Abstract
The chemokine receptor CXCR4 plays important roles in the immune and nervous systems. Abnormal expression of CXCR4 contributes to cancer and inflammatory and neurodegenerative disorders. Although ligand-dependent CXCR4 ubiquitination is known to accelerate CXCR4 degradation, little is known about counter mechanisms for receptor deubiquitination. CXCL12, a CXCR4 agonist, induces a time-dependent association of USP14 with CXCR4, or its C terminus, that is not mimicked by USP2A, USP4, or USP7, other members of the deubiquitination catalytic family. Co-localization of CXCR4 and USP14 also is time-dependent following CXCL12 stimulation. The physical interaction of CXCR4 and USP14 is paralleled by USP14-catalyzed deubiquitination of the receptor; knockdown of endogenous USP14 by RNA interference (RNAi) blocks CXCR4 deubiquitination, whereas overexpression of USP14 promotes CXCR4 deubiquitination. We also observed that ubiquitination of CXCR4 facilitated receptor degradation, whereas overexpression of USP14 or RNAi-induced knockdown of USP14 blocked CXCL12-mediated CXCR4 degradation. Most interestingly, CXCR4-mediated chemotactic cell migration was blocked by either overexpression or RNAi-mediated knockdown of USP14, implying that a CXCR4-ubiquitin cycle on the receptor, rather than a particular ubiquitinated state of the receptor, is critical for the ligand gradient sensing and directed motility required for chemokine-mediated chemotaxis. Our observation that a mutant of CXCR4, HA-3K/R CXCR4, which cannot be ubiquitinated and does not mediate a chemotactic response to CXCL12, indicates the importance of this covalent modification not only in marking receptors for degradation but also for permitting CXCR4-mediated signaling. Finally, the indistinguishable activation of ERK by wild typeor 3K/R-CXCR4 suggests that chemotaxis in response to CXCL12 may be independent of the ERK cascade.
The CXCR4 (CXC chemokine receptor 4) is a member of the chemokine receptor family, which belongs to the superfamily of G protein-coupled receptors (GPCRs)2 (1). Its ligand, CXCL12, also known as SDF-1α, also binds to RDC1, another chemokine receptor that is being proposed to be renamed as CXCR7 (2). CXCR4 mediates CXCL12-induced migration of peripheral blood lymphocytes (3), CD34+ progenitor cells (4), and pre- and pro-B cell lines (5). CXCR4 also plays an important role in the development of the immune system, because mouse embryos lacking either expression of the CXCR4 receptor or of its CXCL12 ligand are embryonic lethal and also manifest abnormalities in B cell lymphopoiesis and bone marrow myelopoiesis (3, 6, 7). The altered cerebellar neuron migration in mice null for the CXCR4 receptor also suggests a role for this receptor in central nervous system development. Abnormal expression and/or function of CXCR4 have been implicated in a number of diseases, including human immunodeficiency virus infection (8), cardiovascular disease (9), allergic inflammatory disease (10), neuroinflammation (11), neurodegenerative diseases (12, 13), and cancers (14-24).
Stimulation of CXCR4 triggers various intracellular signaling cascades (1, 14, 25-27), such as extracellular signal-regulated kinase (ERK), which likely contribute to CXCR4-induced cell proliferation, differentiation, and/or migration. Ligand stimulation of CXCR4 also induces endocytosis of these receptors, which are targeted to lysosomes for degradation through a pathway involving ubiquitination of the C-terminal lysine residues (28). CXCR4 ubiquitination can be catalyzed by a member of the HECT family of E3 ligases, AIP4 (atrophin-interacting protein 4) (29, 30). The ubiquitinated CXCR4 is delivered to the endosomal compartments via a regulated pathway involving several adaptor proteins (31).
It has been noted that deubiquitination also regulates the fate and function of ubiquitin-conjugated proteins. Deubiquitinating enzymes, which catalyze the removal of ubiquitin from ubiquitin-conjugated proteins, represent the largest family of enzymes in the ubiquitin system, implying the possibility that substrate selectivity is even greater for these enzymes than for those that catalyze ubiquitin ligation. Little is known about the mechanisms of CXCR4 deubiquitination and their regulation by receptor ligands. A proteomics study revealed that the steady state level of USP14 was increased upon CXCL12 stimulation of target cells (32), and preliminary studies revealed that ligand stimulation led to enhanced association of USP14 with the CXCR4. The present studies were undertaken to ascertain the functional consequences of this interaction, the selectivity of CXCR4 for USP14, when compared with three other deubiquitinating enzymes, USP2a, USP4, and USP7, and the impact of modifying the ubiquitinated state of the receptor on CXCR4 turnover, CXCL12-evoked chemotaxis, and CXCL12-induced activation of ERK.
EXPERIMENTAL PROCEDURES
Plasmids and siRNAs—Plasmids encoding Myc-CXCR4 and glutathione S-transferase (GST)-conjugated CXCR4 C terminus were obtained from Dr. Gang Pei (Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences). The enhanced green fluorescent protein (EGFP)-conjugated CXCR4 was constructed by amplifying the cDNAs of CXCR4 from HA-CXCR4 in pcDNA3 vector using PCR and inserting the cDNA into the XhoI and BamHI sites of the pEGFP/N1 vector (Clontech). The epitope-tagged CXCR4 has been tested by radioligand binding assay and cyclic AMP assay and was confirmed to function similarly as the nontagged receptor (data not shown). The mutant CXCR4 that cannot be recognized for ubiquitination (HA-3K/R CXCR4) was obtained from Dr. Jeffrey L. Benovic (Thomas Jefferson University, Philadelphia) (28). The USP14 plasmid was constructed by PCR amplification of USP14 cDNA using lymphocyte cDNAs as template and inserted into the pcDNA3 vector. Pre-designed USP14-specific siRNA and pre-designed control (scramble) siRNA were purchased from Ambion. The USP2a plasmid was obtained from Dr. Massimo Loda (Dana Farber Cancer Institute, Harvard Medical School, Boston). The USP4 plasmid was obtained from Dr. Michael Freissmuth (Medical University of Vienna, Vienna, Austria). The pCI-USP7 plasmid was a gift from Dr. Roger Everett (Institute of Virology, University of Glasgow, Scotland, UK). The pcDNA3-USP7 was constructed by amplifying the cDNAs of USP7 in pCI vector using PCR and inserting the cDNA into the XhoI and BamHI sites of the pcDNA3 vector.
Cell Culture and Transfection—Human embryonic kidney (HEK293) and HeLa cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin in 5% CO2, 95% air at 37 °C. Cells were cultured in 100-mm dishes. For studies involving fluorescence microscopy, 22-mm square glass coverslips were placed in the culture dishes before transfection. Transient transfection of recipient HEK293 cells was performed using Lipofectamine 2000 (Invitrogen). Cells stably expressing Myc-CXCR4 or EGFP-CXCR4 were selected with 560 μg/ml geneticin (G418).
