Abstract
α-Accessory factor (AAF) stimulates the activity of DNA polymerase-α·primase, the only enzyme known to initiate DNA replication in eukaryotic cells (Goulian, M., Heard, C. J., and Grimm, S. L. (1990) J. Biol. Chem. 265 ,13221 -13230). We purified the AAF heterodimer composed of 44- and 132-kDa subunits from cultured cells and identified full-length cDNA clones using amino acid sequences from internal peptides. AAF-132 demonstrated no homologies to known proteins; AAF-44, however, is evolutionarily related to the 32-kDa subunit of replication protein A (RPA-32) and contains an oligonucleotide/oligosaccharide-binding (OB) fold domain similar to the OB fold domains of RPA involved in single-stranded DNA binding. Epitope-tagged versions of AAF-44 and -132 formed a complex in intact cells, and purified recombinant AAF-44 bound to single-stranded DNA and stimulated DNA primase activity only in the presence of AAF-132. Mutations in conserved residues within the OB fold of AAF-44 reduced DNA binding activity of the AAF-44·AAF-132 complex. Immunofluorescence staining of AAF-44 and AAF-132 in S phase-enriched HeLa cells demonstrated punctate nuclear staining, and AAF co-localized with proliferating cell nuclear antigen, a marker for replication foci containing DNA polymerase-α·primase and RPA. Small interfering RNA-mediated depletion of AAF-44 in tumor cell lines inhibited [methyl-3H]thymidine uptake into DNA but did not affect cell viability. We conclude that AAF shares structural and functional similarities with RPA-32 and regulates DNA replication, consistent with its ability to increase polymerase-α·primase template affinity and stimulate both DNA primase and polymerase-α activities in vitro.
In eukaryotic cells, DNA replication is initiated at multiple origins internal to each chromosome; the origin recognition complex recruits cell division cycle and minichromosome maintenance proteins to form a preinitiation complex (1). At the G1-S phase transition, the latter complex is activated by cyclin-dependent protein kinases leading to formation of an initiation complex that alters local DNA structure through DNA helicase activity (1, 2). The replication protein A (RPA)2 is recruited to bind and stabilize single-stranded DNA (ssDNA) produced by the initiation complex (3, 4). RPA serves as an auxiliary factor for DNA polymerase-α (pol-α)·primase: it stabilizes the protein complex by direct interaction with both pol-α and primase subunits, and it reduces the misincorporation rate of pol-α, acting as a “fidelity clamp” (5, 6). The pol-α·primase complex consists of four subunits, including the catalytic pol-α subunit (p185), a regulatory B subunit (p70), and two primase subunits (p49 and p58). On an ssDNA template, the primase synthesizes short RNA primers from ribonucleoside triphosphates (rNTPs), which are elongated by pol-α in the presence of deoxyribonucleoside triphosphates (dNTPs) to form short DNA fragments. Through mechanisms requiring other replication factors, pol-α·primase is replaced by the more processive DNA polymerases pol-δ and pol-ε (7). Pol-ε synthesizes the leading strand, whereas pol-δ completes each Okazaki fragment initiated by pol-α·primase on the lagging strand and proofreads errors made by pol-α (7). The initiator RNA and DNA fragments are later removed by nucleases, and the Okazaki fragments are sealed by DNA ligase (7).
The pol-α·primase complex is the only eukaryotic DNA polymerase able to initiate DNA synthesis de novo. In addition to initiating DNA replication and synthesizing Okazaki fragments, it appears to be one of the final targets of cell cycle checkpoint pathways that couple DNA replication to DNA damage response (2, 8). The role of RPA in initiation, elongation, and completion of lagging strand DNA synthesis has been thoroughly investigated (3, 9), but in vitro studies suggest that some additional factors that promote the rapidity of DNA replication in vivo are still lacking (2).
In the course of purifying pol-α·primase from extracts of cultured mouse L1210 cells, we identified a factor we named α-accessory factor (AAF) that stimulates pol-α·primase activity in vitro (10, 11). The protein has a native molecular mass of ∼150 kDa as determined from its sedimentation coefficient and Stokes radius and is composed of two subunits of ∼132 and ∼44 kDa. AAF stimulates pol-α·primase activity with several different templates and types of reactions: (i) It stimulates selfprimed reactions with poly(dT), poly(dI·dT), or single-stranded circular DNA; (ii) it stimulates primed reactions with poly(dA)·oligo(dT) and multiply primed DNA in the absence of rNTPs, indicating that it affects pol-α activity when no primers are being made; and (iii) it stimulates primase activity on ssDNA in the absence of dNTPs, showing that it can enhance RNA primer synthesis in the absence of DNA synthesis (11). AAF increases the template affinity and processivity of pol-α·primase (12). AAF is highly specific for pol-α·primase and has no effect on the other mammalian DNA polymerases β, γ, or δ or on the DNA polymerase·primase complexes from Drosophila and Saccharomyces cerevisiae (11).
The cloning of both AAF subunits based on peptide sequences obtained from the purified protein allowed us now to further characterize the AAF-44·AAF-132 complex structurally and functionally. Based on siRNA experiments in cancer cell lines, AAF appears to regulate DNA replication in vivo.
EXPERIMENTAL PROCEDURES
Purification of AAF and Sequencing of Tryptic Peptides—AAF was purified from 80 g of cultured mouse L1210 lymphoblastic cells as described previously, and its activity was measured following stimulation of pol-α·primase on unprimed poly(dT) using 0.1 unit of purified pol-α·primase (11). Fractions from the S-300 column (step V) containing AAF activity were pooled, concentrated, and analyzed by SDS-PAGE, and the 132- and 44-kDa subunit bands were excised and sent to the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University. Individual tryptic peptides were separated by reverse phase high pressure liquid chromatography and sequenced by mass spectrometry using an electrospray/quadrupole/time-of-flight spectrometer. The N terminus of the 44-kDa band was sequenced by the University of California San Diego Biology Department Protein Sequencing Facility, but the N terminus of AAF-132 was found to be blocked.
cDNA Cloning of AAF-132 and AAF-44—Peptide sequences were used to search available data bases with the BLAST program (13), and initially two expressed sequence tag (EST) clones were found, GenBank™ accession numbersAA655197 and AA039012, containing sequences corresponding to several peptides from the 132- and 44-kDa subunits, respectively. Both clones were obtained from the ATCC and were sequenced. A probe for AAF-132 was produced with the primers 5′-CCAAACAGAGTCTGCAAGCAAAAC-3′ and 5′-CCTTAGGCAGTGTCAAGACG-3′; it was labeled with [α-32PO4]dCTP by random hexamer priming (14) and used to screen an oligo(dT)-primed mouse testis cDNA library in λGT10 (Stratagene). Two overlapping AAF-132 clones were isolated that encoded an uninterrupted open reading frame of 1199 amino acids including the 3′-end but were missing a proper start codon. To clone the 5′-end of both AAF-132 and AAF-44, we used rapid amplification of cDNA 5′-ends (15) with L1210 RNA serving as a template. The gene-specific primers for AAF-132 were 5′-CTTTGGGTCAAAAGTAGCCCAAGCC-3′ and 5′-TCGGAGGTAGGTGGTTCGCCC-3′. The primers for AAF-44 were 5′-TCGGATGATATCTCCAATCCC-3′ and 5′-TCTGCTCGATGGTCTCCTGC-3′. For each subunit, two independent PCR products were sequenced.
Northern Blot Analyses—Mouse tissue Northern blots (Clontech) were hybridized with cDNA probes encoding murine AAF-44 (nucleotides 164-544), AAF-132 (nucleotides 3401-3692), or human β-actin (Clontech) as described previously (14). Probes were generated by PCR and labeled by random hexamer priming using either [α-32PO4]dCTP (AAF-132 and β-actin) or both [α-32PO4]dCTP and [α-32PO4]dATP (AAF-44).
In Vitro Transcription/Translation of Epitope-tagged AAF-44 and AAF-132—5′ fusion sites were created using PCR and the following sets of primers: 5′-GGATCCATGGCTGCTTGCCGAGCACAAC-3′ and 5′-CTTTGGGTCAAAAGTAGCCCAAGCC-3′for AAF-132 and 5′-CTCGAGGAGATGCCACAGCCCTGCCTCTTG-3′ and 5′-TCTGCTCGATGGTCTCCTGC-3′ for AAF-44. The PCR products were sequenced and spliced into the full-length coding sequences in pcDNA3-Myc, pcDNA3-HA, or pXJ40-FLAG, creating an in-frame fusion with an N-terminal Myc, hemagglutinin (HA), or FLAG epitope tag (16, 17). AAF-44 and/or AAF-132 in pcDNA3-Myc was used as a template for a coupled transcription/translation reaction using T7 RNA polymerase, [35S]methionine, and the TnT™ reticulocyte lysate system according to the manufacturer's instructions (Promega).
