Abstract
The APOBEC3 cytidine deaminases are part of the intrinsic defense of cells against retroviruses. Lentiviruses and spumaviruses have evolved essential accessory proteins, Vif and Bet, respectively, which counteract the APOBEC3 proteins. We show here that Bet of the Prototype foamy virus inhibits the antiviral APOBEC3C activity by a mechanism distinct to Vif: Bet forms a complex with APOBEC3C without inducing its degradation. Bet abolished APOBEC3C dimerization as shown by coimmunoprecipitation and cross-linking experiments. These findings implicate a physical interaction between Bet and the APOBEC3C. Subsequently, we identified the Bet interaction domain in human APOBEC3C in the predicted APOBEC3C dimerization site. Taken together, these data support the hypothesis that Bet inhibits incorporation of APOBEC3Cs into retroviral particles. Bet likely achieves this by trapping APOBEC3C protein in complexes rendering them unavailable for newly generated viruses due to direct immobilization.
The APOBEC3 (A3)3 genes form part of the intrinsic immunity against retroviruses (1), are under a high adaptive selection (2), and have undergone a unique evolutionary expansion from three to seven genes in primates (APOBEC3A (A3A), -B, -C, -DE, -F, -G, and -H) (3). In humans, A3F and A3G, can terminally edit human immunodeficiency virus type 1 (HIV-1) by deamination of cytidines to uridines during reverse transcription, in addition to inhibiting cDNA synthesis and integration (4-8).
The virion infectivity factor (Vif) of HIV actively counteracts this host-mediated restriction and prevents encapsidation of A3s (9-13). The binding of Vif to A3 proteins is species-specific, and thus may limit cross-species virus transmission (7). It was shown that Vif protects HIV-1 by binding human A3G and inducing its proteasomal degradation (14-18). In this system Vif acts as an adaptor protein, connecting A3G to an E3 ubiquitin ligase complex comprising ElonginB, ElonginC, Cullin5, and Ring box-1 (17). The Vif protein achieves this counterdefense via the SLQ motif within a SOCS box that mediates the interaction with ElonginC, and an HCCH motif to mediate the interaction with Cullin5 (19-21). The domains in Vif that mediate the interactions with A3G and A3F have also been characterized recently (22, 23).
Contrary to the well characterized A3-neutralizing activities of Vif proteins, the knowledge about A3-counteracting strategies used by retroviruses that do not encode a Vif protein is low. It was reported that murine leukemia virus can exclude murine (mu) A3 from virions without requiring an accessory protein (24, 25), although this remains controversial (26-28). In human T cell leukemia virus type 1, resistance to A3G is mediated by a cis-acting mechanism involving the nucleocapsid protein (29).
Simian foamy viruses are ubiquitous, non-pathogenic retroviruses that infect many primates. A low number of accidental infections of humans have been described, and the prototype foamy virus (PFV) originally isolated from an African cancer patient has been shown to be the end product of the zoonotic transmission of a chimpanzee simian foamy virus (30-33). Because simian foamy viruses have co-speciated with Old World primates for at least 30 million years (34), it was expected that, during the foamy virus/host co-evolution, viral defense strategies evolved to counteract A3 proteins. Indeed, several A3 proteins restrict foamy viruses by editing the viral genome, and it was shown that the FV accessory protein Bet is able to overcome this A3-mediated replication block (3, 35, 36). Löchelt et al. (35) demonstrated editing of feline foamy virus (FFV) genomes in feline (fe) A3C-positive cat cells only when the FFV Bet protein was functionally inactivated and showed that Bet binds feline A3Ca and thereby excludes it from virions. Russell et al. (36) demonstrated that human A3F and A3G proteins inhibit PFV due to a specific Gag-A3 interaction and induce guanine to adenine mutations in reverse transcripts. They also showed binding of PFV Bet to human A3F and -G, but not to muA3. In contrast, Delebecque et al. (37) did not find that Bet expression of PFV had any effect on the inhibitory activity of several A3 proteins.
In this study, we clearly demonstrate that Bet of PFV is an inhibitor of the antiviral activity of human A3C protein. Using PFV- and lentiviral-based reporter vectors, we found that Bet expressed in trans efficiently suppresses the A3-mediated inhibition of PFV and lentiviruses. Furthermore, we show that, unlike Vif, Bet does not induce degradation of A3C but instead inhibits dimerization of A3C. Chimeric A3C proteins of susceptible human and resistant rhesus A3C demonstrated that six amino acids (aa) in the region between aa 22 and aa 85 of A3C species-specifically modulate the binding of Bet to A3C.