Co-immunoprecipitation and Western Blot for Determining CXCR4 Complexes—HEK293 cells stably expressing Myc-CXCR4 were transiently transfected with HA-USP14 or USP14-specific siRNA, treated with CXCL12 (10 nm; Pepro-Tech, Inc.) for various time intervals, and lysed in 1 ml of RIPA buffer containing PBS (pH.7.0), 0.1% sodium deoxycholate, 0.01% SDS, and 1% Nonidet P-40 to which additional SDS was added to bring the final concentration to ∼10%. The cell debris was removed by centrifugation (13,000 × g, 15 min). The supernatant was pre-cleared by incubation with 40 μl of protein A/G-agarose (Pierce) for 1 h at 4 °C to reduce nonspecific binding. After removing the protein A/G-agarose by centrifugation (13,000 × g, 1 min), the cleared supernatant was collected, and 10 μl of mouse monoclonal anti-Myc antibody (Santa Cruz Biotechnology) was added for an overnight incubation at 4 °C. Protein A/G (40 μl) was then added to this mixture, and the incubation was continued for 2 h at 4 °C. The protein A/G-antibody-antigen complex was collected by centrifugation and washing the pelleted resin three times with ice-cold immunoprecipitation buffer. The final pellets were resuspended in 40 μl of SDS sample buffer containing 5% β-mercaptoethanol and heated to 50 °C for 10 min. Forty microliters of this preparation were separated by 10% SDS-PAGE. The PAGE-embedded proteins were transferred to nitrocellulose membranes (Bio-Rad) by electrophoretic transfer at 60 V for 60 min at 4 °C in a buffer (25 mm Tris, 192 mm glycine, 20% methanol). Proteins were identified on the nitrocellulose filter by Western blotting, using the following antibodies. CXCR4 was detected using a mouse monoclonal antibody directed against the Myc epitope in the N-terminal domain of the receptor. USP14 was detected using a rabbit polyclonal antibody (Abgent). USP2a and USP7 were detected using a rabbit polyclonal antibody (Santa Cruz Biotechnology, Inc.) directed against the HA epitope engineered into the N terminus of the protein. USP4 was detected using a rabbit polyclonal antibody (Santa Cruz Biotechnology) directed against the His epitope engineered into the N terminus of the protein.
In Vitro Binding Assay for Protein Association with the C Terminus of CXCR4—For isolating proteins that interact with the CXCR4 C terminus, we exploited a GST-CXCR4 C-terminal fusion protein, using GST alone as a control. For the purification of the GST or GST-CXCR4 C-terminal fusion proteins, DH5α bacteria transformed with plasmids encoding GST or GST-CXCR4 C-terminal fusion proteins were cultured overnight at 37 °C. Isopropyl d-thiogalactopyranoside was added to the culture to induce protein expression, and incubation was continued for another 3 h. The bacteria were lysed in PBST buffer (10 mm sodium phosphate, 2 mm potassium phosphate (pH 7.4), 140 mm NaCl, 3 mm KCl, 0.1% (v/v) Tween 20) and then probe-sonicated on ice for 10 s. The supernatant of the bacterial lysate was incubated with glutathione-Sepharose (Pierce/Thermo Scientific, Inc.) for 30 min at 4 °C to isolate GST or the GST-CXCR4 C-terminal fusion protein. After washing three times with PBST buffer, the purified GST- or GST-CXCR4 C-terminal fusion protein-bound beads were resuspended in PBST buffer and stored on ice until use that day.
Cell lysates that were to be incubated with Sepharose-GST or Sepharose-GST-CXCR4 C terminus were prepared in the following way. HEK293 cells not expressing CXCR4 (and not stimulated with CXCL12) but transiently expressing HA-USP14, HA-USP2a, HIS-USP4, or HA-USP7 (with parental cells serving as controls) were lysed using PBST buffer (10 mm sodium phosphate, 2 mm potassium phosphate (pH 7.4), 140 mm NaCl, 3 mm KCl, 0.1% (v/v) Tween 20) 48 h after transfection. Cell lysates were pelleted to remove debris (13,000 × g, 10 min at 4 °C in a microcentrifuge), and the supernatants of this centrifugation were used for the GST pulldown assay.
For the GST pulldown assay, the supernatant of the cell lysates was incubated with aliquots of the purified GST or GST-CXCR4 C-terminal fusion proteins (to bring the GST fusion protein concentration to 50 μg/ml) for 2 h at 4 °C with rotation. To terminate the incubation, Sepharose beads were pelleted by centrifugation (13,000 × g, 2 min) and washed four times with PBST buffer. Proteins bound to the C-terminal CXCR4 receptor-GST fusions protein (or control GST) were released by boiling in SDS-PAGE sample buffer containing 5% β-mercaptoethanol for 10 min, resolved by SDS-PAGE, and analyzed by Western blot, as outlined above.
Confocal Microscopy—Our initial studies revealed a selectively greater association of CXCR4 with USP14 compared with other family members; thus, most of our studies focused on CXCR4 and its association with USP14. To examine the localization of USP14 compared with CXCR4 in cells expressing both, HEK293 cells stably expressing EGFP-CXCR4 were transiently transfected with HA-USP14 and grown on coverslips for 1 or 2 days. Cells were treated with CXCL12 (10 nm) for various time intervals and fixed with 100% methanol at room temperature before immediate transfer to 4 °C until further processing. Fixed cells were washed with PBS and incubated with a mouse monoclonal HA antibody (Santa Cruz Biotechnology) for 30 min at room temperature. Cells were washed with PBS and incubated with a CY3-conjugated anti-mouse antibody (Molecular Probes, Eugene, OR) for 30 min at room temperature and then washed twice with PBS and then briefly with deionized water to remove buffer salts. Coverslips were mounted on the microscope slide with a mounting solution.
Confocal microscopy was performed on an LSM-510 laser scanning microscope (Carl Zeiss, New York) with a 63 × 1.3 numerical aperture oil immersion lens using dual excitation (488 nm for EGFP and 568 nm for Cy3) and emission (515-540 nm for EGFP and 590-610 nm for Cy3) filter sets. All digital images were captured at the same settings to allow direct quantitative comparison of staining patterns. Final images were processed using Adobe Photoshop software.
Time Course of CXCR4 Ubiquitination and Deubiquitination—To assess whether ligand activation of CXCR4 altered the ubiquitination of this receptor, HEK293 cells stably expressing Myc-CXCR4 were incubated with 10 nm CXCL12 for the time intervals indicated in the figures and figure legends. The cells were then lysed in 1 ml of ice-cold RIPA buffer. The cell debris was removed by centrifugation (13,000 × g, 15 min). The supernatant was pre-cleared by incubation with 40 μl of protein A/G-agarose (Pierce) for 1 h at 4 °C to reduce nonspecific binding. After removing the protein A/G-agarose by centrifugation (13,000 × g, 1 min), the cleared supernatant was collected, and 10 μl of mouse monoclonal Myc antibody (Santa Cruz Biotechnology) was added for overnight incubation at 4 °C. Protein A/G (40 μl) was then added, and the incubation was continued for 2 h at 4 °C. The protein A/G-antibody-antigen complex was then collected by centrifugation, as above, and was washed three times with ice-cold immunoprecipitation buffer. The final pellets were resuspended in 40 μl of SDS sample buffer containing 5% β-mercaptoethanol and heated to 50 °C for 10 min. Forty microliters of this preparation were separated by 10% SDS-PAGE, and the proteins transferred to nitrocellulose membranes (Bio-Rad), as described in detail above for Western blotting. The state of ubiquitination of the CXCR4 receptor was detected by Western blot analysis using a rabbit polyclonal anti-ubiquitin antibody (Santa Cruz Biotechnology).
Two complementary strategies were used to determine the effect of USP14 on CXCR4 modification, i.e. overexpression of USP14 in CXCR4-expressing HEK293 cells and RNA silencing of endogenous USP14 in cells expressing CXCR4. For USP14 overexpression, cells were transiently transfected with vector (control) or HA-USP14 (see under “Cell Culture and Transfection”) and incubated with ligand 48 h later. For knockdown of USP14, cells were transiently transfected with scrambled siRNA (control) or USP14-siRNA (Ambion, Inc.). In either case, cells were incubated with CXCL12 for 10 min to assess the impact of ligand on the ubiquitination of CXCR4. To assess the reversibility of the ubiquitination, another set of cells was incubated for 10 min with CXCL12 followed by a 60-min “recovery from stimulation” period (after removal of ligand and replacement of fresh medium after the 10 min of stimulation). Incubations were terminated by cell lysis in 1 ml of ice-cold RIPA buffer. The cells were processed as described above for assessment of CXCR4 ubiquitination.