Site-directed Mutagenesis—Site-directed mutagenesis of AAF-44 was performed using the QuikChange™ mutagenesis II XL kit from Stratagene with the following primers (sense strand with mutant nucleotides is underlined): 5′-GTTATAAACTGCGTCTGCGCAAAGAAGCTGAGCAAC-3′ (W96A) and 5′-GTCCGAGGCTCTGTGCGTATGGCCCGAGAAGAGCGAGA-G-3′ (F151A). The mutagenesis product was sequenced.
Cell Culture—Cells were grown in the following media: human embryonal kidney 293T cells in Iscove's modified Dulbecco's medium, MDA-MB-231 and MCF7 breast cancer cells in Dulbecco's modified Eagle's medium/F-12 medium, PC3 prostate cancer cells in RPMI 1640 medium, and HeLa cervical cancer cells in Dulbecco's modified Eagle's medium. All media were supplemented with 10% fetal bovine serum (FBS) unless otherwise specified.
DNA Transfections and Co-immunoprecipitation Experiments—239T cells were transfected in full growth medium with 2 μg of DNA and 5 μl of Lipofectamine 2000™/6-well cluster dish. Cells were lysed 24 h later in 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Triton X-100, and protease inhibitor mixture (Calbiochem) for 10 min at 4 °C. The lysates were cleared by centrifugation at 14,000 × g for 10 min, and a monoclonal mouse anti-HA epitope antibody (Santa Cruz Biotechnology, SC7392) or a polyclonal rabbit anti-Myc antibody (SC789) were added (16). Immunoprecipitates were collected on protein G-agarose, washed six times in lysis buffer, and analyzed by SDS-PAGE/Western blotting (16).
Immunoaffinity Purification of AAF-44—239T cells were transfected with an expression vector encoding FLAG epitope-tagged AAF-44 and either Myc-tagged AAF-132 or empty vector as described above. Cells were lysed in buffer containing 50 mm Tris-HCl, pH 7.5, 50 mm NaCl, 0.3% Nonidet P-40, and protease inhibitor mixture; cleared lysates were incubated with anti-FLAG M2-agarose beads (Sigma) for 1 h. The beads were washed six times in lysis buffer, and AAF-44 was eluted in buffer containing 0.03% Nonidet P-40 and 0.1 mg/ml FLAG peptide (Sigma).
DNA Binding Assay—For DNA binding assays, cells were transfected as described above, and 20-200 μg of cell lysate protein were incubated for 20 min at 4 °C in binding buffer (10 mm K-HEPES, pH 7.9, 25 mm KCl, 0.25 mm dithiothreitol, 0.1 mm EDTA, and 5% glycerol) with 0.1 mg/ml bovine serum albumin and the indicated amount of biotinylated (dC)30 oligodeoxyribonucleotide (oligo(dC); Invitrogen) in a total volume of 70 μl. In some experiments, cell lysates were preincubated with non-biotinylated oligo(dC)30 for 15 min prior to addition of biotinylated probe. Streptavidin-agarose beads (Thermo Scientific) were added for an additional 20 min, the beads were washed three times in binding buffer with 0.1% Triton X-100, and bound proteins were analyzed by SDS-PAGE/Western blotting using the appropriate anti-epitope antibody.
Primer-dependent DNA Synthesis Assay—Oligoribonucleotides synthesized by purified pol-α·primase were assayed by measuring their ability to serve as primers for the Escherichia coli pol I Klenow fragment. The four-subunit DNA pol-α·primase complex was immunoaffinity-purified from human leukemic myeloblasts or calf thymus using SJK132-20 antibody as described previously (18, 19); both preparations were devoid of detectable exo- or endonuclease activity. Reactions were carried out in a 20-μl mixture containing 50 mm K-HEPES, pH 7.6, 50 mm KCl, 8 mm MgCl2, 2 mm dithiothreitol, 0.1 mm EDTA, 0.1 mg/ml bovine serum albumin, 0.01% Nonidet P-40, 5% glycerol, 2 mm rATP, 20 μm [α-32PO4]dATP (6 μCi/nmol), 500 ng of poly(dT) template (Sigma), 1 unit of Klenow fragment (New England Biolabs), and 0.1 unit of purified pol-α·primase (11, 18). To this assay were added variable amounts of immunoaffinity-purified AAF-44·AAF-132 complex or AAF-44 purified in the absence of AAF-132; FLAG elution buffer served as a control. After a 15-min incubation at 37 °C, acid-insoluble radioactivity was collected on glass fiber filters and quantified by scintillation counting.
Immunofluorescence Studies—HeLa cells were plated on glass coverslips and 16-20 h later were transfected with vectors encoding epitope-tagged AAF-44 or AAF-132 using Lipofectamine 2000. Cells were transferred to serum-free medium for 48 h and then treated with 10 μg/ml aphidicolin (Sigma) in 10% serum-containing medium for an additional 24 h to stimulate them to reach the G1/S border. To release the aphidicolin block, cells were washed three times and incubated in complete growth medium to allow semisynchronous progression through S phase (20). At the indicated times, cells were fixed in methanol at -20 °C, permeabilized in 0.1% Triton X-100, and stained with antibodies specific for proliferating cell nuclear antigen (PCNA; Santa Cruz Biotechnology) and Myc or HA epitopes as described previously (16, 21).
siRNA Transfections and [3H]Thymidine Uptake—Cells were plated at 2 × 105 cells/well in 6-well cluster dishes and 18 h later were transfected with 100 pmol of siRNA and 4 μl of Lipofectamine 2000. After 20 h, they were plated at 7 × 104 cells/well in 24-well dishes and transferred to media containing either 10% dialyzed FBS or 0.5% FBS for 24 h prior to incubation with 5 μCi/ml [methyl-3H]thymidine (PerkinElmer Life Sciences; 20 Ci/mmol) for 4 h. Cells were lysed in 0.1% SDS and 0.1 n NaOH, nucleic acids were precipitated in 10% trichloroacetic acid, and precipitates were collected on glass fiber filters; radioactivity was quantified by liquid scintillation counting. The assay was linear with time and cell number. The target sequences for AAF-44 siRNAs were 1) 5′-UGGAUCCUGUGUUUCUAGCCU-3′ and 2) 5′-CAGCUUAACCUCACAACUUAA-3′; siRNA oligoribonucleotides were synthesized by Qiagen, including the AllStars Negative Control siRNA (1027281). A second control siRNA was from Dharmacon and was targeted at green fluorescent protein (GFP; D-001300-01).
Quantitative Reverse Transcription-PCR—One microgram of total cytoplasmic RNA was subjected to reverse transcription with random hexamer primers, and quantitative PCR was performed using an Mx3000 real time PCR detection system (Stratagene) with IQ™ SYBR Green Supermix (Bio-Rad) (22). The following primers were used: 5′-GACCAGCCTTTTCACAGCTC-3′ (AAF-44, sense) and 5′-GCAAGGGACAGCAAAGACTC-3′ (AAF-44, antisense) and 5′-AACGGATTTGGTCGTATTGGG-3′ (glyceraldehyde-3-phosphate dehydrogenase, sense) and 5′-TGGAGGATGGTGATGGGATTTC-3′ (glyceraldehyde-3-phosphate dehydrogenase, antisense). Relative changes in mRNA expression were calculated with glyceraldehyde-3-phosphate dehydrogenase serving as an internal reference (22).
Data Presentation—Results presented in bar graphs represent the means ± S.E. of at least three independent experiments performed in duplicate. Student's t test was used for pairwise comparisons; a p value of <0.05 was considered to indicate statistical significance.
RESULTS
Purification and Peptide Sequencing of AAF-132 and AAF-44 Subunits—AAF was purified from L1210 murine lymphoblasts through phosphocellulose, hydroxylapatite, DNA cellulose, and gel filtration chromatography (11). Amino acid sequences from several internal peptides of the 132- and 44-kDa subunits were obtained by tandem mass spectrometry; in addition, Edman degradation resulted in an N-terminal sequence of the small subunit (Table 1).
TABLE 1.