EXPERIMENTAL PROCEDURES
Plasmids—Reporter virus for SIVagmTAN-1 (pSIVagm-Luc-R-E-Δvif) has been described (7). The PFV vector pczDWP002 is a variant of the PFV Gag/Pol-expressing vector pczDWP001 (38) where NLS-LacZ replaces the enhanced green fluorescent protein open reading frame. The PFV Env expression construct pczHFVenvEM002 has been described (39). The human (hu), African green monkey (agm), and chimpanzee (cpz) A3G-HA and muA3-HA expression constructs were provided by N. R. Landau (7). The pHuA3DE-V5 was a gift from Y.-H. Zheng (40). Plasmids expressing HA-tagged huA3C, huA3B, huA3F, huA3H, and feA3Ca have been described (35, 41). cDNAs of agmA3C, rhA3C, rhA3H, and ptA3G were obtained by reverse transcription-PCR on RNA of PHA/IL2-activated peripheral blood mononuclear cells from AGM, Rhesus (rh) and Pigtailed macaques (pt), respectively. To generate the pRhA3H-HA the forward (fw) primer JZ-ARP-10Fw.2 (5′-TAAGCGGAATTCGTGGCCAGAAGCACAGATCA-3′) and the reverse (rv) primer JZ-ARP-10Rv.3Tag (5′-AGAGCTCGAGTCAAGCGTAATCTGGAACATCGTATGGATATCTTGAGTTGTGTGTTGACG-3′) were used. To generate the pPtA3G-HA the fw primer CEM15-CM12 (5′-TAAGCGGAATTCCTTAGTCGGGACTAGCCGGC-3′) and the rv primer CEM15-HA-CII (5′-AGCTCGAGTCAAGCGTAATCTGGAACATCGTATGGATAGTTTTCCTGATTCTGGAGAATGGC-3′) were used. The amplicons were cloned into the EcoRI and XhoI sites of pcDNA3.1(+) (Invitrogen). Both the pRhA3C-HA and pAgmA3C-HA were generated using fw primer CEM15-BPKC2 (5′-TATGCGGCCGCTGAACATGAATCCACAGATCAGAAACCCG-3′) and rv primer CEM15-BPKC1 (5′-GTACTCGAGTCAAGCGTAATCTGGAACATCGTATGGATACTGAAGAATCTCCCGTAGGCGTC-3′). The amplicons were cloned into the NotI and XhoI sites of pcDNA3.1(+). V5-tagged rhA3C was generated using fw primer RhAPO3C (5′-TATAAGCTTTGAAGAGGAATGAATCCACAGATCAGAAACC-3′) and rv primer rhA3C-V5-XhoI rc (5′-GGCTCGAGTCACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCCTGAAGAATCTCCCGTAGGCG-3′). The amplicon was cloned into the HindIII and XhoI sites of pcDNA3.1(+). V5-tagged huA3C has been described previously (10). pBetPFV was generated by insertion of the PFV Bet sequence, which was obtained from HEL299 cells infected with the PFV isolated by Achong et al. (32) into the HindIII and SmaI sites of pBC12-CMV (42). pVifHIV-1 was provided by N. R. Landau (43). pVifagmTAN-1 plasmid, also a gift from N. R. Landau, was created in an analogous manner. huTRIM5α expression plasmid pLNCX2-huTRIM5αHA was provided by T. Hatziioannou (44). To generate the Bet bacterial expression plasmid pEXP5-BetHFV-6xHis, PCR was performed with primer BetHFV-start and BetHFV-end and pBetPFV as template. The amplicon was cloned into pEXP5-CT/TOPO (Invitrogen). To generate the bacterial expression plasmid for huA3C-HA, pEXP5-6xHIS-TEV-hA3C-HA, PCR was performed with primer hA3C-start and HA-stop_r and phuA3C-HA as template. The amplicon was cloned into pEXP5-NT/TOPO (Invitrogen).
Construction of rh/huA3C Chimeras—The HA-tagged rh/hu chimeras were constructed by PCR amplification of the corresponding part of huA3C or rhA3C, gel purification of the resulting products, followed by a second PCR amplification primed by complementary sequences within each product and flanking primers fw RhAPO3C (5′-TATAAGCTTTGAAGAGGAATGAATCCACAGATCAGAAACC-3′) and rv RhAPOend (5′-AGCTCGAGTCAAGCGTAATCTGGAACATCGTATGGATACTGAAGAATCTCCCGTAGGCG-3′). To generate prh/hu17, the N-terminal rhA3C part was amplified using the fw primer RhAPO3C and the rv primer Rhehu 1.1 (5′-tccacggtgaagcacagc-3′), the central huA3C part was amplified using the fw primer Rhehu 1.2 (5′-GCTGTGCTTCACCGTGGA-3′) and the rv primer Rhehu 9.1 (5′-aagggctccaagatgtgtac-3′) and the C-terminal rhA3C part was amplified using the fw primer Rhehu 9.2 (5′-GTACACATCTTGGAGCCCTT-3′) and the rv primer RhAPOend. To generate prh/hu19, the N-terminal rhA3C part was amplified using the fw primer RhAPO3C and the rv primer Rhehu 1.1, the central huA3C part was amplified using the fw primer Rhehu 1.2 and the rv primer Rhehu 7.1 (5′-AGGAAGCACCTTTCTGCATG-3′), and the C-terminal rhA3C part was amplified using the fw primer Rhehu 7.2 (5′-catgcagaaaggtgcttcct-3′) and the rv primer RhAPOend. To generate V5-tagged prh/hu19 HA-tagged prh/hu19 served as template and RhAPO3C was used as fw primer and rhA3C-V5-XhoI rc as rv primer. To generate prh/hu29, the N-terminal rhA3C part was amplified using the fw primer RhAPO3C and the rv primer Rhehu 5.1 (5′-TTCGGAAGACGCCCGTCT-3′), the central huA3C part was amplified using the fw primer Rhehu 5.2 (5′-AGACGGGCGTCTTCCGAA-3′) and the rv primer Rhehu 7.1 and C-terminal rhA3C part was amplified using the fw primer Rhehu 7.2 and the rv primer RhAPOend. The full-length amplicons were cloned into the HindIII and XhoI sites of pcDNA3.1(+).