Receptor Degradation Assay—To assess the impact of USP14 and reversal of ubiquitination on CXCR4 turnover and degradation, HEK293 cells expressing CXCR4 and transiently expressing HA-USP14 (see figure legends) were pretreated with cycloheximide (5 μg/ml) for 15 min at 37 °C to prevent new protein synthesis during the course of our experiments. Cells were incubated with CXCL12 (10 nm) for 8 h at 37 °C to maximally induce receptor ubiquitination and degradation. To terminate the incubation, cells were transferred to ice and then lysed in ice-cold RIPA buffer. Lysates containing equal amounts of proteins were subjected to 10% SDS-PAGE. CXCR4 was detected by Western blot analysis using an anti-CXCR4 antibody (Abcam). The blots were stripped and reprobed with anti-tubulin antibody to confirm equal loading (Santa Cruz Biotechnology).
To assess the effect of USP14 modulation on endogenous CXCR4 turnover and degradation, HeLa cells, which endogenously express CXCR4, were transiently transfected with HA-USP14 or USP14-specific siRNA. Cells were pretreated with cycloheximide (5 μg/ml) for 15 min at 37 °C, to prevent new protein synthesis during the course of our experiments, and incubated with CXCL12 (10 nm) for varying time points at 37 °C (see figure legends). To terminate the incubation, cells were transferred to ice and then lysed in ice-cold RIPA buffer. Lysates containing equal amounts of proteins were subjected to 10% SDS-PAGE. CXCR4 was detected by Western blot analysis using an anti-CXCR4 antibody (Abcam). The blots were stripped and reprobed with anti-tubulin antibody to confirm equal loading (Santa Cruz Biotechnology).
Densitometric Analysis of Western Blot Bands—The relative amount of all Western blot bands was measured using UNSCAN-IT gel version 6.1 (Silk Scientific Corp.). The relative density of the protein bands was calculated in the area encompassing the immunoreactive protein band following subtraction of the density of the background signal detected in an adjacent area without protein signal in the same lane as the protein of interest.
Statistical Analysis—Student's t tests were performed to test statistical significance for the paired data comparisons. Analysis of variance tests were used to test the significant differences for the group data comparisons.
Chemotaxis Assay—The migration of HEK293 cells stably expressing Myc-CXCR4 and transiently expressing vector alone (control), HA-USP14, USP14-specific siRNA, or scrambled (control) siRNA was evaluated using a chemotaxis assay (36). Briefly, polycarbonate filters (10-μm pore size) coated with 20 μg/μl human collagen type IV were placed between the upper and lower compartments of the Boyden chambers (Neuroprobe, Gaithersburg, MD). The lower compartment of the chamber was loaded with 400-μl aliquots of 1 mg/ml ovalbumin/Dulbecco's modified Eagle's medium (chemotaxis buffer) or CXCL12 (0.01-100 nm) diluted in chemotaxis buffer. Cells (5 × 105/100 μl) were loaded into the upper compartment and incubated for 4 h at 37 °C in a 5% CO2 atmosphere. Cells that had migrated through the filter into the bottom chamber of medium or medium containing CXCL12 were counted under the microscope (×20 objective) after being stained with a Diff-Quik kit. The migration index was calculated and was defined as number of cells crossing the filter toward CXCL12 (various concentrations)/number of cells migrating toward medium alone (control). Each experiment was performed at least three times in duplicate.
To confirm that the modulation of chemotaxis was being mediated via the receptor itself, and not some other USP14-interacting protein, and to confirm the necessity of a ubiquitination/deubiquitination cycle, we assessed the effect of CXCL12 stimulation on mutant CXCR4-mediated, HA-3K/R CXCR4, cell migration. As such, the chemotactic index of HEK293 cells transiently transfected with HA-WT CXCR4 or HA-3K/R CXCR4 was calculated as described above.
CXCL12-induced CXCR4-mediated ERK Activation—To exploit a possible mechanism for the inhibition of CXCL12-induced CXCR4-mediated cell chemotaxis, we explored possible changes in the activation of ERK, a signaling molecule located downstream of CXCR4 activation. HEK293 cells transiently transfected with HA-WT CXCR4 or HA-3K/R CXCR4 were stimulated with CXCL12 (10 nm) for varying time points (see figure legends) at 37 °C. Stimulation was terminated by transferring cells to ice and then lysed with ice-cold RIPA buffer. Lysates containing equal amounts of protein were subjected to 10% SDS-PAGE, and activated ERK was detected using an anti-P-ERK antibody (Santa Cruz Biotechnology). The blots were stripped and reprobed with an antibody (anti-ERK2 antibody; Santa Cruz Biotechnology) that recognizes ERK in both its phosphorylated and nonphosphorylated states to confirm equal loading. An anti-HA antibody (Santa Cruz Biotechnology) was also employed to detect the level of expression of HA-CXCR4.
RESULTS
CXCL12 Activation Leads to a Time-dependent Association of USP14 with CXCR4—A previous series of experiments revealed the possibility that USP14 was a CXCR4-interacting protein (37). This study verified that CXCL12 incubation of HEK293 cells stably expressing the receptor and transiently expressing HA-USP14 leads to a time-dependent association of USP14 with CXCR4 (Fig. 1A). USP14 association with CXCR4 was not readily detected before ligand stimulation, but exposure of CXCR4-expressing cells to CXCL12 led to detection of USP14 association with the receptor at the earliest time point evaluated (2 min). Association continued for ∼5 min, after which time detectable receptor-dependent association declined (Fig. 1B), despite sustained receptor expression in the cells (Fig. 1A). It was not necessary to overexpress USP14 to detect this interaction, because a similar time course for CXCL12-induced association of USP14 with myc-CXCR4 was also detected in HEK cells expressing endogenous USP14 (data not shown). Of further interest was our finding that USP14 interaction with CXCR4 is relatively selective, compared with other members of the USP family, including USP2a, USP4, and USP7, which were evaluated using the same experimental strategy (Fig. 1C).
FIGURE 1.
CXCR4 selectively interacts with USP14 via the C terminus of the receptor. HEK293 cells stably expressing Myc-CXCR4 were exposed to CXCL12 (10 nm) for the indicated time intervals (A-C). Myc-CXCR4 was immunoprecipitated (IP) from cell lysates using a mouse anti-Myc antibody (see “Experimental Procedures”). A, CXCL12 causes a time-dependent association of CXCR14 with USP14. The amount of co-precipitated USP14 protein was detected by Western blotting for the HA epitope on USP14. The membrane was stripped and reprobed using a rabbit anti-Myc antibody to evaluate Myc-CXCR4 loading. The migration of molecular weight markers is shown to the left of the gel. Data shown are representative of one experiment performed six times. IB, immunoblot. B, quantitation of the relative amount of USP14 co-precipitated with CXCR4 was determined by densitometric scanning as outlined under “Experimental Procedures”; n = 6. C, selectivity of USP-isoform interaction with Myc-CXCR4. Experiments were performed as in A and described in detail under “Experimental Procedures.” There was no detectable interaction of the CXCR4 with USP7 (data not shown). Co-precipitated HA-USP2a (▴, n = 4) and His-USP4 (▪, n = 3) were detected using anti-HA and anti-His antibodies (see “Experimental Procedures”). D, USP14 interacts with the C terminus of CXCR4; GSH-Sepharose-bound GST (lane 2, control) or GST-CXCR4 C-terminal fusion protein (lane 3) was incubated with HEK293 cell lysates prepared from control (i.e. not stimulated by CXCL12 ligand), as described under “Experimental Procedures.” Upper panel, HA-USP14 was detected by Western blotting using an anti-HA antibody. Lower panel, GST was detected using a rabbit anti-GST antibody. An aliquot of the cell lysate is shown in lane 1. Data shown in B and C are mean ± S.E. from the number of independent experiments outlined above. *, p < .05; **, p < .01; ***, p < .001, compared with cells not stimulated with CXCL12 (control).
