J. Phys. Chem. B 104 10678–10691 [Google Scholar]
- 27.Tajkhorshid, E., Paizs, B., and Suhai, S. (1997) J. Phys. Chem. B 101 8021–8028 [Google Scholar]
- 28.Tajkhorshid, E., and Suhai, S. (1999) J. Phys. Chem. B 103 5581–5590 [Google Scholar]
- 29.Tajkhorshid, E., Baudry, J., Schulten, K., and Suhai, S. (2000) Biophys. J. 78 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baudry, J., Crouzy, S., Roux, B., and Smith, J. C. (1997) J. Chem. Inf. Comput. Sci. 37 1018–1024 [Google Scholar]
- 31.Nina, M., Roux, B., and Smith, J. C. (1995) Biophys. J. 68 25–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang, L., and Hermans, J. (1996) Proteins 24 433–438 [DOI] [PubMed] [Google Scholar]
- 33.Murakami, M., and Kouyama, T. (2008) Nature 453 363–367 [DOI] [PubMed] [Google Scholar]
- 34.Porter, M. L., Cronin, T. W., McClellan, D. A., and Crandall, K. A. (2007) Mol. Biol. Evol. 24 253–268 [DOI] [PubMed] [Google Scholar]
- 35.Zuker, C. S., Cowman, A. F., and Rubin, G. M. (1985) Cell 40 851–858 [DOI] [PubMed] [Google Scholar]
- 36.Hunt, D. M., Fitzgibbon, J., Slobodyanyuk, S. J., and Bowmaker, J. K. (1996) Vision Res. 36 1217–1224 [DOI] [PubMed] [Google Scholar]
- 37.Hunt, D. M., Dulai, K. S., Partridge, J. C., Cottrill, P., and Bowmaker, J. K. (2001) J. Exp. Biol. 204 3333–3344 [DOI] [PubMed] [Google Scholar]
- 38.Sugawara, T., Terai, Y., Imai, H., Turner, G. F., Koblmuller, S., Sturmbauer, C., Shichida, Y., and Okada, N. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 5448–5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yokoyama, S., Tada, T., Zhang, H., and Britt, L. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 13480–13485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altun, A., Yokoyama, S., and Morokuma, K. (2008) J. Phys. Chem. B 112 6814–6827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dryja, T. P., Berson, E. L., Rao, V. R., and Oprian, D. D. (1993) Nat. Genet. 4 280–283 [DOI] [PubMed] [Google Scholar]
- 42.Gross, A. K., Rao, V. R., and Oprian, D. D. (2003) Biochemistry 42 2009–2015 [DOI] [PubMed] [Google Scholar]
- 43.Nakayama, T. A., and Khorana, H. G. (1991) J. Biol. Chem. 266 4269–4275 [PubMed] [Google Scholar]
- 44.Fasick, J. I., Lee, N., and Oprian, D. D. (1999) Biochemistry 38 11593–11596 [DOI] [PubMed] [Google Scholar]
- 45.Feiler, R., Bjornson, R., Kirschfeld, K., Mismer, D., Rubin, G. M., Smith, D. P., Socolich, M., and Zuker, C. S. (1992) J. Neurosci. 12 3862–3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feiler, R., Harris, W. A., Kirschfeld, K., Wehrhahn, C., and Zuker, C. S. (1988) Nature 333 737–741 [DOI] [PubMed] [Google Scholar]
- 47.Hara-Nishimura, I., Kondo, M., Nishimura, M., Hara, R., and Hara, T. (1993) FEBS Lett. 317 5–11 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.