Tryptic peptides of AAF-132 and AAF-44 and an N-terminal peptide of AAF-44
AAF was purified from L1210 cells as described previously (11). After SDS-PAGE, the bands corresponding to AAF-132 and AAF-44 were excised, and tryptic fragments were analyzed by tandem mass spectrometry or N-terminal Edman degradation (AAF-44 peptide 1) as described under “Experimental Procedures.”
| Identified as amino acid residues | |
|---|---|
| AAF-132 peptide fragments | |
| 1 GGRPGDGVLPR | 98-108 |
| 2 QLILLGTLVDLLGDSEQE | 110-127 |
| 3 AWATFDPK | 561-568 |
| 4 LVAPGPSTPVFETEGSSGT | 814-832 |
| 5 XHFSQLEK | 962-968 |
| 6 ALGLSPSEWSSILEHAR | 1090-1106 |
| 7 VALQFTGLGGQTXSASK | 1111-1127 |
| 8 THEPLTLLLR | 1128-1137 |
|
9 PTDISPR |
1159-1165 |
| AAF-44 peptide fragments | |
| 1 PQPXLXME | 2-9 |
| 2 IGIGDIIR | 136-143 |
| 3 XIXANIYYK | 157-164 |
| 4 XDDPVWNMQIAR | 166-176 |
| 5 LQEEEALNNKDNLDLAGLTSLLSEK | 197-221 |
| 6 VLELLEDQSDIVSTADHYY | 357-375 |
cDNA Cloning and Genomic Organization of AAF-132—Data base searches with the BLAST program (13) yielded a single EST-based nucleotide sequence comprising peptides 7 and 8 of AAF-132 (Table 1) (GenBank accession number AA655197). We used this sequence to isolate two overlapping clones from a mouse cDNA library, and we used rapid amplification of cDNA 5′-ends (15) to extend the open reading frame to a total of 1211 amino acids. The first methionine is in the context of a good Kozak sequence and is preceded by 41 presumably untranslated nucleotides. The open reading frame contains the exact amino acid sequences of all nine tryptic peptides derived from purified AAF-132 (Table 1), and the calculated molecular weight for the encoded protein is 133,917, which is in good agreement with the apparent molecular weight observed on SDS-PAGE (11).
After completion of the above described work, full-length cDNA clones of unidentified proteins became available through the NCBI data bases; clones NM_026889 and NM_025099 encode murine and human AAF-132, respectively, which share 69% amino acid sequence identity (GenBank accession numbers of AAF sequences are summarized in supplemental Table I). Several minor sequence differences between the two murine cDNAs are listed in supplemental Table II; most differences did not result in amino acid changes, but in three of four cases where it did, our sequence corresponds to amino acids found in the human protein. Two genomic clones from mouse chromosome 11 demonstrate that the AAF-132 gene contains 23 exons spanning 15 kb. Several mouse EST clones were found that encode exons I and II and extend the 5′-untranslated region to a total of 208-222 nucleotides (supplemental Table I provides a list of the EST and genomic clones).
cDNA Cloning and Genomic Organization of AAF-44—BLAST searches with the peptide sequences obtained for AAF-44 yielded an EST clone containing perfect matches for peptides 2-6 of AAF-44 (GenBank accession number AA039012) (Table 1). Rapid amplification of cDNA 5′-ends extended the open reading frame to a total of 378 amino acids (calculated molecular weight, 43,482), including peptide 1 obtained from the N-terminal sequencing of AAF-44 (Table 1); the latter included 197 nucleotides of the 5′-untranslated region with a termination codon upstream and in-frame with the ATG. Two genomic clones from mouse chromosome 19 (supplemental Table I) show that the AAF-44 gene contains 10 exons over a distance of 36 kb. Recently BLAST searches also identified a full-length cDNA clone encoding murine AAF-44 (GenBank accession number NM_175360); compared with our sequence, this clone contains one nucleotide change but identical amino acid sequence and 229 nucleotides of 5′-untranslated region. Three EST clones were found that extend further 5′ (corresponding to the genomic sequence); these clones suggest that the 5′-untranslated region may be unusually long, consisting of 327-349 nucleotides (supplemental Table I provides accession numbers). Based on a human cDNA sequence, human and murine AAF-44 share 71% amino acid sequence identity.
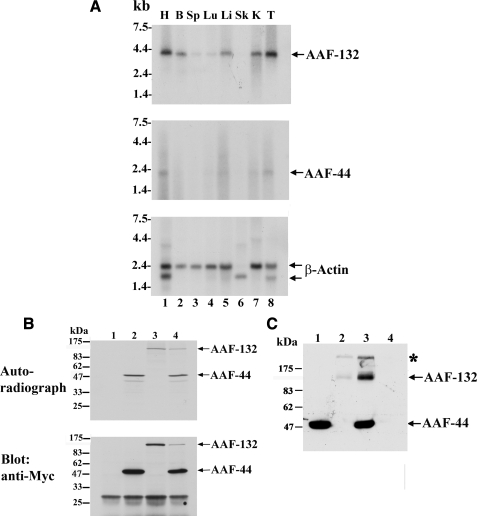
AAF mRNA Analysis—Using probes encoding parts of AAF-132 or AAF-44 coding sequences, we performed Northern blot analysis of poly(A)+ RNA from adult mouse tissues and found single messenger RNA species of ∼4 and 2 kb for AAF-132 and AAF-44, respectively (Fig. 1A). The sizes of the mRNAs correspond approximately to the length of their cDNAs (4 and 1.9 kb for AAF-132 and -44, respectively). The amount of AAF-44 message was low but correlated with the amount of AAF-132 mRNA with highest levels (compared with β-actin) found in testis and undetectable levels in skeletal muscle.
FIGURE 1.
AAF-44 and AAF-132 mRNA expression in murine tissues and expression of Myc epitope-tagged AAF-44 and AAF-132 proteins. A, a Northern blot prepared with 2 μg of poly(A)+ RNA/lane was hybridized to radioactively labeled cDNA probes encoding murine AAF-132 (top panel), AAF-44 (middle panel), or human β-actin (lower panel). RNA molecular weight markers (in kb) are shown on the left. Lanes 1-8 show RNA isolated from adult murine heart (H), brain (B), spleen (Sp), lung (Lu), liver (Li), skeletal muscle (Sk), kidney (K), and testis (T). B, cDNA sequences encoding AAF-44 (lanes 2 and 4) and/or AAF-132 (lanes 3 and 4) with an N-terminal Myc epitope tag were subjected to in vitro transcription and translation in a coupled T7 RNA polymerase/reticulocyte lysate system using [35S]methionine. Lane 1 shows a control reaction with “empty vector” template. Aliquots of the reaction product were analyzed by SDS-PAGE/autoradiography (upper panel) and Western blotting using a Myc epitope-specific antibody (lower panel). C, 293T cells were transfected with equal amounts of expression vectors encoding Myc-tagged AAF-44 (lanes 1 and 3), AAF-132 (lanes 2 and 3), or empty vector (lane 4), and cell lysates were analyzed by SDS-PAGE/Western blotting using an anti-Myc antibody. AAF-132 appears to form high molecular weight aggregates that do not enter the gel properly (marked with an asterisk).
Expression of Epitope-tagged AAF-44 and AAF-132 Proteins—In vitro transcription and translation of cDNAs encoding Myc epitope-tagged versions of AAF-44 and/or AAF-132 yielded products with apparent molecular masses corresponding to those of the native AAF subunits, and [35S]methionine-labeled proteins reacted with antibodies directed against the Myc epitope (Fig. 1B; the Myc epitope increases the apparent molecular weight). We observed high levels of the 44-kDa subunit but lower levels of the 132-kDa subunit in 293T cells transfected with expression vectors encoding Myc-tagged AAF-44 and/or AAF-132 (Fig. 1C). Co-expression of AAF-44 always improved recovery of the large subunit, whereas storing samples at 4, -20, or -80 °C caused dramatic losses of the 132-kDa protein. The 132-kDa subunit generally migrated as a blurry band, suggesting the presence of post-translational modification(s). In addition, variable amounts of high molecular weight protein aggregates were observed at the top of the gel (marked with an asterisk in Fig. 1C). This was also observed during AAF purification where a portion of AAF-132 appeared to form large aggregates that migrated only a short distance on SDS-PAGE (11).
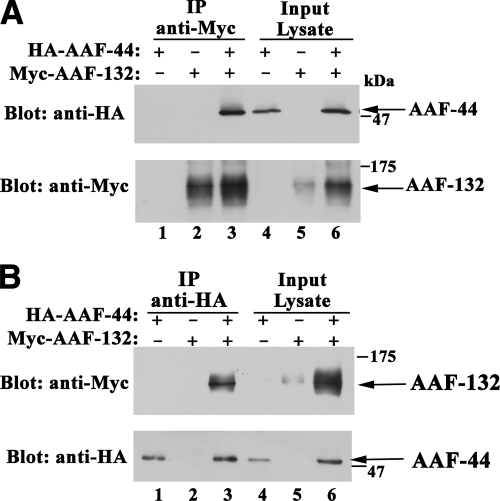
Co-immunoprecipitation of AAF-132 and AAF-44 from Intact Cells—To determine whether the two AAF proteins associate in intact cells, 293T cells were transfected with vectors encoding Myc epitope-tagged AAF-132 and/or HA-tagged AAF-44. When AAF-132 was immunoprecipitated from lysates of cells co-expressing both proteins, AAF-44 was found in the immunoprecipitates, but no AAF-44 was observed in anti-Myc immunoprecipitates obtained from cells expressing either AAF-44 or AAF-132 alone (Fig. 2A, top panel, lanes 1-3; lanes 4-6 show 10% of input lysate, and lanes 1-3 of the bottom panel show the amount of AAF-132 present in anti-Myc immunoprecipitates). Similar results were obtained in reciprocal experiments when AAF-44 was immunoprecipitated and the anti-HA immunoprecipitates were examined for the presence of AAF-132 (Fig. 2B). We conclude that AAF-44 and -132 form a complex in intact cells.