Cells, Transfections, and Protein Expression—Cell lines BHK-LL, HOS, and 293T, were maintained in Dulbecco's modified Eagle's medium complete (Invitrogen) supplemented with 10% fetal bovine serum, 0.29 mg/ml l-glutamine, and 100 units/ml penicillin/streptomycin. Plasmid transfection into 293T and BHK-LL cells was done with LipofectamineLTX (Invitrogen); only PFV plasmid transfection was done with Polyfect (Qiagen) with a DNA:Polyfect ratio of 1:2.5. PFV stocks were generated by transfection of 293T cells in 6-well plates with 1 μg of pczHSRV. PFV containing supernatant was harvested 2 days later, serially diluted and used for infection on BHK-LL cells. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside staining of cells 3 days after infection obtained titer. Reporter lentiviruses were generated by transfection in 6-well plates with 1.5 μg of pSIVagm-Luc-E-R-Δvif, 0.5 μg of VSV-G expression plasmid pMD.G, 1 μg of A3 expression plasmid, and 2 μg of pVifHIV or pVifagmTAN-1 or pBetPFV. PFV reporter vectors were generated in 12-well plates with 1 μg of pczDWP002, 1 μg of pczHFVenvEM02, 0.33 μg of A3 expression plasmid, and 0.66 μg of pBetPFV. Total plasmid DNA was maintained at 4.5 or 3 μg, respectively, by the addition of pcDNA3.1 as needed. Reverse transcriptase (RT) activity of SIV-containing supernatants was determined using the Cavidi HS kit Lenti RT or the C-type RT Activity Kit (Cavidi Tech) for PFV vectors. For infectivity assays HOS cells were transduced in 96-well plates in triplicate with a virus amount equivalent to 10 pg of RT for SIV vectors and 20 milliunits for PFV. Three days post infection (dpi), luciferase activity was measured using the Steadylite HTS kit (PerkinElmer) or β-galactosidase activity by the Galacto-Star kit (Applied Biosystems). Data are presented as counts/s normalized to virus obtained in the absence of A3 and Bet. All investigations using reporter viruses were performed in at least three independent experiments. For in vitro expression of pEXP5-6xHIS-TEV-hA3C-HA and pEXP5-BetHFV-6xHis an Expressway Cell-Free E. coli Expression System (Invitrogen) was used. 30 min after initiation of the protein synthesis reaction, feed buffer was added. The samples were incubated for 6 h at 30 °C and clarified by centrifugation (10 min, 15,000 × g).
Immunoblot Analysis—To generate protein lysates, cells were lysed in radioimmune precipitation assay buffer for 5 min on ice. Lysates were cleared by centrifugation. For cross-linking experiments lysates were generated with phosphate-buffered saline-radioimmune precipitation assay buffer (phosphate-buffered saline instead of Tris), and cleared samples were subsequently treated with NEM at a final concentration of 31 mm for 2 h at room temperature. For cross-linked samples, non-reducing SDS-PAGE was performed. For protein turnover experiments, 293T cells were incubated prior to lysis with cell culture medium containing 100 μg/ml cycloheximide. To generate virus lysates, supernatant samples were layered on a 20-ml 20% w/v sucrose cushion (in phosphate-buffered saline), and virions were pelleted at 35,000 rpm (SW-41 rotor) for 2 h at 4 °C. Pellets were resuspended with radioimmune precipitation assay buffer and normalized to reverse transcriptase values of 500 pg determined from original supernatants. The levels of p27 and HA-tagged A3 proteins were analyzed by immunoblots. For co-immunoprecipitation experiments, 293T cells were transfected with pBetPFV and HA-tagged or V5-tagged A3 expression plasmids. After 2 days, cells were lysed and lysates were cleared by centrifugation, and supernatants were incubated with anti-HA beads (Roche Diagnostics) for 60 min at 4 °C and washed (5×) with radioimmune precipitation assay buffer. For co-immunoprecipitation of in vitro expressed Bet and huA3C, samples were incubated with anti-HA-beads (Roche Applied Science) for 60 min at 4 °C. RNA digestion was performed using a final concentration of 50 μg/ml RNase A (AppliChem). For immunoblot analysis, samples were boiled in NuPAGE LDS Sample Buffer and NuPAGE Sample Reducing Agent (Invitrogen) and subjected to SDS-PAGE followed by transfer to a polyvinylidene difluoride membrane. Proteins were detected using an anti-HA antibody (Ab) (1:104 dilution, MMS-101P, Covance) or an α-V5 Ab (1:5,000 dilution, MCA1360, ABDserotek). For the detection of BetPFV, a Bel2/Bet-specific hyper-immune serum (1:50,000 dilution) was used (45). Equal loading of cell lysates was confirmed on immunoblots probed with α-tubulin Ab (1:104 dilution, B5-1-2, Sigma) and equal loading of virions with α-p24/p27 mAb AG3.0 (46). Antimouse and anti-rabbit horseradish peroxidase (Amersham Biosciences) were used as secondary Abs. Signals were visualized using ECL (Amersham Biosciences).
Data Deposition—The following sequences were deposited in the GenBank™ data base: rhA3H, NM_001042372; ptA3G, DQ093376; and BetPFV, EU381420.