Because the C terminus of CXCR4 has been shown to interact with a variety of intracellular proteins after ligand stimulation (32-35, 37), we explored whether USP14 association with CXCR4 might occur via interactions with the C terminus of the receptor. To test this hypothesis we created a cDNA encoding a fusion of the C-terminal domain of the receptor with GST. As shown in Fig. 1D, the GST-CXCR4 C-terminal domain fusion proteins interacted with USP14 when cell lysates expressing USP14 were incubated with GSH-Sepharose-GST-CXCR C-terminal fusion protein, whereas no interaction of USP14 occurred with GST alone (control). We did not need to stimulate the HEK cells with CXCL12 prior to harvesting the cell lysates to see interaction of the C terminus with USP14 in those lysates, presumably because in these in vitro assays, in contrast to the intact cell assays shown in Fig. 1, A and B, the concentration of the C terminus was high enough to detect this interaction even in the absence of ligand. It is probable that CXCL12 activation of the CXCR4 induces a receptor conformation that preferentially interacts with USP14 via the receptor C terminus. In nonstimulated cells, the C terminus of the membrane-bound receptor may be limiting in concentration, perhaps by steric hindrance in an inactive receptor conformation. However, in in vitro GST-pulldown experiments, the C terminus is freely available and presumably in excess concentrations, such that it is not necessary to have pre-stimulated the cells from which the lysate is derived to see USP14 association with the C terminus of the CXCR4. The finding, however, that the C terminus of CXCR4 can directly interact with USP14 provides additional confirmation of a direct interaction between this enzyme and CXCR4. Consistent with the data shown in Fig. 1C, the CXCR4 C terminus does not interact in a detectable fashion with USP2a, USP4, or USP7 (data not shown).
CXCL12 Stimulation Induces USP14 Apparent Co-localization with CXCR4—To date, the subcellular distribution of USP14 has been limited to proteasomes (38) and synaptic vesicles (39). We were curious whether CXCL12 activation of CXCR4 might lead to redistribution of USP14 to CXCR4-containing membrane compartments, consistent with the ligand- and time-dependent association of USP14 with CXCR4 noted in Fig. 1. CXCL12 treatment of HEK293 cells stably expressing EGFP-CXCR4 and transiently expressing HA-USP14 led to a time-dependent co-localization of these proteins both on the cell surface and in apparently internalized membrane compartments (Fig. 2). This co-localization was more readily detected at 5 min when compared with a 60-min exposure to CXCL12, which is also consistent with the time course for CXCL12-induced association of CXCR4 with USP14 described in Fig. 1, A and B. In studies not shown here, we observed that ligand stimulation leads to detection of CXCR4 in a variety of Rab GTPase-associated compartments, including Rab 7 (late endosomal) and Rab 11a (recycling endosome) compartments.
FIGURE 2.
CXCL12 enhances apparent USP14 co-localization with CXCR4. HEK293 cells stably expressing EGFP-CXCR4 and transiently transfected with HA-USP14 were treated without (control) or with CXCL12 (10 nm) for the indicated time intervals. The cells were fixed and evaluated using immunohistochemistry via confocal microscopy as detailed under “Experimental Procedures.” Representative laser-scanning confocal micrographs demonstrating the distribution of EGFP-CXCR4 (green), USP14 (red), and overlay (yellow) are shown. Images were processed using Photoshop software.
Reversal of CXCL12-induced CXCR4 Ubiquitination Is USP14-dependent—Although CXCR4 ubiquitination has been detected in cells heterologously expressing HA-ubiquitin, we wanted to establish whether detectable ubiquitination of CXCR4 occurs when only endogenous ubiquitin is available, and what impact USP14 has on this covalent modification. Cells stably expressing Myc-CXCR4 were treated with CXCL12 for varying times before immunoisolation of Myc-CXCR4; the extent of ubiquitination of the receptor at these various time points was assessed using an anti-ubiquitin antibody. As shown in Fig. 3A, CXCL12 increased the extent of detectable receptor-associated ubiquitin (upper gel panel) at a migration in the SDS-PAGE that corresponds to ∼45,000, as well as the laddering of CXCR4 (bracketing the 54-kDa molecular mass marker), characteristic of ubiquitinated proteins, including GPCRs (40). When we blotted for the Myc-CXCR4 with an anti-Myc antibody, these laddered proteins were not detectable, suggesting that only a fraction of the expressed receptor was being modified.
FIGURE 3.
USP14 modulates CXCR4 ubiquitination. A, antibody directed against endogenous ubiquitin reveals the time-dependent ubiquitination of Myc-CXCR4 (upper panel) in response to CXCL12 (10 nm) treatment of HEK293 cells (see “Experimental Procedures”). The lower panel, obtained by reprobing the Western blot with an anti-Myc antibody directed against the Myc-CXCR4, readily detects the receptor protein migrating at ∼45,000, but not the higher molecular mass “ladders” at 54 kDa and above. IP, immunoprecipitation; IB, immunoblot. B, quantitation of the relative density of bands representing CXCR4-endogenous Ub complexes was determined by densitometric scanning (•; see “Experimental Procedures”). Superimposed on these data are the findings from the time course of USP14 association with the CXCR4 in CXCL12-exposed cells (○), reported in Fig. 1B, for comparison. C, overexpression of USP14 eliminates detectable CXCR4 ubiquitination in response to CXCL12. HEK293 cells stably expressing Myc-CXCR4 and transiently transfected with vector alone (Vector) or HA-USP14 were incubated for 10 min with CXCL12 as in A. This incubation was terminated immediately for some samples (10 min, no recovery) but allowed to continue after washing away the CXCL12, for 60 min (10 min, +60 min recovery). The gel data shown below the bar graph, from one representative experiment, confirm that transfection of the HEK293 cells with the cDNA encoding HA-USP14 indeed leads to overexpression of this enzyme. D, RNA interference knockdown of endogenous USP14 eliminates deubiquitination of CXCL12-evoked ubiquitination of Myc-CXCR4. HEK293 cells transfected with scrambled (control) siRNA or USP14 siRNAs were treated without CXCL12 (none) or with 10 nm CXCL12 for 10 min (10 min, no recovery) or 10 min followed by a 60-min recovery period (10 min, + 60 min recovery) as described under “Experimental Procedures.” The panel below the bar graph provides a representative gel that confirms the ability of the siRNA construct to successfully reduce the expression of the USP14 protein in these cells under these conditions. Myc-CXCR4 was isolated by immunoprecipitation with an anti-Myc antibody, and the CXCR4-Ub complexes were quantified by Western blot using an antibody against endogenous ubiquitin. Blots were stripped and reprobed using a Myc antibody to evaluate loading of Myc-CXCR4. Data in B-D represent the mean ± S.E. from three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001, compared with controls cells (no CXCL12 treatment).
Ubiquitination of the CXCR4 is time-dependent, as summarized for multiple experiments in Fig. 3B (•). Fig. 3B also superimposes the time course for CXCR4 ubiquitination with the time course for CXCR4 association with USP14 (○), as already reported above in Fig. 1B. This comparison indicates that when the receptor association with USP14 begins to decline, receptor ubiquitination continues to increase, as might be expected in a continuing ubiquitination/deubiquitination cycle on the receptor molecule.