FIGURE 2.
Association of AAF-44 with AAF-132 in intact cells. A, 293T cells were transfected with expression vectors encoding HA epitope-tagged AAF-44 and/or Myc epitope-tagged AAF-132 as indicated, and cell lysates were subjected to immunoprecipitation (IP) with anti-Myc antibody (lanes 1-3). Proteins present in the immunoprecipitates were analyzed by SDS-PAGE/Western blotting using an anti-HA epitope antibody (upper panel) or an anti-Myc epitope antibody (lower panel). Ten percent of total cell lysate input was analyzed in parallel (lanes 4-6). B, the experiment was similar to that described in A, but cell lysates were subjected to immunoprecipitation with anti-HA antibody (lanes 1-3).
AAF-44 Is a Member of the OB Fold Protein Superfamily with Homology to RPA-32—When the predicted amino acid sequence derived from the large open reading frame of AAF-132 was used to search available data bases with the BLAST program (13), no significant homology to known proteins was found, and no specific conserved domains were identified. In contrast, BLAST analysis of the AAF-44 sequence predicted the presence of an oligonucleotide/oligosaccharide-binding (OB) fold domain and demonstrated homology to RPA-32, the 32-kDa subunit of the replication protein A complex. RPA is the most abundant ssDNA-binding protein in mammalian cells. It is a heterotrimer composed of a 70-, 32-, and 14-kDa subunit (RPA-70, -32, and -14) and plays a central role in DNA replication, DNA damage recognition, and DNA repair (9, 23, 24). The RPA heterotrimer binds ssDNA via four DNA-binding OB fold domains located on the RPA-70 and -32 subunits; in addition, RPA directly interacts with pol-α·primase and diverse DNA processing proteins (3, 23). Therefore, AAF and RPA-32 may share not only structural but also functional homology (discussed below).
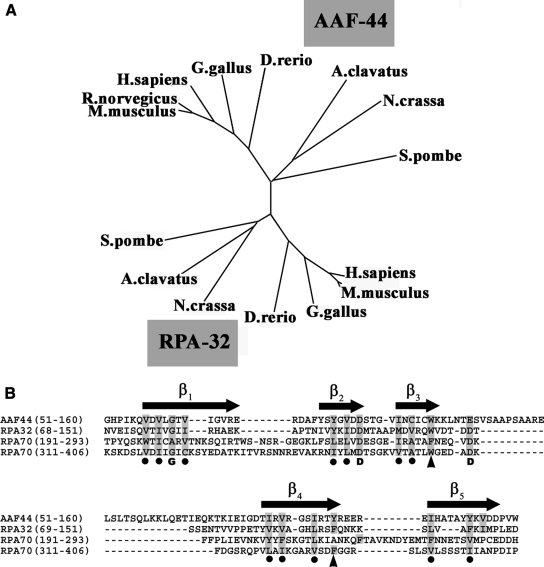
To determine the evolutionary relationship between AAF-44 and RPA-32, we used ClustalW (25) for progressive alignment of the predicted amino acid sequences of AAF-44 and RPA-32 from different species, including human, rat, mouse, chicken, zebrafish, Schizosaccharomyces pombe, and other fungi. From the sequence alignment, an unrooted phylogenetic tree was constructed (Fig. 3A). The data suggest that AAF-44 and RPA-32 are evolutionarily related and may have arisen from gene duplication before the divergence of fungi and animals. No AAF-44 orthologs could be identified in Drosophila melanogaster, Caenorhabditis elegans, or S. cerevisiae. Murine AAF-44 and RPA-32 share 21% sequence identity compared with 28% sequence identity between murine and S. pombe RPA-32. The relationship between AAF and RPA-32 was confirmed using the progressive alignment procedure of Feng and Doolittle (26). The positions of sequence identity are distributed over the length of the AAF-44 protein with some highly conserved clusters of several residues. The highest density of conserved residues is found in the centrally located, single OB fold domain of RPA-32 involved in ssDNA binding (Fig. 3B).
FIGURE 3.
Phylogenetic relationship of AAF-44 and RPA-32 and alignment of AAF and RPA OB fold domains. A, AAF-44 sequences were aligned with RPA-32 sequences of human, rat, mouse, chicken, zebrafish, and fungal origin using the ClustalW program (25). An unrooted phylogenetic tree was drawnby the neighbor-joining method using the program Phylodendron. GenBank accession numbers for AAF-44 are as follows: Homo sapiens, NP_079204; Rattus norvegicus, NP_001011943; Mus musculus, NP_780569; Gallus gallus, XP_421742; Danio rerio, NP_956683; Neurospora crassa, XP_960343; Aspergillus clavatus, XP_001275592; and S. pombe, CAL44730. GenBank accession numbers for RPA-32 are as follows: H. sapiens, NP_002937; R. norvegicus, NP_067593; M. musculus, NP_035414; G. gallus, NP_001026063; D. rerio, NP_571786; N. crassa, XP_961967; A. clavatus, XP_001268210; and S. pombe, NP_588227. B, human AAF-44 was aligned with the sequences of the OB fold domains of human RPA-70 and RPA-32 using ClustalW with slight manual modifications; RPA-70 and RPA-32 were aligned according to published crystal structures, and regions of β-strands conserved among the OB folds are shown above as black arrows (28, 30). Numbers in parentheses refer to amino acid positions in the human sequences, and residues that are identical or conserved between AAF-44 and RPA are shaded in gray. Conserved hydrophobic residues in alternating amino acid positions within the β-sheets are indicated by black dots, and conserved glycine and aspartate residues are marked by their bold letter symbols. Two arrowheads indicate aromatic amino acid residues that are thought to be important for ssDNA binding of RPA-32 and are conserved in members of the S1 family of OB fold proteins (30).
Crystal structures of the three main RPA ssDNA binding domains have been solved with two of the domains residing on RPA-70 and one residing on RPA-32 (27, 28). All three domains contain a central OB fold, which is a five-stranded closed β-barrel capped by an α-helix and forms the binding surface for ssDNA (23, 29). DNA binding is sequence-independent and occurs primarily via non-polar interactions between amino acid side chains and the nucleic acid bases or ribose moieties (29, 30). Fig. 3B shows a structure-based sequence alignment of the RPA-70 and RPA-32 OB fold domains with the putative OB fold domain of AAF-44 (27, 28, 30). The nucleic acid-binding OB family has low sequence similarity among its members; however, structure-based alignment has demonstrated a pattern of conserved amino acid residues within β-sheets (30). AAF-44 contains several conserved features consistent with an OB fold topology (Fig. 3B): (i) hydrophobic residues in alternating amino acid positions, especially in β1, β2, and β4; (ii) a conserved glycine/alanine in β1; (iii) a conserved aspartate at the end of β2; and (iv) two aromatic amino acids at the end of β3 and β4, which in RPA-70 stack with the DNA bases (27, 29). These results suggest that AAF-44 contains an OB fold domain that is highly related to the OB fold domains of RPA-32 and RPA-70.