RESULTS
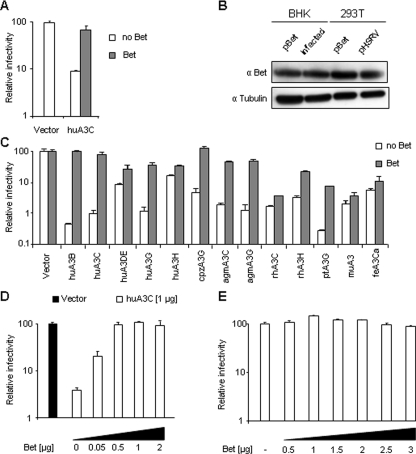
Broad and Pan-species Anti-APOBEC3 Activity of the Bet Protein—Our studies on FFV revealed that A3C but not A3H or A3CH of the cat reduced the infectivity of Bet-deleted but not of wt FFV (3, 35). The huA3C is more widely and higher expressed than huA3F and huA3G (47, 48) and represents one of the three evolutionary oldest A3 genes (3, 49). Therefore, we questioned if PFVΔbet like SIVΔvif (50) is targeted by huA3C and whether Bet can counteract huA3C, as shown for huA3F and huA3G (36). As shown in Fig. 1A, huA3C reduced the infectivity of PFV Δbet 10-fold. Bet provided in trans restored the infectivity to levels of A3-free samples. In this experiment, bet-deficient β-galactosidase reporter viruses were generated by co-transfection of a PFV and a huA3C expression plasmid into 293T cells in combination with a Bet expression plasmid. HOS cells were then transduced with reverse transcription-activity normalized virions and 3 dpi the infectivity of the viruses was determined by quantitation of intracellular β-galactosidase. The cytomegalovirus promoter-driven Bet expression plasmid generated similar amounts of Bet protein compared with PFV-infected or -transfected cells (Fig. 1B). For infection experiments (multiplicity of infection 0.5) BHK cells had to be used because FV proteins are weakly expressed in 293T cells upon infection (51) and were compared with transfected BHK cells (transfection efficiency of 50%, data not shown). Bet has likely additional functions in the foamy virus replication (52-54). Therefore, we tested Bet also in a heterologous viral system (SIVagmΔvif-Luc (7)) to study the mechanism of A3 counteraction.
FIGURE 1.
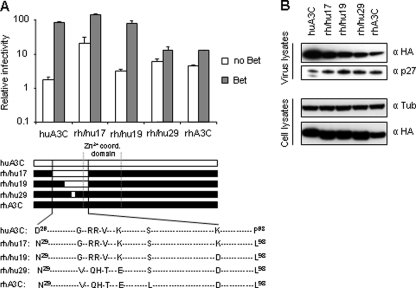
Broad-range protection of SIV-based vectors by Bet to A3-mediated restriction. A, Bet-deficient lacZ-reporter viruses derived from PFV were produced in the absence (Vector) or presence of huA3C and Bet. Infectivity of normalized amounts of viruses was determined by quantification of β-galactosidase expression 3 days post infection (dpi). Infectivity, relative to the virus generated in absence of A3 and Bet, is given. B, the expression level of Bet in baby hamster kidney (BHK) cells was compared after transfection with Bet expression plasmid pBet and after infection with PFV at multiplicity of infection of 0.5 as well as in 293T cells after transfection with pBet and pHSRV (plasmid encoding full-length PFV genome). Cell lysis and immunoblot analysis were performed 2 days after transfection and infection, respectively. Bet was detected by using αBet antiserum. Equal loading of cell lysate samples was confirmed with α-tubulin antibody. C, Vif-deficient luciferase reporter viruses derived from SIVagm were produced in the absence (Vector) or presence of the indicated A3 and Bet proteins. Infectivity of equal amount of viruses was determined by quantification of luciferase activity 3 dpi. Infectivity, relative to the virus generated in absence of A3 and Bet, is shown. D, Δvif SIVagm-luc viruses were produced in the absence (Vector) or presence of huA3C and increasing amounts of Bet, or Bet alone (E). Infectivity of equal amount of viruses was determined by quantification of luciferase activity 3 dpi.
Because a pilot experiment supported a virus-type-independent activity of Bet against A3C, all further experiments on the mechanism of Bet-mediated A3 inhibition were performed with the Δvif SIVagm-derived reporter virus. Screening of several A3 proteins with Δvif SIV for Bet susceptibility demonstrated that Bet counteracted most primate A3s. In detail, Bet increased the infectivity of SIV in the presence of huA3B by 221-fold, huA3C by 81-fold, huA3G by 31-fold, cpzA3G by 28-fold, agmA3C by 24-fold, agmA3G by 38-fold, and ptA3G by 27-fold, but only weakly affected the infectivity in the presence of rhA3H by 6-fold, huA3DE by 3-fold, and rhA3C, huA3H, muA3 and feA3Ca by 2-fold each (Fig. 1C). These results show that Bet can protect not only PFV, but also nonrelated lentiviruses like SIV against many primate A3s.
The inhibition of huA3C by the Bet protein was clearly dose-dependent: 0.05 μg of Bet plasmid significantly enhanced the infectivity of SIV, and the antiviral activity of huA3C was completely neutralized using 0.5 μg of Bet-expression plasmid (0.5-fold of huA3C expression plasmid) (Fig. 1D). Increasing amounts of Bet without co-transfection of A3 did not change the infectivity of SIV (Fig. 1E) or influenced the release of virions (data not shown).