The data in Fig. 3C provide additional evidence for the reversibility of CXCR4 ubiquitination. When stimulation of the receptor with CXCL12 for 10 min was followed by ligand removal, cell washing, and an additional 60-min incubation, we observed decreased ubiquitination of CXCR4, suggesting an ongoing and parallel process of CXCR4 ubiquitination and deubiquitination (Fig. 3C, left panel, control, vector alone). When USP14 was overexpressed in these cells, no accumulated ubiquitinated CXCR4 was detected, even at the end of the first 10-min incubation with CXCL12. These data are consistent with the interpretation that not only does USP14 associate with CXCR4 in a ligand-modulated fashion (as in Fig. 1), but also that USP14 recognizes this receptor as a substrate for deubiquitination.
We used a complementary experimental strategy to test whether endogenous USP14 catalyzes the deubiquitination of CXCR4. For this purpose, we exploited silencing RNA to reduce the expression of endogenous USP14 in our HEK293 cells stably expressing the CXCR4 receptor (Fig. 3D). Cells expressing USP14-specific siRNA manifest a greater basal ubiquitination of CXCR4, as well as a greater ligand-induced ubiquitination of the receptor following a 10-min stimulation with CXCL12. These data suggest that the USP14 gene product indeed serves as a catalyst to deubiquitinate the CXCR4, because when its expression is reduced there is a reciprocal increase in CXCR4 ubiquitination. Reduced expression of endogenous USP14 (following siRNA exposure) eliminated the reduction of ligand-evoked ubiquitination of the receptor during the 60-min under “recovery” phase following CXCL12 removal. Collectively, the data in Fig. 3, C and D, support the conclusion that the USP14 gene product endogenously contributes to a ligand-enhanced CXCR4-Ub cycle.
Receptor Degradation Is Regulated by USP14 Expression—As for other proteins, ubiquitin has been implicated as a “tag” identifying CXCR4 for proteasome- or lysosome-mediated degradation (41). Deubiquitination of CXCR4 by USP14 would thus be expected to reduce the rate of ligand-accelerated receptor degradation and result in an increased steady state level of receptors. To test this hypothesis, HEK293 cells stably expressing EGFP-CXCR4 and overexpressing USP14 were stimulated with CXCL12 (10 nm) for 0 or 8 h; we chose this 8-h time point because previous studies have shown that prolonged incubation of the CXCR4 with CXCL12 permits more facile detection of ligand-accelerated receptor degradation (28). As shown in Fig. 4A, sustained incubation with CXCL12 led to an ∼35% decrease in the steady state level of CXCR4. (Preliminary studies using cycloheximide to block new receptor synthesis resulted in quantitatively indistinguishable findings.) Overexpression of USP14, however, led to two readily detectable consequences in CXCR4-expressing cells. First, the steady state level of CXCR4 was increased by ∼40%. Second, overexpression of USP14 entirely eliminated CXCL12-induced CXCR4 degradation.
FIGURE 4.
USP14 prevents CXCL12-mediated EGFP-CXCR4 degradation and increases the steady state level of the receptor. A, HEK293 cells stably expressing EGFP-CXCR4 were treated with CXCL12 for 8 h (+); this prolonged incubation allowed detection of CXCL12-evoked receptor down-regulation. EGFP-CXCR4 levels were detected by Western blot using an anti-EGFP antibody. B, quantitation of the relative amount of CXCR4 was determined by densitometric scanning as outlined under “Experimental Procedures.” Data are mean ± S.E. from three independent experiments. *, p < 0.05; compared with control cells (no CXCL12 treatment (-)). ns, not statistically significant.
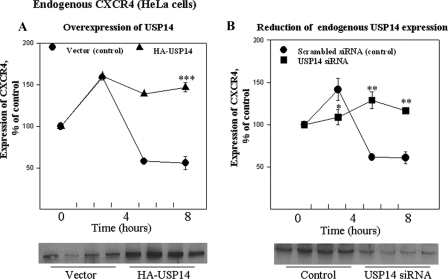
Degradation of Endogenous CXCR4 Receptor Also Is Regulated by USP14 Expression—To expand our findings and assess the effects of USP14 modulation on the turnover and degradation of endogenous CXCR4, we extended our studies to HeLa cells, which endogenously express the CXCR4 receptor. HeLa cells were transiently transfected with HA-USP14 or USP14-specific siRNA and stimulated with CXCL12 (10 nm) at 37 °C for varying time points (see figure legend). As shown in Fig. 5A, overexpression of HA-USP14 (confirmed in the accompanying SDS-PAGE) increased steady state levels of CXCR4 and also blocked CXCL12-evoked down-regulation of CXCR4 levels characteristic of control cells (vector alone). We also explored the impact of reducing USP14 expression in HeLa cells by transfection with USP14-specific siRNA (confirmed in the accompanying SDS-PAGE). Unexpectedly, reduction of USP14 expression also led to slightly increased steady state levels of CXCR4 and elimination of the ability of CXCL12 to down-regulate the receptor, even after 8 h of exposure to CXCL12. As mentioned above, these findings of sustained or slightly increased levels of CXCR4 following suppression of USP 14 expression were unexpected, as we had expected a decline in receptor levels under conditions where the level of USP14, a deubiquitinating enzyme for this receptor, was reduced. One interpretation of these data is an ongoing cycle of ubiquitination/deubiquitination is critical for CXCL12-modulated changes in receptor density.
FIGURE 5.
Degradation of endogenous CXCR4 in HeLa cells is disrupted by modulation of USP14 expression. A, HeLa cells transiently transfected with vector or HA-USP14 were treated with CXCL12 for 0, 3, 5, or 8 h as indicated. CXCR4 levels were detected by Western blot using an anti-CXCR4 antibody. Quantitation of relative amount of CXCR4 was determined by densitometric scanning as outlined under “Experimental Procedures.” Data obtained at time 0 and 8 h Data are mean ± S.E. from three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with control cells (no CXCL12 treatment, i.e. time 0); the data obtained at 3 and 5 h after CXCL12 incubation are from only two independent experiments, and thus statistical analyses were not performed for these time points. B, HeLa cells transiently transfected with scrambled siRNA (control) or USP14-specific siRNA were treated with CXCL12 for 0, 3, 5, or 8 h, as indicated. CXCR4 levels were detected by Western blot using an anti-CXCR4 antibody. Quantitation of relative amount of CXCR4 was determined by densitometric scanning as outlined under “Experimental Procedures.” Data are mean ± S.E. from three independent experiments. *, p < 0.05; **, p < 0.01.
CXCR4 Ubiquitination/Deubiquitination Cycle Appears to Be Critical for CXCL12-induced Cell Chemotaxis—We were curious whether ubiquitination of CXCR4 would have functional consequences beyond altering CXCL12-induced receptor down-regulation. Consequently, we examined CXCL12-mediated chemotaxis in HEK293 cells stably expressing Myc-CXCR4. As shown in Fig. 6A, CXCL12 led to a concentration-dependent movement of cells across the filter of the Boyden chamber, with a maximal chemotactic index being achieved between 1 and 10 nm CXCL12, and declining at higher concentrations, presumably because of a desensitization response (36). Overexpression of USP14 dramatically reduced CXCR4-mediated chemotaxis (Fig. 6A). Interestingly, siRNA knockdown of endogenous USP14 also diminished CXCL12-induced chemotaxis (Fig. 6B). These findings are extremely interesting, because they are consistent with the hypothesis that it is not the ubiquitinated versus nonubiquitinated receptor that is critical for receptor-mediated signaling, but rather the ability of the receptor to undergo an accelerated Ub cycle as part of the chemotactic process. This requirement for a CXCR4-Ub cycle for chemotaxis is reminiscent of the requirement for a GTPase cycle for cellular migration/chemotaxis.