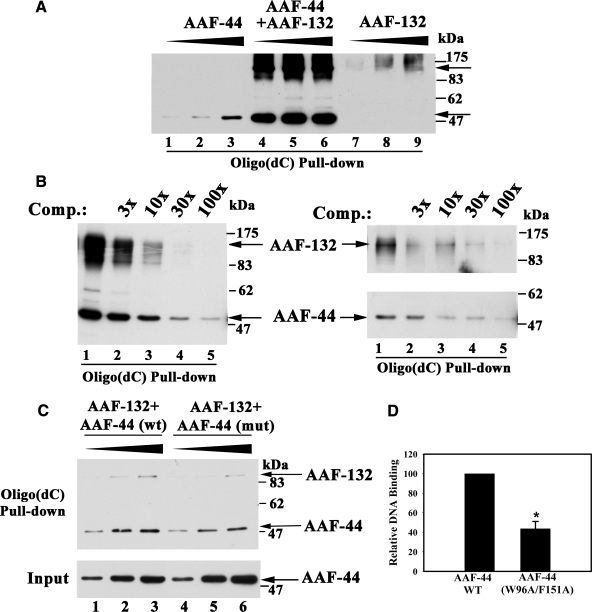
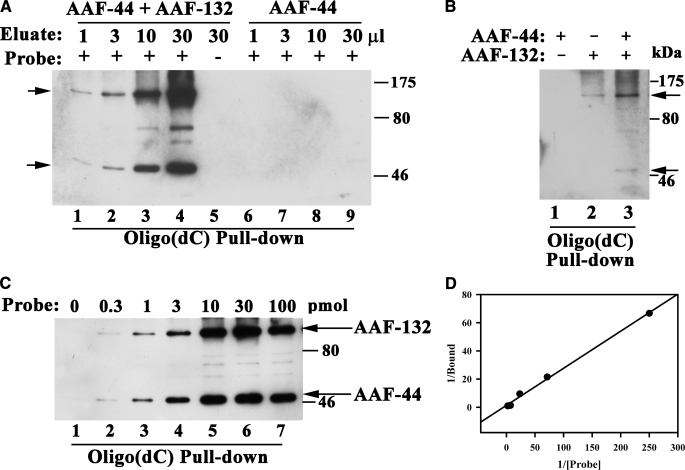
AAF Binds to Single-stranded DNA—To determine whether AAF binds to single-stranded DNA, we transfected 293T cells with expression vectors encoding Myc epitope-tagged AAF-44 and/or AAF-132 and incubated increasing amounts of cell lysate protein with biotinylated oligo(dC)30. The oligo(dC) was captured on streptavidin-coated beads, and after extensive washing, bound proteins were analyzed by SDS-PAGE and Western blotting with anti-Myc antibody. Fig. 4A shows that AAF-44 and AAF-132, when expressed separately, bound weakly to the single-stranded oligo(dC), but when both subunits were expressed together, strong DNA binding was observed. The differences in the amount of DNA-bound AAF-44 were not explained by differences in the amount of AAF-44 expressed in the presence or absence of AAF-132, although AAF-132 was less abundant when expressed by itself as mentioned before (supplemental Fig. 1A). No binding of AAF to streptavidin beads was detectable in the absence of biotinylated probe (data not shown).
FIGURE 4.
ssDNA binding of AAF and effects of mutations in the putative OB fold domain of AAF-44. A, 293T cells were transfected with equal amounts of expression vectors encoding Myc epitope-tagged AAF-44 (lanes 1-3), Myc-tagged AAF-132 (lanes 7-9), or both (lanes 4-6), and 20, 60, or 200 μg of cell lysate protein (designated by the triangle above) were incubated with 10 pmol of biotinylated oligo(dC)30 as described under “Experimental Procedures.” Biotinylated oligo(dC)30 was isolated using streptavidin-agarose beads, and proteins bound to the washed beads were analyzed by SDS-PAGE/Western blotting with anti-Myc antibody. The arrows indicate migration of full-length AAF-132 and -44. B, cells were transfected with equal amounts of Myc-tagged AAF-44 and -132 (left panel) or with each subunit separately (right panels), and cell lysates (20 and 100 μg of protein in the left or right panels, respectively) were incubated with 30 pmol of biotinylated oligo(dC)30 in the absence (lane 1) and presence (lanes 2-5) of increasing amounts of non-biotinylated competitor (Comp.) oligo(dC) (90, 300, 900, and 3000 pmol) as indicated. Protein·DNA complexes were isolated on streptavidin-agarose beads and analyzed as described in A. C, cells were co-transfected with vectors expressing Myc-tagged AAF-132 and FLAG-tagged wild type AAF-44 (wt; lanes 1-3) or mutant AAF-44 containing alanine substitutions for Trp-96 and Phe-151 (mut; lanes 4-6). Binding to oligo(dC)30 was assessed as described in A except that bound proteins were detected by immunoblotting with anti-Myc plus anti-FLAG antibodies (upper panel). The lower panel shows a Western blot of cell lysates demonstrating similar amounts of wild type and mutant AAF-44 present in the lysates used for the DNA pulldown assay. D, three independent experiments were performed as described in C, and relative amounts of wild type versus mutant AAF-44 bound to oligo(dC)30 were quantified by densitometry scanning. Shown are the mean ± S.D.; asterisk, p < 0.05 for the comparison between wild type and mutant AAF-44.
To examine binding specificity, we performed competition experiments in which increasing amounts of non-biotinylated oligo(dC)30 were added to cell lysates prior to adding biotinylated probe. As shown in Fig. 4B, left panel, binding of the AAF-44·AAF-132 complex to biotinylated probe was almost completely prevented in the presence of a 100-fold excess of non-biotinylated oligo(dC). Similar competition experiments were performed with AAF-44 and AAF-132 expressed separately (Fig. 4B, right panels); although DNA binding was much weaker, it was competed to a similar extent by the non-biotinylated oligo(dC), suggesting similar binding affinities. It is possible that recombinant AAF-44 and AAF-132 expressed separately bind DNA by forming complexes with endogenous AAF subunits present in limiting amounts (this question is addressed below). Myc-tagged AAF-44 also bound to single-stranded DNA-cellulose but not to control cellulose beads (supplemental Fig. 1B).
Effect of Mutations in the Putative OB Fold Domain of AAF-44—To test the hypothesis that AAF-44 contains a functional OB fold involved in ssDNA binding, we performed site-directed mutagenesis and replaced two conserved aromatic amino acids with alanine residues (Trp-96 and Phe-151 in the murine sequence; the corresponding residues in the human sequence are marked with arrowheads in Fig. 3B). When 293T cells were transfected with vectors encoding FLAG epitope-tagged wild type or double mutant AAF-44 together with Myc epitope-tagged AAF-132, similar amounts of wild type and mutant proteins were expressed (Fig. 4C; the lower panel shows an anti-FLAG Western blot of cell lysates; the anti-Myc blot is not shown but demonstrated equal amounts of AAF-132 in both types of lysate). When increasing amounts of cell lysates were incubated with biotinylated oligo(dC)30, significantly less mutant AAF-44W96A/F151A·AAF-132 complex bound to the probe compared with wild type AAF (Fig. 4C; the top panel shows a typical DNA pulldown assay with DNA-bound proteins detected using anti-Myc plus anti-FLAG antibodies). The W96A/F151A mutation in AAF-44 reduced DNA binding to 44 ± 7% of wild type AAF (Fig. 4D summarizes results of three independent experiments). The difference was also apparent when wild type or mutant AAF-44 was expressed in the absence of recombinant AAF-132 and cell lysates were incubated with increasing amounts of biotinylated oligo(dC)30 probe (supplemental Fig. 1C). These results suggest that mutation of the two conserved aromatic amino acids within the putative OB fold domain of AAF-44 interferes with DNA binding of the protein. Although the effect of the mutations was moderate, a similar reduction in RPA DNA binding is observed when the corresponding two aromatic amino acids in the OB fold domain of RPA-32 are mutated to alanine (31).
AAF DNA Binding Requires Both Subunits—To determine whether AAF-44 can bind ssDNA by itself or whether binding requires association with the large subunit, we immunoaffinitypurified FLAG epitope-tagged AAF-44 from 293T cells. The protein was isolated from cells expressing FLAG-tagged AAF-44 in the presence or absence of Myc-tagged AAF-132. AAF-44 was eluted from the immunoaffinity matrix using FLAG peptide, and eluted proteins were analyzed by SDS-PAGE/silver staining. AAF-44 appeared as a major band together with two contaminating bands that were also present in eluates derived from empty vector-transfected cells, and Myc-tagged AAF-132 co-purified with AAF-44 from cells co-transfected with both subunits (supplemental Fig. 2A). When increasing amounts of these eluates were incubated with biotinylated oligo(dC)30, strong DNA binding of both subunits was detected in samples containing AAF-44 with co-purified AAF-132, but no binding was apparent in samples containing purified AAF-44 alone although similar amounts of AAF-44 were present whether or not AAF-132 was co-purified (Fig. 5A shows DNA-bound proteins detected by probing with anti-FLAG plus anti-Myc antibodies, and supplemental Fig. 2B shows a Western blot of AAF-44 present in the eluates). Nonspecific binding of the AAF-44·AAF-132 complex to streptavidin beads in the absence of biotinylated probe was excluded (Fig. 5A, lane 5). To determine whether addition of AAF-132 could rescue DNA binding of purified AAF-44, we incubated purified FLAG-tagged AAF-44 with biotinylated oligo(dC)30 and added cell lysates from 293T cells transfected with either empty vector or Myc-tagged AAF-132. Although no binding of AAF-44 was apparent in the presence of control lysate, AAF-44 binding was detectable in the presence of lysate containing AAF-132 (Fig. 5B, compare lane 1 (AAF-44 plus control lysate) with lane 3 (AAF-44 plus AAF-132-containing lysate)). These results suggest that AAF-44 requires the presence of AAF-132 for DNA binding.