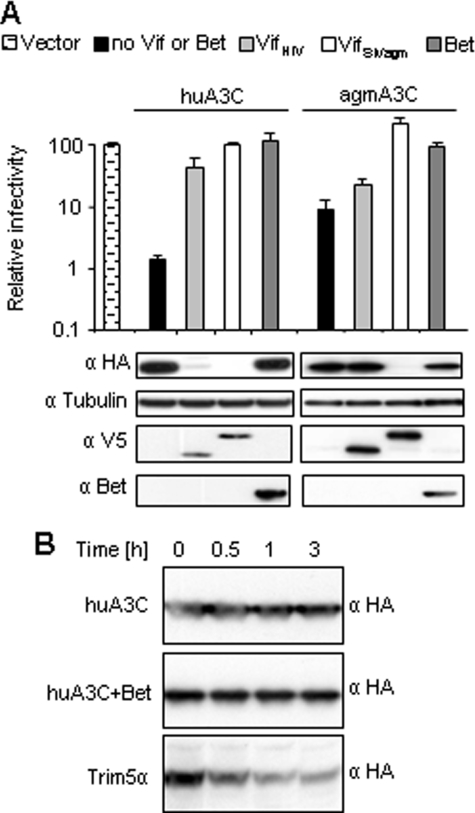
Because Vif promotes the depletion of A3 by inducing its proteasomal degradation (14-18), we directly compared the effect of Vif and Bet co-transfection on the A3C protein steady-state level. Vif of SIVagm and HIV-1 induced the depletion of susceptible A3Cs, which fully correlated with restored virus titers (Fig. 2A). In contrast, Bet did not induce degradation, despite counteracting the A3 restriction (Fig. 2A). The huA3C was degraded by both Vif proteins and agmA3C only by VifSIVagm. Equal protein loading was controlled by anti-tubulin staining.
FIGURE 2.
Bet inhibits A3C antiviral activity without leading to A3C degradation. A, Δvif SIVagm-luc viruses were produced in the presence or absence (Vector) of the indicated HA-tagged A3, V5-tagged VifHIV-1, and VifSIVagm and Bet expression vectors. Immunoblot analysis was performed with lysates of virus-producing cells. Expression of A3 proteins was detected with α-HA mAb, tubulin with α-tubulin mAb, Vif with α-V5 mAb, and Bet with Bet antiserum. Relative infectivity (relative to the virus generated in absence of A3 and Bet) of equal amounts of the reporter viruses is shown, demonstrating a close correlation of Vif-mediated degradation of A3 and protection, whereas Bet-mediated rescue is not associated with decreased A3 levels. Infectivity of viruses was determined by quantification of luciferase activity 3 dpi. B, 293T cells transiently expressing HA-tagged huA3C alone or together with Bet, and HA-tagged huTRIM5α were treated with cycloheximide. At the indicated time points after cycloheximide-induced suppression of protein biosynthesis, the cells were lysed and analyzed by immunoblotting using α-HA mAb.
To further confirm that Bet does not promote degradation of huA3C, we investigated the stability of huA3C in a time-course experiment by treating cells with cycloheximide, an inhibitor of translational elongation. Immunoblot analysis at the indicated time points revealed that huA3C is stable for at least 3 h (Fig. 2B), and the presence of Bet did not alter this stability. Proper poisoning of the protein biosynthesis was confirmed by analysis of huTRIM5α that showed the expected short half-life (55). Together the experiments demonstrate that Bet of PFV counteracts huA3C protein without inducing its degradation.
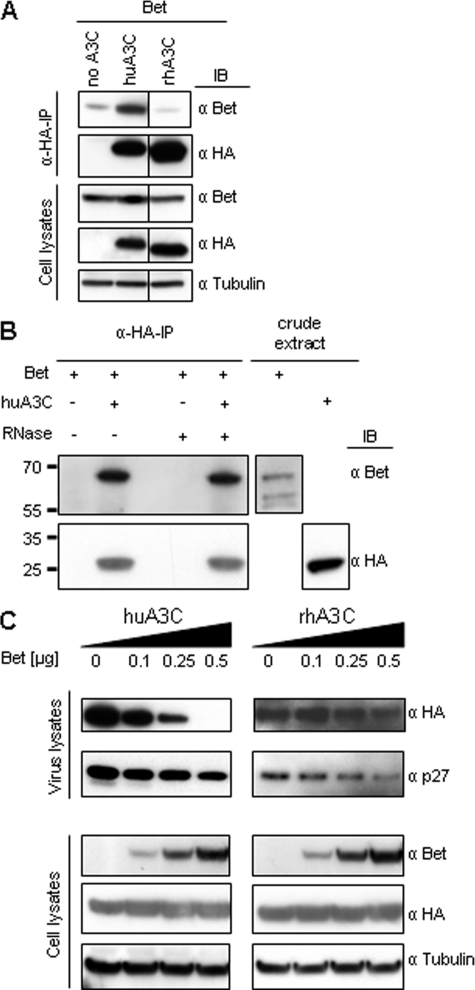
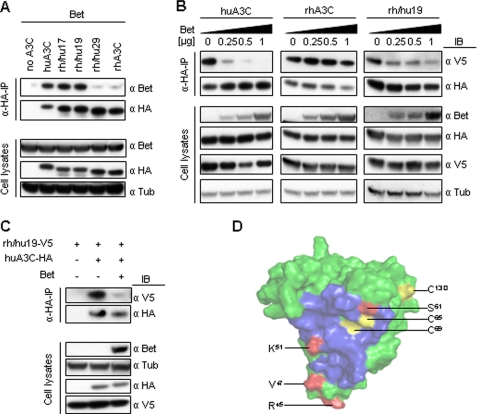
Bet Binds A3C—To test whether interaction of A3C with Bet is crucial for the abolishment of its anti-viral activity, we performed pulldown assays. We compared huA3C and rhA3C that are both proteins with strong anti-SIVagm activity but differ in their susceptibility to Bet. Immunoblot analyses were performed with samples after HA-immunoprecipitation and on whole cell lysates (Fig. 3A). The signal of co-precipitated Bet, detected with Bet-specific antibodies, was significantly stronger in the sample with huA3C compared with the sample with rhA3C that was similar to the background level of nonspecifically bound Bet (Fig. 3A). Both A3C proteins bound equally to the anti-HA beads (Fig. 3A). Immunoblot analysis of cell lysates showed the presence of Bet, the indicated A3 proteins, and equal protein loading by the anti-tubulin staining (Fig. 3A). These data demonstrate a clear correlation between physical interaction of Bet and the ability to neutralize the antiviral activity of huA3C. To test whether Bet binds directly to huA3C without host cellular proteins, we performed co-immunoprecipitations with Bet and huA3C expressed in vitro using Escherichia coli extracts. To additionally exclude the involvement of RNA, the samples were treated with RNase A. Immunoblot analyses performed with extracts and after HA-immunoprecipitation show that Bet precipitates only in presence of huA3C in a RNA-independent manner (Fig. 3B).