FIGURE 6.
CXCR4-Ub cycle is essential for CXCR4-mediated chemotaxis. A, chemotaxis was evaluated in HEK293 cells stably expressing Myc-CXCR4 (and endogenous levels of USP14, i.e. “vector alone”) or in cells overexpressing HA-USP14 as described in detail under “Experimental Procedures.” Overexpression of USP14, which dramatically reduces CXCL12-evoked CXCR4 ubiquitination (cf. Fig. 3C), also dramatically attenuates CXCL12-evoked chemotaxis. B, knockdown of endogenous USP14 expression with USP14-directed siRNA, which leads to enhanced CXCL12-induced CXCR4 ubiquitination (cf. Fig. 3D), also significantly reduces CXCL12-induced chemotaxis. C, chemotaxis in response to CXCL12 was evaluated in cells expressing an HA-WT CXCR4 and in cells expressing a 3K/R mutant receptor, as described under “Experimental Procedures.” Taken together, A and B suggest that the CXCR4-Ub cycle, and not a particular ubiquitinated state of CXCR4, is essential for CXCL12-mediated chemotaxis, and the data in C confirm that it is the CXCR4 molecule itself that must undergo a ubiquitination/deubiquitination cycle for chemotaxis to occur. Values represent the mean ± S.E. from three independent experiments performed in duplicate. All chemotaxis data are expressed as the chemotactic index, which is calculated as the ratio of the number of cells that migrate across the Boyden chamber in the presence of CXCL12 at a given concentration compared with the number of cells migrating in the absence of CXCL12; the value of 1 means that there was no migration greater than that observed in control, nonstimulated cells. Data were analyzed using Student's unpaired t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001, and compared with control cells, e.g. cells expressing endogenous USP14 (A), scrambled siRNA (B), or HA-WT CXCR4 (C).
Mutation of the CXCR4 to Eliminate Receptor Ubiquitination Eliminates CXCL12-induced Cell Chemotaxis—One of the limitations of directing all of our experimental efforts to manipulating the levels of expression of USP14 is that the ubiquitination of a plethora of proteins could be altered in parallel with the CXCR4 as a consequence of these experimental alterations in USP14 levels. To assess whether CXCL12-induced chemotaxis requires a ubiquitination cycle that involves the receptor protein itself, we compared the ability of CXCL12 to evoke chemotaxis of HEK293 cells expressing either a WT receptor or the 3K/R mutant CXCR4 that cannot be ubiquitinated, as described previously (28). As shown in Fig. 6C, elimination of the ability of the CXCR4 to undergo ubiquitination also eliminates the ability of CXCL12 to evoke chemotaxis of the cells expressing this 3K/R mutant receptor. Thus, taken together, these findings provide the first evidence that CXCR4 mediates chemotaxis in a fashion that requires a ubiquitination/deubiquitination cycle on the receptor protein itself.
Mutation of the CXCR4 to Eliminate Receptor Ubiquitination Does Not Alter the Time Course or Extent of CXCL12-induced ERK Activation—A commonly monitored consequence of CXCR4 activation by CXCL12 is the increase in tyrosine phosphorylation on ERK. As shown in Fig. 7, the time course of this stimulation by CXC12 is indistinguishable in HEK293 cells expressing WT versus 3K/R CXCR4. As an increase in threonine phosphorylation on ERK also can be initiated by CXCL12-induced CXCR4 activation, it is possible that changes in Thr(P) phosphorylation of ERK occurred in response to CXCL12 activation of the 3K/R CXCR4, which we would not have detected using only an anti-Tyr antibody. Thus, we assessed ERK phosphorylation in response to CXCL12 activation of WT and 3K/R CXCR4 using a p-ERK p42/p44 Thr/Tyr antibody (Cell Signaling Technologies). Interestingly, even with this antibody, we observed no difference in the time course or the extent of activation of ERK upon CXCL12 exposure to cells expressing WT versus 3K/R CXCR4. With this p-ERK p42/p44 Thr/Tyr antibody, we detected ∼3-fold increase in ERK activation at 5 min following CXCL12 (the time of peak stimulation detected by this antibody, as was detected with the anti-Tyr(P) antibody) in WT and 3K/R CXCR4-expressing cells. After continued stimulation, ERK activation diminished, returning to basal levels at 60 min of stimulation (data not shown). The finding that equivalent ERK activation occurs in HEK cells expressing the WT versus 3K/R CXCR4 provides additional functional evidence that a comparable level of CXCR4 expression is likely occurring in both circumstances, and thus the loss of chemotaxis observed for the 3K/R CXCR4 in Fig. 6C cannot be due to diminished expression of the mutant receptor. Finally, these data provide evidence that although ERK is indeed a downstream target of CXCL12 activation of CXCR4, this pathway is not necessarily essential for CXCR4-evoked chemotaxis.
FIGURE 7.
ERK activation by CXCR4 occurs independently of the ability of the CXCR4 to be ubiquitinated. A, ERK activation was evaluated in HEK293 cells transiently transfected with HA-WT CXCR4 or HA-3K/R CXCR4 and stimulated with CXCL12 (10 nm) for 0, 5, 15, 30, or 60 min, and ERK activity in cell lysates assessed was as described under “Experimental Procedures.” B, quantitation of ERK activation was based on the amount of ERK detected using an anti-P-ERK antibody; total ERK, assessed using the ERK2 antibody, was indistinguishable in all conditions, and thus the data were not normalized to total ERK. Data are mean ± S.E. from three independent experiments.
DISCUSSION
Although ubiquitination mechanisms and the resulting degradative fates of proteins, including cell surface receptors, have been exhaustively studied, the mechanisms and functional consequences of protein deubiquitination are less understood. Our present studies reveal that CXCL12 activation of CXCR4, a member of the G protein-coupled receptor superfamily, results in reversible ubiquitination of the receptor (Fig. 3). Receptor deubiquitination is paralleled by CXCL12-dependent association of USP14 with CXCR4, an interaction that occurs, at least in part, via the C terminus of the receptor. USP14 interaction with CXCR4 is not mimicked by other deubiquitinating enzymes of the USP family evaluated, including USP2a, USP4, and USP7 (Fig. 1, A-C). CXCL12 activation of cells also leads to the redistribution of USP14 to cellular compartments shared by CXCR4 (Fig. 2). Similar redistribution of GPCR-interacting proteins has been observed previously, including the redistribution of arrestin (44) and of spinophilin (44, 45).
Our findings also are consistent with the interpretation that CXCR4 undergoes a ligand-modulated ubiquitination-deubiquitination cycle that regulates the steady state level of CXCR4. Based on pre-existing data in the literature, it would have been expected that CXCL12 incubation of receptor-expressing cells would lead to receptor ubiquitination, receptor internalization, and lysosomal degradation, which over time would lead to CXCL12-evoked down-regulation of CXCR4 density. Consequently, we expected that overexpression of USP14, a deubiquitinating enzyme, would lead to an increase in the steady state level of the receptor and blockade of CXCL12-induced down-regulation of receptor density. Although this is what we observed, we also observed that suppression of USP14 expression using siRNA, which should favor accelerated receptor degradation, does not. Instead, we observed a loss of the ability of CXCL12 to evoke down-regulation of CXCR4 following either overexpression or suppression of USP14 expression. These findings indicate that when the ubiquitination cycle is perturbed, the ability of CXCL12 stimulation of the CXCR4 to lead to receptor down-regulation also is perturbed, and suggest that ubiquitination of the receptor is perhaps involved in more than simply marking the receptor for degradation.