FIGURE 5.
DNA binding of AAF requires both subunits. A, 293T cells were co-transfected with FLAG-tagged AAF-44 and Myc-tagged AAF-132 (lanes 1-5) or received vector encoding AAF-44 plus empty vector (lanes 6-9). Cell lysate proteins were passed over an anti-FLAG antibody affinity matrix, and AAF-44 was eluted with FLAG peptide as described under “Experimental Procedures.” Analysis of eluates by SDS-PAGE/silver staining and Western blotting is shown in supplemental Fig. 2. Increasing amounts of the eluates containing purified FLAG-AAF-44 with (lanes 1-5) or without (lanes 6-9) co-purified Myc-AAF-132 were incubated with 30 pmol of biotinylated oligo(dC)30 probe as indicated by +. Protein·DNA complexes were isolated on streptavidin-agarose, and bound proteins were detected by Western blotting with anti-FLAG plus anti-Myc antibodies as described under “Experimental Procedures.” In lane 5, the eluate was incubated with streptavidin-agarose in the absence of biotinylated probe. The arrows indicate migration of full-length AAF-132 and -44. B, purified FLAG-tagged AAF-44 was incubated with cell lysates from 293T cells transfected with either empty vector (lane 1) or Myc-tagged AAF-132 (lane 3). Lane 2 shows the cell lysate containing AAF-132 in the absence of purified AAF-44. Proteins bound to biotinylated oligo(dC)30 were analyzed as described in A. C, FLAG-tagged AAF-44 and Myc-tagged AAF-132 were co-purified as described in A and incubated with increasing amounts of biotinylated oligo(dC)30 probe as indicated. Protein·DNA complexes were analyzed as described in A. D, the relative amounts of DNA-bound AAF-44 were determined by densitometry scanning of two independent experiments performed as described in C. A double reciprocal plot of 1/[bound AAF-44] versus 1/[probe] is shown (with the probe concentration in μm).
To determine the DNA binding affinity of AAF, we incubated a constant amount of purified AAF-44·AAF-132 complex with increasing amounts of biotinylated oligo(dC)30 probe, isolated the protein·DNA complex on streptavidin-coated beads, and quantified the amount of DNA-bound AAF-44 by Western blotting (Fig. 5C). DNA binding was saturated at probe concentrations >0.14 μm (10 pmol of probe). Fig. 5D shows a double reciprocal plot of 1/[relative DNA-bound AAF-44] versus 1/[probe]; from these data we estimated the apparent association constant to be Ka. 2.8 × 106 m-1. For comparison, the apparent association constant of the RPA heterotrimer varies depending on the length of the oligodeoxynucleotide probe from ∼7 × 107 m-1 for oligo(dT)10 to ∼1.5 × 109 m-1 for oligo(dT)50 (as determined by electrophoretic mobility shift assay (32)). Thus, AAF appears to bind ssDNA with lower affinity than RPA.
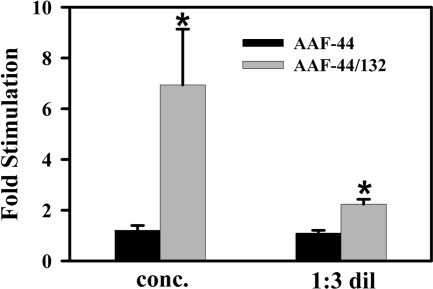
Recombinant AAF Stimulates DNA Primase Activity—We examined the effect of purified recombinant AAF on DNA primase activity using purified pol-α·primase from calf thymus with poly(dT) template. Oligoribonucleotides synthesized by purified pol-α·primase in the absence and presence of AAF-44 or AAF-44·AAF-132 complexes were assayed by measuring their ability to serve as primers for E. coli pol I Klenow fragment. Adding concentrated immunoaffinity-purified AAF-44·AAF-132 to pol-α·primase stimulated primase activity about 7-fold compared with the activity measured in the presence of FLAG peptide elution buffer (Fig. 6; the gray bars show results for concentrated and 1:3 diluted AAF-44·AAF-132; because of variable degrees of AAF-132 proteolysis, the amount of intact AAF-44·AAF-132 complex in each preparation could not be accurately determined). In contrast, adding purified AAF-44 by itself had no significant effect on primase activity (Fig. 6, black bars). Similar results were obtained with pol-α·primase purified from human myeloid leukemic blasts (data not shown). Neither AAF-44 by itself nor the AAF-44·AAF-132 complex affected Klenow activity (data not shown). Thus, consistent with the finding that the intact AAF-44·AAF-132 complex is required for DNA binding (Fig. 5C), both subunits are required for stimulation of DNA primase activity.
FIGURE 6.
Recombinant AAF stimulates DNA primase activity. Immunoaffinity-purified FLAG-AAF-44 with (gray bars) or without (black bars) co-purified Myc-AAF-132 was incubated with 0.1 unit of purified pol-α·primase complex, and primase activity was determined as described under “Experimental Procedures.” Primase activity measured in the presence of an equal volume of FLAG peptide-containing elution buffer was assigned a relative value of 1. Asterisk, p < 0.05 for the comparison between AAF-44 and the AAF-44·AAF-132 complex. conc., concentrated; dil, diluted. Shown are the means ± S.D. of three experiments.
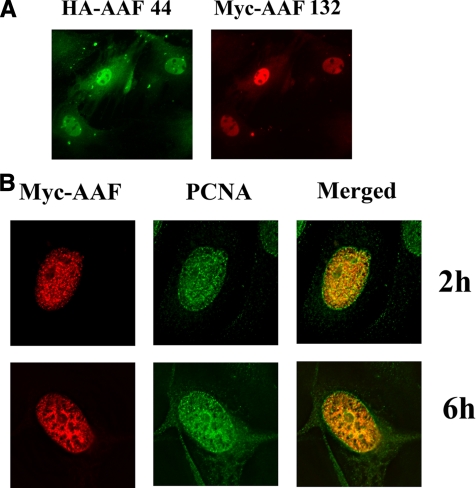
Nuclear Localization of AAF Subunits, and Co-localization of AAF with PCNA—To examine the subcellular localization of AAF in intact cells, we used rabbit anti-HA and murine antiMyc epitope-specific antibodies to perform double immunofluorescence staining in HeLa cells transfected with HA epitope-tagged AAF-44 and Myc-tagged AAF-135. AAF-44 appeared to be largely nuclear with sparing of nucleoli; some cytoplasmic immunofluorescence was apparent perhaps due to high expression levels (Fig. 7A, left panel). AAF-132 staining was exclusively nuclear also with exclusion of nucleoli (Fig. 7A, right panel).
FIGURE 7.
Subcellular localization of AAF subunits and co-localization with PCNA. A, HeLa cells were co-transfected with expression vectors encoding HA epitope-tagged AAF-44 and Myc epitope-tagged AAF-132 and analyzed by double immunofluorescence staining as described under “Experimental Procedures.” B, HeLa cells were co-transfected with Myc epitope-tagged AAF-44 and AAF-132, serum-starved, exposed to aphidicolin in full growth media, and released from aphidicolin blockade to allow semisynchronous progression through S phase. Two and 6 h later, cells were fixed and stained with antibodies specific for PCNA (green) and the Myc-epitope (red) as indicated; yellow fluorescence indicates co-localization in the merged images. Cells were sectioned at 2-μm intervals and midnuclear sections are shown; images were obtained using a Delta Vision deconvolution microscope.
During S phase, chromosomal replication occurs at distinct sites in the nucleus termed replication foci, which contain multiple proteins required for DNA replication (20). Pol-α, RPA, and PCNA co-localize with bromodeoxyuridine-labeled nascent DNA within replication foci (20, 21, 33, 34). To determine whether AAF localized in replication foci, we transfected HeLa cells with Myc epitope-tagged AAF-44 and AAF-132, enriched cells for S-phase by releasing them from aphidicolin blockade, and subjected them to double immunofluorescence staining using rabbit anti-PCNA and murine anti-Myc antibodies (20). Deconvolution microscopy demonstrated co-localization of AAF with PCNA in discrete, punctate regions of the nucleus with PCNA also present on the nuclear membrane (Fig. 7B). The punctate and nuclear membrane staining of PCNA in S phase nuclei has been observed previously (20, 21, 35).