FIGURE 3.
Bet binds to huA3C and prevents its incorporation in virus particles. A, cells were transfected with Bet alone or with expression plasmids for HA-tagged huA3C and rhA3C. Cell lysates were prepared and α-HA-immunoprecipitation (α-HA-IP) was performed. A3-associated Bet was detected by immunoblot (IB) analysis using Bet antiserum. A3 proteins were detected in α-HA-IP samples and in cell lysates using α-HA mAbs, as well as Bet antiserum and α-tubulin antibodies in cell lysates. B, Bet-huA3C interaction occurs directly in an RNA-independent manner: Bet and huA3C were expressed in vitro using E. coli extracts, and α-HA-immunoprecipitations (α-HA-IP) were performed in the presence and absence of RNase A. Bet- and HA-tagged huA3C were detected in crude extracts and in immunoprecipitates by immunoblot analysis using Bet antiserum and α-HA mAb, respectively. C, virions were generated by co-transfection of Δvif SIVagm-luc, HA-tagged huA3C, or rhA3C and increasing amounts of Bet expression vector. Virions and cells were analyzed by immunoblot. A3C was detected with α-HA mAb, Bet with Bet antiserum, tubulin with α-tubulin mAb, and Capsid p27 protein with mAb to p24/p27.
To confirm that binding of Bet excludes huA3C from viral particles, immunoblot analysis was performed on virions produced with huA3C protein and increasing amounts of Bet. The results show that Bet strongly reduced huA3C-specific bands in virus samples, did not affect A3C levels in virus-producing cells and did not inhibit the viral packaging of rhA3C (Fig. 3C). We conclude, the interaction of Bet with huA3C prevents its incorporation into viral particles.
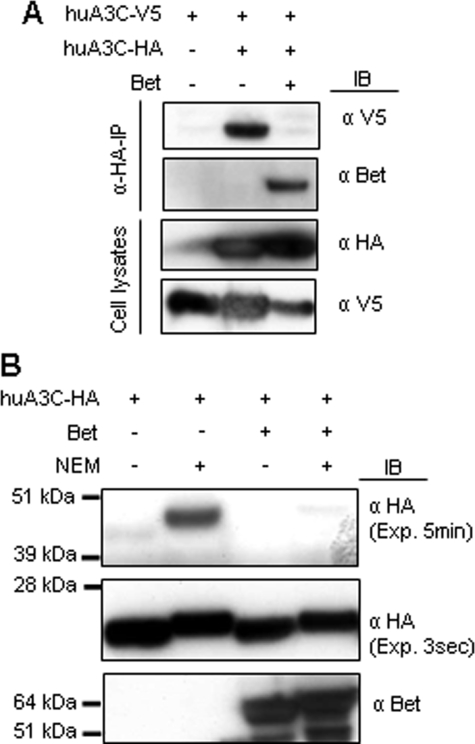
Bet Inhibits A3C-A3C Interaction—To analyze if Bet inhibits A3C-A3C interactions, we studied the dimer formation of huA3C in the presence of Bet. Co-precipitation experiments were done with HA-tagged and V5-tagged huA3C. The results show that A3C-HA co-precipitated A3C-V5 (Fig. 4A). Interestingly, these A3-A3 interactions were not detectable when Bet was co-expressed. To address the question of whether the A3-A3 complexes shown in Fig. 4A are in fact dimers of huA3C, we cross-linked the A3 proteins and analyzed the cell lysates by immunoblotting. Clarified lysates of 293T cells, transiently expressing huA3C in combination with Bet, were treated with N-ethylmaleimide (NEM), a sulfhydryl alkylating reagent. NEM cross-links proteins via cysteine residues provided they are sterically close to each other. Immunoblots of untreated cell lysates displayed the expected A3 band of ∼23 kDa for A3C (Fig. 4B). NEM-treated samples showed an additional band with the molecular weight of ∼46 kDa, which is in agreement with an A3C dimer (Fig. 4B). This presumed huA3C-dimer band was only faintly detectable in the presence of Bet (Fig. 4B). Based on this, we conclude that Bet can disrupt or prevent A3C dimers.