A second initially unexpected finding was that eliminating the ability of the CXCR4 to be ubiquitinated, i.e. by expressing the 3K/R mutant of the CXCR4, as well as by overexpressing or blocking USP14 expression (which modifies the ability of this enzyme to modify all of its targets in the cell), leads to inhibition of receptor-activated chemotaxis. This may mean, in addition to the known role of ubiquitination in targeting receptors for lysosomal degradation, that the covalent modification of the receptor by ubiquitin is also important for receptor signaling. A number of possible molecular events might explain this role for a ubiquitination cycle on the CXCR4. For example, the ubiquitinated state of the receptor could dictate a particular conformation of the receptor, unique protein-protein interactions of the CXCR4, or particular receptor localization on the surface that favors CXCR4 involvement in chemotaxis. Because chemotaxis requires a sustained “sensing” of a concentration gradient, often involving cycling of receptors between activated and un-activated states, it is possible that the ubiquitination-deubiquitination cycle on the CXCR4 also is causally involved in this gradient sensing role. Our findings that a ubiquitination/deubiquitination cycle involving the CXCR4 receptor appears to be essential for receptor-mediated chemotaxis elevate the role of receptor ubiquitination beyond a mechanism simply to target the receptor for a particular degradative fate.
The selective role of USP14 for interaction with CXCR4 is also of interest (Fig. 1). Other members of the deubiquitinating enzyme family have been shown to interact with a variety of receptor families, including the association of USP2a with the androgen receptor (42) and USP4 with the adenosine receptor (43). However, we are the first to show a preferential interaction of USP14 with CXCR4. Our finding that the C terminus of CXCR4, on its own, can interact with USP14 may be related to the existence of a purported “degradation motif” in this region of CXCR4 necessary for ubiquitin-mediated receptor degradation (28). It is also important to note that the C terminus of the CXCR4 receptor, like the holoreceptor, can distinguish among several USP family members studied, and selectively recognize and bind USP14 (data not shown).
There is one aspect of our findings that differs from previous reports regarding the ubiquitination of CXCR4. Our studies show a laddering of the CXCR4 receptor in response to CXCL12 that can be detected with an antibody to endogenous ubiquitin. However, our data also suggest that this laddering involves only a small fraction of the receptor population, because an antibody against the Myc-CXCR4 used to assess protein loading on gels also did not reliably detect higher molecular weight “ladders” of the CXCR4. Previous studies did not report a “laddering” of the CXCR4 receptor in response to CXCL12 stimulation, but those studies were examining the incorporation of HA-tagged ubiquitin following transient transfection of a cDNA encoding this epitope-tagged ubiquitin molecule (28). We suspect that the difference in appearance/detection of the ubiquitinated CXCR4 receptor following CXCL12 stimulation is likely because of our differences in experimental strategies.
Another interesting finding of our study was the observation that the time course of ERK activation by CXCL12-induced activation of CXCR4 was indistinguishable for the WT and 3K/R receptor structures, providing evidence that a signaling pathway independent of ERK activation links CXCL12 activation of the receptor to chemotaxis.
In summary, our findings demonstrate that CXCL12 activation of the CXCR4 leads to a dynamic ubiquitination/deubiquitination cycle and that USP14 preferentially interacts with and deubiquitinates CXCR4. The functional consequences of this CXCR4-Ub cycle not only modulate Ub-targeted receptor degradation but also CXCL12-evoked chemotaxis. Thus, our findings suggest the exciting possibility that regulation of the CXCR4-Ub cycle will have a plethora of signaling as well as turnover consequences that may be a generalized property of a variety of other GPCRs.
Acknowledgments
We thank Dr. Gang Pei (Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences) for the generous gift of Myc-CXCR4 and GST-conjugated CXCR4 plasmids. We thank Dr. Jeffrey L. Benovic (Thomas Jefferson University, Philadelphia) for providing us with the 3K/R mutant CXCR4 plasmid; Dr. Massimo Loda (Dana Farber Cancer Institute, Harvard Medical School, Boston) for providing us with the USP2a plasmid; Dr. Michael Freissmuth (Medical University of Vienna, Vienna, Austria) for providing us with the USP4 plasmid; and Dr. Roger Everett (Institute of Virology, University of Glasgow, Scotland, UK) for the gift of pCI-USP7 plasmid.
This work was supported, in whole or in part, by National Institutes of Health grants and Grants U54NS41071 (NINDS) and RR0303-19. This work was also supported by a career investigator award from the Veterans Affairs Hospital (to G.-H. F.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: GPCR, G protein-coupled receptor; HEK293 cells, human embryonic kidney 293 cells; RIPA, radioimmunoprecipitation assay; siRNA, short interference RNA; USP, ubiquitin-specific protease; EGFP, enhanced green fluorescence protein; Ub, ubiquitin; ERK, extracellular signal-regulated kinase; GST, glutathione S-transferase; PBS, phosphate-buffered saline; WT, wild type; HA, hemagglutinin.
References
- 1.Haribabu, B., Richardson, R. M., Fisher, I., Sozzani, S., Peiper, S. C., Horuk, R., Ali, H., and Snyderman, R. (1997) J. Biol. Chem. 272 28726-28731 [DOI] [PubMed] [Google Scholar]
- 2.Balabanian, K., Lagane, B., Infantino, S., Chow, K. Y., Harriague, J., Moepps, B., Arenzana-Seisdedos, F., Thelen, M., and Bachelerie, F. (2005) J. Biol. Chem. 280 35760-35766 [DOI] [PubMed] [Google Scholar]
- 3.Nagasawa, T., Hirota, S., Tachibana, K., Takakura, N., Nishikawa, S., Kitamura, Y., Yoshida, N., Kikutani, H., and Kishimoto, T. (1996) Nature 382 635-638 [DOI] [PubMed] [Google Scholar]
- 4.Aiuti, A., Webb, I. J., Bleul, C., Springer, T., and Gutierrez-Ramos, J. C. (1997) J. Exp. Med. 185 111-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Apuzzo, M., Rolink, A., Loetscher, M., Hoxie, J. A., Clark-Lewis, I., Melchers, F., Baggiolini, M., and Moser, B. (1997) Eur. J. Immunol. 27 1788-1793 [DOI] [PubMed] [Google Scholar]