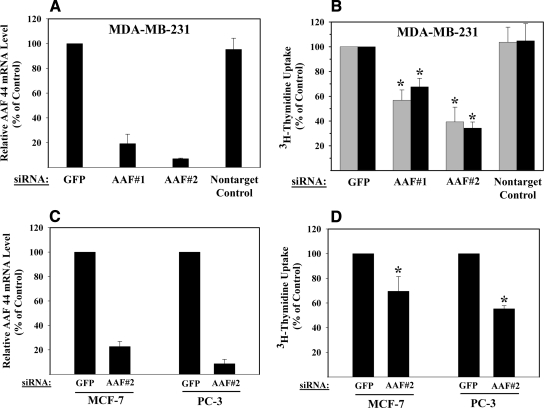
siRNA-mediated Knockdown of AAF-44 Inhibits DNA Replication—Because AAF appears to work as a template affinity protein that assists pol-α·primase in synthesis of the lagging strand of DNA replication forks (12), we asked whether AAF is required for DNA replication in intact cells. We used siRNA oligoribonucleotides targeting two different sequences in human AAF-44, which reduced AAF-44 mRNA levels in MDA-MB-231 breast cancer cells by ∼80 and >90% compared with mRNA levels in cells transfected with two control siRNAs (Fig. 8A). In MDA-MB-231 cells grown in the presence of 10% dialyzed FBS, the AAF-44 siRNAs reduced [methyl-3H]thymidine uptake into DNA by 32 ± 7 and 65 ± 5% compared with thymidine uptake measured in control siRNA-transfected cells (Fig. 8B, black bars; p < 0.05 for the comparison between cells transfected with AAF siRNA versus GFP siRNA). These results indicate that reducing the intracellular AAF concentration affects the rate of DNA replication. Similar results were obtained when cells were grown in low serum-containing medium, indicating that AAF levels were limiting for DNA synthesis even in serum-deprived cells with low DNA synthesis rates (Fig. 8B, gray bars). We also examined MCF-7 breast cancer and PC-3 prostate cancer cells and found that the AAF-44 siRNA inhibited DNA synthesis significantly (p < 0.05) albeit less than observed in MDA-MB-231 cells (Fig. 8, C for AAF-44 mRNA levels and D for [methyl-3H]thymidine uptake). The AAF-44 siRNA did not affect cell viability as measured by trypan blue staining or by flow cytometry examination of propidium iodine-stained cells for sub-G1 DNA content even when cells were treated repeatedly for up to 96 h (data not shown).
FIGURE 8.
Effect of AAF-44 siRNAs on DNA replication. A, MDA-MB-231 cells were transfected with siRNAs targeting two different sequences in AAF-44 or with control siRNAs (one control siRNA targeting GFP and the other not targeting any known mRNA). Forty-eight hours later, AAF-44 mRNA levels were quantified by real time reverse transcription-PCR. Expression levels were normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA levels, and the relative AAF-44 mRNA level in cells transfected with GFP siRNA was assigned a value of 100%. B, MDA-MB-231 cells were transfected as described in A and grown in medium containing either 0.1% (gray bars) or 10% (black bars) dialyzed FBS. Forty-eight hours after transfection, cells were incubated with [methyl-3H]thymidine for 4 h, and thymidine uptake into DNA was measured as described under “Experimental Procedures”; the uptake observed in GFP siRNA-transfected cells was assigned a value of 100%. For GFP siRNA-transfected cells grown in 0.1 or 10% FBS, [methyl-3H]thymidine uptake into DNA was 42,040 ± 14,545 and 149,175 ± 48,030 cpm/105 cells/h, respectively. C, MCF-7 and PC-3 cells were transfected with siRNAs targeting GFP or AAF-44, and AAF-44 mRNA levels were quantified as described in A. D, MCF-7 and PC-3 cells were transfected as in C and transferred to medium containing 0.1% FBS, and [methyl-3H]thymidine uptake into DNA was determined as in B. Asterisk, p < 0.05 for the comparison between cells transfected with AAF siRNA versus GFP siRNA. Shown are the means ± S.D. of at least three independent experiments.
DISCUSSION
DNA replication starts at many origins on each chromosome, and DNA primase in the pol-α·primase complex is the only enzyme known to prime DNA replication in eukaryotic cells (2). In addition to starting the leading strand of the nascent fork at each replication origin, pol-α·primase also initiates each of the vast number of Okazaki fragments on the lagging strand. More than 20 million Okazaki fragments have to be initiated during a single S phase in mammalian cells (2). The rates of primer synthesis and pol-α extension rates measured in vitro fall significantly short of the assumed rapidity of the in vivo process, indicating a key role of factors that enhance pol-α·primase activity in vivo (2). The RPA complex, which enhances processivity and fidelity of primer extension by pol-α, is one of those factors (3). AAF appears to be a second such factor because AAF improves the efficiency of pol-α·primase in initiating and extending DNA fragments by three mechanisms: (i) it keeps the enzyme associated with a ssDNA template; (ii) it facilitates the movement of pol-α·primase over double-stranded regions; and (iii) it increases the processivity of pol-α (12). All three AAF functions serve to enhance the activities of pol-α·primase on the lagging strand of the replication fork. Because the effects of AAF are seen at a stoichiometry of ∼1 molecule of AAF/molecule of DNA template, it is unlikely that AAF functions simply by “coating” single-stranded DNA (12). However, AAF and RPA may have coordinated functions in initiating DNA synthesis.
The function of RPA has been studied extensively in the simian virus 40 (SV40) DNA replication system where all three RPA subunits are essential for replication (36, 37). In this system, the SV40 large T antigen directs assembly of a replisome through physical interaction with RPA, pol-α·primase, and topoisomerase I (36, 37). AAF cannot replace RPA in this in vitro system (11). Others in previous work raised the possibility that AAF may share some functional characteristics and antigenic determinants with the SV40 large T antigen (38). However, we compared AAF-132 and AAF-44 sequences directly with those of SV40 large T antigen using the BLAST 2 program (39) and detected no significant similarities.
AAF-44 is homologous to the 32-kDa subunit of RPA; multiple key residues important for the OB fold structure and DNA binding of RPA are conserved in AAF-44. DNA binding of RPA occurs in several stages that involve sequential participation of four different OB fold domains, three in the RPA-70 subunit and one in the RPA-32 subunit (3, 23, 40). Although the intact RPA heterotrimer binds ssDNA with high affinity, the three OB fold domains in RPA-70 are thought to account for most if not all of this binding affinity, and the interaction of the RPA-32·RPA-14 subcomplex with ssDNA is only detectable when the N and C termini of RPA-32 are truncated to produce a “core” domain bound to RPA-14 (4, 31). AAF contains a single OB fold domain on the 44-kDa subunit. We found that recombinant AAF-44 bound ssDNA only in the presence of AAF-132 and that the DNA binding affinity of AAF was lower than that reported for the intact RPA complex (31, 32). However, a mutant RPA complex containing alanine substitutions in four conserved aromatic residues in the first and second OB fold domains of RPA-70 has an apparent association constant similar to that of AAF; this mutant RPA complex still contains one intact “low affinity” OB fold domain each in the 70- and 32-kDa subunits (31). During purification of AAF from L1210 cells (11), pol-α·primase co-purified through the first three steps, but by the fourth step over ssDNA-cellulose, AAF fractions were free of DNA polymerase activity; this suggests that intact AAF binds to ssDNA-cellulose in the absence of pol-α·primase, and we did not detect any DNA polymerase·primase activity in immunoaffinity-purified recombinant AAF-44·AAF-132 preparations.3 Moreover elution of AAF from ssDNA-cellulose columns required much higher salt concentrations than elution of pol-α·primase, suggesting that the affinity of AAF for ssDNA is greater than that of pol-α·primase (11, 12).
AAF-44 and AAF-132 mRNAs are relatively widely expressed in different tissues with the exception of skeletal muscle. AAF activity was originally identified in L1210 cells, which have a population doubling time of <20 h; however, it remains to be determined whether AAF levels vary with the cell cycle and whether levels are higher in cells with a short doubling time compared with slow growing cells.
We found that AAF co-localized with PCNA in the nuclei of replicating cells, suggesting that AAF localizes to replication centers, i.e. areas of active DNA replication. This observation, together with the stimulatory effect of AAF on pol-α·primase activity in vitro, suggests that AAF might regulate DNA replication in intact cells. Indeed, siRNA-mediated suppression of AAF-44 expression decreased DNA synthesis in several cell types. In MDA-MB-231 breast cancer cells, two different AAF-44-specific siRNAs were tested, and they inhibited [methyl-3H]thymidine uptake into DNA. However, AAF-44 depletion had no significant effect on cell viability. These findings are similar to the phenotype observed in cells treated with siRNA targeting the catalytic subunit of pol-α that show a delay in S phase entry and decreased DNA synthesis with little change in cell viability (41). This is in contrast to siRNA-mediated suppression of RPA-70 or RPA-32 expression that causes little decrease in DNA synthesis but significant cell death due to a high level of spontaneous DNA damage (42, 43).