FIGURE 4.
Bet inhibits dimerization of huA3C. A, cells were transfected with V5-tagged huA3C alone or additionally with HA-tagged huA3C in presence or absence of Bet. Cell lysates were prepared, and α-HA-immunoprecipitation (α-HA-IP) was performed. huA3C-V5 co-precipitated with HA-tagged A3; Bet was detected by immunoblot (IB) analysis using Bet antiserum, huA3C-V5 using α-V5 mAb, and huA3C-HA using α-HA mAbs. B, lysates of cells expressing HA-tagged huA3C in the presence or absence of Bet were prepared and cross-linked with NEM. Monomeric and dimeric forms of huA3C were detected with α-HA mAb, whereas Bet was detected using Bet antiserum. Exp: exposure time.
Bet Interacts with the Predicted Dimerization Site of A3C—The observation that Bet interacts with and counteracts huA3C but displayed only minor reactivity against rhA3C (Figs. 1C and 3A), allowed us to map the Bet-A3C interaction site using hu/rhA3C chimeras. A total of 40 chimeric A3Cs was generated and tested for inhibition of the Δvif SIV-Luc reporter virus and susceptibility to Bet. Only chimeras with protein expression levels comparable to parental huA3C and rhA3C and a strong capacity to reduce infectivity were used for further analysis (examples given in Fig. 5, A and B). The chimera with the shortest section of huA3C that possessed full activity against Δvif SIV-Luc and was susceptible to Bet was the A3C-chimera rh/hu19 (Fig. 5A). In this chimera, the huA3C insert (aa 40-86) differs from the corresponding rhA3C only in 6 amino acids (G41V, R44Q, R45H, V47T, K51E, and S61L). Mutant rhA3C-L61S (rh/hu29) exhibits the same behavior as rhA3C with a strong antiviral activity and low level susceptibility to Bet (Fig. 5A). All chimeras were expressed and packaged into virions at similar level as parental huA3C and rhA3C (Fig. 5B). The chimera rh/hu19 showed interaction with Bet-like huA3C and rh/hu17, whereas rh/hu 29 failed to interact with Bet-like rhA3C (Fig. 6A). Homo-dimerization of the chimera rh/hu19 was sensitive to Bet as shown for huA3C, whereas the homo-dimerization of rhA3C was not affected by the co-expression of Bet (Fig. 6B). Finally, the hetero-dimerization of chimera rh/hu19 with the huA3C was also inhibited by Bet (Fig. 6C). The same observations were made using rhA3C-E51K (data not shown). In line with these chimeras, efforts to abolish the functional huA3C-Bet interaction by single amino acid exchanges of R44Q, R45H, V47T, or K51E failed (data not shown). Hence, neither Lys-51 and Ser-61 in the background of rhA3C nor Gln-44, His-45, Thr-47, and Glu-51 in the background of huA3C can individually modulate the interaction with Bet. However, all these residues are located on the surface close to the predicted dimerization interface of huA3C (blue area in Fig. 5D)4. Together our data indicate that Bet prevents dimerization of huA3C due to its binding close to or within the A3 dimerization site.
FIGURE 5.
Mapping of the Bet binding site within huA3C. A, Δvif SIVagmluc viruses were produced in the presence or absence of the indicated wt and rh/hu chimeric A3 and Bet proteins. Infectivity of equal amount of viruses was determined by quantification of luciferase activity, and is shown relative to virus generated in absence of A3 and Bet. Alignment of huA3C, rhA3C, and chimeric protein sequences (rh/hu17, -19, and -29) between aa 29 and 98 relative to the linear protein sequence with indicated Zn2+ coordination domain are shown. Identical aa to human and rhesus A3C are represented by a dashed line, aa that differ are shown, and human A3C-derived aa are highlighted in bold font. B, chimeric proteins are expressed and packaged into virions at similar level as parental huA3C and rhA3C. Expression and packaging were tested by immunoblot analysis of virus and cell lysates by using α-HA mAbs. Equal loading of samples was confirmed by detecting tubulin with α-tubulin mAb and Capsid p27 with mAb to p24/p27.
FIGURE 6.
Bet interacts within the predicted dimerization site of huA3C. A, cells were transfected with Bet alone or with expression plasmids for HA-tagged huA3C, rhA3C, and indicated chimeras. Cell lysates were prepared, and α-HA-immunoprecipitation (α-HA-IP) was performed. A3-associated Bet was detected by immunoblot (IB) analysis using Bet antiserum. A3 proteins were detected in α-HA-IP samples and in cell lysates using α-HA mAbs, filters of cell lysates were also stained for Bet and tubulin. B, V5-tagged and HA-tagged huA3C (left panel), rhA3C (middle panel), and chimera rh/hu19 (right panel) were expressed in 293T cells in the presence of increasing amounts of Bet. Cell lysates were prepared, and α-HA-immunoprecipitation (α-HA-IP) was performed. Bet was detected by immunoblot (IB) analysis using Bet antiserum, V5-tagged proteins were detected using α-V5 mAb and HA-tagged proteins using α-HA mAbs. C, V5-tagged rh/hu19 was co-expressed with HA-tagged huA3C in the presence and absence of Bet. D, surface of huA3C modeled on the crystal structure of a complex of dimeric APOBEC2, including the predicted dimerization interface (blue), cysteine residues (yellow), and residues believed to mediate direct physical interaction with Bet (red).