- 6.Ma, Q., Jones, D., Borghesani, P. R., Segal, R. A., Nagasawa, T., Kishimoto, T., Bronson, R. T., and Springer, T. A. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 9448-9453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou, Y. R., Kottmann, A. H., Kuroda, M., Taniuchi, I., and Littman, D. R. (1998) Nature 393 595-599 [DOI] [PubMed] [Google Scholar]
- 8.Feng, Y., Broder, C. C., Kennedy, P. E., and Berger, E. A. (1996) Science 272 872-877 [DOI] [PubMed] [Google Scholar]
- 9.Walter, D. H., Haendeler, J., Reinhold, J., Rochwalsky, U., Seeger, F., Honold, J., Hoffmann, J., Urbich, C., Lehmann, R., Arenzana-Seisdesdos, F., Aicher, A., Heeschen, C., Fichtlscherer, S., Zeiher, A. M., and Dimmeler, S. (2005) Circ. Res. 97 1142-1151 [DOI] [PubMed] [Google Scholar]
- 10.Abu El-Asrar, A. M., Struyf, S., Al-Mosallam, A. A., Missotten, L., Van Damme, J., and Geboes, K. (2001) Br. J. Ophthalmol. 85 1357-1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rauer, M., Pagenstecher, A., Schulte-Mönting, J., and Sauder, C. (2002) J. Neurovirol. 8 168-179 [DOI] [PubMed] [Google Scholar]
- 12.Mines, M., Ding, Y., and Fan, G. H. (2007) Curr. Med. Chem. 14 2456-2470 [DOI] [PubMed] [Google Scholar]
- 13.Xia, M. Q., and Hyman, B. T. (1999) J. Neurovirol. 5 32-41 [DOI] [PubMed] [Google Scholar]
- 14.Burger, M., Glodek, A., Hartmann, T., Schmitt-Graff, A., Silberstein, L. E., Fujii, N., Kipps, T. J., and Burger, J. A. (2003) Oncogene 22 8093-8101 [DOI] [PubMed] [Google Scholar]
- 15.Burger, J. A., and Kipps, T. J. (2006) Blood 107 1761-1767 [DOI] [PubMed] [Google Scholar]
- 16.Su, L., Zhang, J., Xu, H., Wang, Y., Chu, Y., Liu, R., and Xiong, S. (2005) Clin. Cancer Res. 11 8273-8280 [DOI] [PubMed] [Google Scholar]
- 17.Saur, D., Seidler, B., Schneider, G., Algül, H., Beck, R., Senekowitsch-Schmidtke, R., Schwaiger, M., and Schmid, R. M. (2005) Gastroenterology 129 1237-1250 [DOI] [PubMed] [Google Scholar]
- 18.Kang, H., Watkins, G., Douglas-Jones, A., Mansel, R. E., and Jiang, W. G. (2005) Breast 14 360-367 [DOI] [PubMed] [Google Scholar]
- 19.Cabioglu, N., Sahin, A., Doucet, M., Yavuz, E., Igci, A., O Yildirim, E., Aktas, E., Bilgic, S., Kiran, B., Deniz, G., and Price, J. E. (2005) Clin. Exp. Metast. 22 39-46 [DOI] [PubMed] [Google Scholar]
- 20.Rempel, S. A., Dudas, S., Ge, S., and Gutiérrez, J. A. (2000) Clin. Cancer Res. 6 102-111 [PubMed] [Google Scholar]
- 21.Oh, J. W., Drabik, K., Kutsch, O., Choi, C., Tousson, A., and Benveniste, E. N. (2001) J. Immunol. 166 2695-2704 [DOI] [PubMed] [Google Scholar]
- 22.Zhou, Y., Larsen, P. H., Hao, C., and Yong, V. W. (2002) J. Biol. Chem. 277 49481-49487 [DOI] [PubMed] [Google Scholar]
- 23.Woerner, B. M., Warrington, N. M., Kung, A. L., Perry, A., and Rubin, J. B. (2005) Cancer Res. 65 11392-11399 [DOI] [PubMed] [Google Scholar]
- 24.Hong, X., Jiang, F., Kalkanis, S. N., Zhang, Z. G., Zhang, X. P., DeCarvalho, A. C., Katakowski, M., Bobbitt, K., Mikkelsen, T., and Chopp, M. (2006) Cancer Lett. 236 39-45 [DOI] [PubMed] [Google Scholar]
- 25.Wysoczynski, M., Reca, R., Ratajczak, J., Kucia, M., Shirvaikar, N., Honczarenko, M., Mills, M., Wanzeck, J., Janowska-Wieczorek, A., and Ratajczak, M. Z. (2005) Blood 105 40-48 [DOI] [PubMed] [Google Scholar]
- 26.Daaka, Y., Luttrell, L. M., and Lefkowitz, R. J. (1997) Nature 390 88-91 [DOI] [PubMed] [Google Scholar]
- 27.Peng, S. B., Peek, V., Zhai, Y., Paul, D. C., Lou, Q., Xia, X., Eessalu, T., Kohn, W., and Tang, S. (2005) Mol. Cancer Res. 3 227-236 [DOI] [PubMed] [Google Scholar]
- 28.Marchese, A., and Benovic, J. L. (2001) J. Biol. Chem. 276 45509-45512 [DOI] [PubMed] [Google Scholar]
- 29.Pickart, C. M. (2001) Annu. Rev. Biochem. 70 503-533 [DOI] [PubMed] [Google Scholar]
- 30.Marchese, A., Raiborg, C., Santini, F., Keen, J. H., StenMark, H., and Benovic, J. L. (2003) Dev. Cell 5 709-722 [DOI] [PubMed] [Google Scholar]
- 31.Bhandari, D., Trejo, J., Benovic, J. L., and Marchese, A. (2007) J. Biol. Chem. 282 36971-36979 [DOI] [PubMed] [Google Scholar]
- 32.Ding, Y., Zhang, L., Goodwin, J. S., Wang, Z., Liu, B., Zhang, J., and Fan, G. H. (2008) Exp. Cell Res. 314 590-602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan, H., Luo, C., Li, R., Qiao, A., Zhang, L., Mines, M., Nyanda, A. M., Zhang, J., and Fan, G. H. (2008) J. Biol. Chem. 283 623-637 [DOI] [PubMed] [Google Scholar]
- 34.Wang, Z., Zhang, L., Qiao, A., Watson, K., Zhang, J., and Fan, G. H. (2008) J. Biol. Chem. 283 3951-3959 [DOI] [PubMed] [Google Scholar]
- 35.Li, R., Luo, C., Mines, M., Zhang, J., and Fan, G. H. (2006) J. Biol. Chem. 281 37616-37627 [DOI] [PubMed] [Google Scholar]
- 36.Luo, C., Pan, H., Mines, M., Watson, K., Zhang, J., and Fan, G. H. (2006) J. Biol. Chem. 281 30081-30093 [DOI] [PubMed] [Google Scholar]
- 37.Ding, Y., Li, M., Zhang, J., Li, N., Xia, Z., Hu, Y., Wang, S., and Fan, G. H. (2006) Mol. Pharmacol. 69 1269-1279 [DOI] [PubMed] [Google Scholar]
- 38.Anderson, C., Crimmins, S., Wilson, J. A., Korbel, G. A., Ploegh, H. L., and Wilson, S. M. (2005) J. Neurochem. 95 724-731 [DOI] [PubMed] [Google Scholar]
- 39.Wilson, S. M., Bhattacharyya, B., Rachel, R. A., Coppola, V., Tessarollo, L., Householder, D. B., Fletcher, C. F., Miller, R. J., Copeland, N. G., and Jenkins, N. A. (2000) EMBO J. 20 5187-5196 [Google Scholar]
- 40.Cook, L. B., Zhu, C. C., and Hinkle, P. M. (2003) Mol. Endocrinol. 17 1777-1791 [DOI] [PubMed] [Google Scholar]
- 41.Varshavsky, A. (1997) Trends Biochem. Sci. 22 383-387 [DOI] [PubMed] [Google Scholar]
- 42.Priolo, C., Tang, D., Brahamandan, M., Benassi, B., Sicinska, E., Ogino, S., Farsetti, A., Porrello, A., Finn, S., Zimmermann, J., Febbo, P., and Loda, M. (2006) Cancer Res. 66 8625-8632 [DOI] [PubMed] [Google Scholar]
- 43.Milojevic, T., Reiterer, V., Stefan, E., Korkhov, V. M., Dorostkar, M. M., Ducza, E., Ogris, E., Boehm, S., Freissmuth, M., and Nanoff, C. (2006) Mol. Pharmacol. 69 1083-1094 [DOI] [PubMed] [Google Scholar]
- 44.Brady, A. E., Wang, Q., Allen, P. B., Rizzo, M., Greengard, P., and Limbird, L. E. (2005) Mol. Pharmacol. 67 1690-1696 [DOI] [PubMed] [Google Scholar]
- 45.Richman, J. G., Brady, A. E., Wang, Q., Hensel, J. L., Colbran, J., and Limbird, L. E. (2001) J. Biol. Chem. 276 15003-15008 [DOI] [PubMed] [Google Scholar]