In conclusion, AAF-44 demonstrates structural and functional homology with RPA-32, but its function is not redundant with RPA because it cannot replace RPA in an SV40 DNA replication system in vitro, and its depletion in intact cells causes a phenotype different from that seen in RPA-depleted cells (11, 42, 43). Future work is needed to determine precisely how AAF participates in the complex processes of DNA replication fork assembly and propagation.
Supplementary Material
Acknowledgments
We are grateful to Dr. Russell F. Doolittle for help with the phylogenetic analysis of AAF and RPA-32 and to Dr. James R. Feramisco for help with deconvolution microscopy at the University of California San Diego Cancer Center Digital Imaging Core Facility. We thank Min Min Sun for expert technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-AR051300 (to R. B. P.) and K22-CA124517 (to D. E. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables I and II and Figs. 1 and 2.
Footnotes
The abbreviations used are: RPA, replication protein A; AAF, α-accessory factor; dNTP, deoxyribonucleoside triphosphate; FBS, fetal bovine serum; OB, oligonucleotide/oligosaccharide-binding; PCNA, proliferating cell nuclear antigen; pol, DNA polymerase; rNTP, ribonucleoside triphosphate; siRNA, small interfering RNA; ss, single-stranded; SV40, simian virus 40; BLAST, basic local alignment search tool; EST, expressed sequence tag; HA, hemagglutinin; GFP, green fluorescent protein.
S. Zhuang and R. B. Pilz, unpublished results.
References
- 1.Bell, S. P., and Dutta, A. (2002) Annu. Rev. Biochem. 71333 -374 [DOI] [PubMed] [Google Scholar]
- 2.Garg, P., and Burgers, P. M. (2005) Crit. Rev. Biochem. Mol. Biol. 40115 -128 [DOI] [PubMed] [Google Scholar]
- 3.Fanning, E., Klimovich, V., and Nager, A. R. (2006) Nucleic Acids Res. 344126 -4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bochkareva, E., Frappier, L., Edwards, A. M., and Bochkarev, A. (1998) J. Biol. Chem. 2733932 -3936 [DOI] [PubMed] [Google Scholar]
- 5.Frick, D. N., and Richardson, C. C. (2001) Annu. Rev. Biochem. 7039 -80 [DOI] [PubMed] [Google Scholar]
- 6.Maga, G., Frouin, I., Spadari, S., and Hubscher, U. (2001) J. Biol. Chem. 27618235 -18242 [DOI] [PubMed] [Google Scholar]
- 7.Burgers, P. M. (October 3, 2008) J. Biol. Chem. 10.1074/jbc.R800062200 [DOI] [PMC free article] [PubMed]
- 8.Osborn, A. J., Elledge, S. J., and Zou, L. (2002) Trends Cell Biol. 12509 -516 [DOI] [PubMed] [Google Scholar]
- 9.Haring, S. J., Mason, A. C., Binz, S. K., and Wold, M. S. (2008) J. Biol. Chem. 28319095 -19111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goulian, M., and Heard, C. J. (1989) J. Biol. Chem. 26419407 -19415 [PubMed] [Google Scholar]
- 11.Goulian, M., Heard, C. J., and Grimm, S. L. (1990) J. Biol. Chem. 26513221 -13230 [PubMed] [Google Scholar]
- 12.Goulian, M., and Heard, C. J. (1990) J. Biol. Chem. 26513231 -13239 [PubMed] [Google Scholar]
- 13.Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990) J. Mol. Biol. 215403 -410 [DOI] [PubMed] [Google Scholar]
- 14.Chen, Y., Zhuang, S., Cassenaer, S., Casteel, D. E., Gudi, T., Boss, G. R., and Pilz, R. B. (2003) Mol. Cell. Biol. 234066 -4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frohman, M. A., Dush, M. K., and Martin, G. R. (1988) Proc. Natl. Acad. Sci. U. S. A. 858998 -9002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casteel, D. E., Zhuang, S., Gudi, T., Tang, J., Vuica, M., Desiderio, S., and Pilz, R. B. (2002) J. Biol. Chem. 27732003 -32014 [DOI] [PubMed] [Google Scholar]
- 17.Zhang, T., Zhuang, S., Casteel, D. E., Looney, D. J., Boss, G. R., and Pilz, R. B. (2007) J. Biol. Chem. 28233367 -33380 [DOI] [PubMed] [Google Scholar]
- 18.Perrino, F. W., and Loeb, L. A. (1989) J. Biol. Chem. 2642898 -2905 [PubMed] [Google Scholar]
- 19.Perrino, F. W., and Mekosh, H. L. (1992) J. Biol. Chem. 26723043 -23051 [PubMed] [Google Scholar]
- 20.Dimitrova, D. S., and Berezney, R. (2002) J. Cell Sci. 1154037 -4051 [DOI] [PubMed] [Google Scholar]
- 21.Bravo, R., and Donald-Bravo, H. (1987) J. Cell Biol. 1051549 -1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng, Y., Zhuang, S., Gloddek, J., Tseng, C. C., Boss, G. R., and Pilz, R. B. (2006) J. Biol. Chem. 28116951 -16961 [DOI] [PubMed] [Google Scholar]
- 23.Zou, Y., Liu, Y., Wu, X., and Shell, S. M. (2006) J. Cell. Physiol. 208267 -273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang, G., and Sancar, A. (2006) Mol. Cell. Biol. 2639 -49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higgins, D. G., Thompson, J. D., and Gibson, T. J. (1996) Methods Enzymol. 266383 -402 [DOI] [PubMed] [Google Scholar]
- 26.Feng, D. F., and Doolittle, R. F. (1996) Methods Enzymol. 266368 -382 [DOI] [PubMed] [Google Scholar]
- 27.Bochkarev, A., Pfuetzner, R. A., Edwards, A. M., and Frappier, L. (1997) Nature 385176 -181 [DOI] [PubMed] [Google Scholar]
- 28.Bochkarev, A., Bochkareva, E., Frappier, L., and Edwards, A. M. (1999) EMBO J. 184498 -4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bochkarev, A., and Bochkareva, E. (2004) Curr. Opin. Struct. Biol. 14 36-42 [DOI] [PubMed] [Google Scholar]
- 30.Theobald, D. L., Mitton-Fry, R. M., and Wuttke, D. S. (2003) Annu. Rev. Biophys. Biomol. Struct. 32115 -133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bastin-Shanower, S. A., and Brill, S. J. (2001) J. Biol. Chem. 27636446 -36453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, C., Paulus, B. F., and Wold, M. S. (1994) Biochemistry 3314197 -14206 [DOI] [PubMed] [Google Scholar]
- 33.Dimitrova, D. S., and Gilbert, D. M. (2000) Exp. Cell Res. 254321 -327 [DOI] [PubMed] [Google Scholar]
- 34.Philimonenko, A. A., Janacek, J., and Hozak, P. (2000) J. Struct. Biol. 132201 -210 [DOI] [PubMed] [Google Scholar]
- 35.Fuss, J., and Linn, S. (2002) J. Biol. Chem. 2778658 -8666 [DOI] [PubMed] [Google Scholar]
- 36.Fanning, E., and Pipas, J. M. (2006) in DNA Replication and Human Disease (DePamphilis, M. L., ed) pp.627 -644, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 37.Hurwitz, J., Dean, F. B., Kwong, A. D., and Lee, S. H. (1990) J. Biol. Chem. 26518043 -18046 [PubMed] [Google Scholar]
- 38.Crouch, E., Miller, S., Wilson, V., and Busbee, D. (1997) Mutat. Res. 374109 -123 [DOI] [PubMed] [Google Scholar]
- 39.Tatusova, T. A., and Madden, T. L. (1999) FEMS Microbiol. Lett. 174247 -250 [DOI] [PubMed] [Google Scholar]
- 40.Bochkareva, E., Korolev, S., Lees-Miller, S. P., and Bochkarev, A. (2002) EMBO J. 211855 -1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chattopadhyay, S., and Bielinsky, A. K. (2007) Mol. Biol. Cell 184085 -4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dodson, G. E., Shi, Y., and Tibbetts, R. S. (2004) J. Biol. Chem. 27934010 -34014 [DOI] [PubMed] [Google Scholar]
- 43.Olson, E., Nievera, C. J., Klimovich, V., Fanning, E., and Wu, X. (2006) J. Biol. Chem. 28139517 -39533 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.