DISCUSSION
Members of the APOBEC3 cytidine deaminase family exhibit a strong antiviral activity against viruses with single strand DNA replication intermediates (retroviruses, hepadnaviruses, and adeno-associated viruses) (57). The viral strategies to counteract A3 proteins are only partially understood and encompass exclusion of A3 proteins from virions by induction of proteasomal A3 degradation by the viral protein Vif (14-18) and by impaired nucleocapsid interaction (29). In addition, Vif might inhibit packaging of A3G by other less characterized mechanisms (7, 16, 58). We describe here and confirm the initial data from Russell et al. (36) that the PFV accessory protein Bet counteracts A3 proteins by yet another distinct mechanism. We evaluated the PFV Bet-mediated protection in the homologous (PFV) and in a heterologous read-out system (Δvif SIV): the Bet protein of PFV is active against most human and non-human primate A3s, in line with the occurrence of zoonotic transmissions of chimpanzee FV to man (59). Of notable exception, rhA3C, huA3DE, and huA3H were resistant, and more distantly related non-primate A3s from mouse and cat were also not inactivated by PFV Bet. Previous and current analyses failed to detect similarities between Bet and Vif or other cellular or viral proteins (data not shown and Ref. 60).
In all known FVs, productively infected cells show very high expression of Bet (61-63). The Bet protein of PFV and FFV is not encapsidated in retroviral particles (data not shown and Refs. 35, 36). Essential high Bet expression in vivo and in vitro could be related to its mode of anti-A3 function. In addition to its A3 inhibitory activity, Bet might fulfill other functions during FV replication: as a regulator of expression and particle release and an inhibitor of superinfection (52-54). During the course of our studies, we found that low level Bet expression plasmids failed to counteract many A3s but not huA3C (data not shown). This observation might explain the negative results of Bet on the A3 activity obtained by others (37).
Our experiments imply a physical interaction between Bet and susceptible A3 proteins, which we showed by co-immunoprecipitations of Bet and A3Cs. Bet, in contrast to Vif, did not induce protein degradation of A3C. However, we demonstrate here that Bet inhibits A3-A3 complex formation of huA3C. The Betinsensitive rhA3C was a helpful tool to map the Bet binding region in huA3C. This Bet binding site in A3C localizes to the predicted A3C dimerization site, thereby explaining the capacity of Bet to disrupt or prevent huA3C homodimers. A3C proteins bound to Bet likely in a monomeric form. This correlated with a lost in the capacity to be incorporated into viral particles. In a simple model, either A3C requires dimerization to get packaged into virions or the binding of Bet to A3C blocks the A3 domain required for virus encapsidation. Alternatively, the presumed intrinsic virus exclusion domain of Bet could be dominant active in a Bet-A3 complex. It remains unknown whether Bet moves A3C to a subcellular compartment, which resists simple detection in the microscope (data not shown) or forms insoluble structural complexes that either inhibit the interaction of bound A3C with the viral Gag protein or sequesters it away from progeny virions.
The dynamic evolution of host-retrovirus interactions resulted in at least two basically different viral defense proteins (Bet and Vif) to inhibit the antiviral cytidine deaminases of the A3 family. Why and how foamy and lentiviruses developed or acquired these unique proteins rather than simply avoiding packaging of A3s into viral particles, like human T cell leukemia virus and MuLV, remain open questions for further studies.
Acknowledgments
We thank Marion Battenberg, Nico Scharpfenecker, and Benjamin Rengstel for expert technical assistance and Nathaniel R. Landau, Theodora Hatziioannou, and Yong-Hui Zheng for the gift of reagents. The following reagents were obtained through the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pcDNA3.1-APOBEC3C-V5-6XHis from Matija Peterlin and Yong-Hui Zheng and monoclonal antibody to HIV-1 p24 (AG3.0) from Jonathan Allan.
The atomic coordinates and structure factors (codes NM_001042372, DQ093376, and EU381420) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grant from the Intramural Research Program of NCI, NIH, Center for Cancer Research. This work was also supported by the DFG (Grant MU 1608/3-1 to C. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: A3, APOBEC3; HIV-1, human immunodeficiency virus, type 1; Vif, virion infectivity factor; mu, murine; PFV, prototype foamy virus; FFV, feline foamy virus; fe, feline; aa, amino acid(s); agm, African green monkey; cpz, chimpanzee; hu, human; HA, hemagglutinin; rh, Rhesus; pt, Pigtailed macaques; RT, reverse transcription; SIV, simian virus; dpi, daps post infection; mAb, NEM, N-ethylmaleimide.
B. Stauch, H. Hofmann, M. Weisel, K. Cichutek, C. Münk, and G. Schneider, unpublished data.
References
- 1.Bieniasz, P. D. (2004) Nat. Immunol. 5 1109-1115 [DOI] [PubMed] [Google Scholar]
- 2.Sawyer, S. L., Emerman, M., and Malik, H. S. (2004) PLoS Biol. 2 E275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Münk, C., Beck, T., Zielonka, J., Hotz-Wagenblatt, A., Chareza, S., Battenberg, M., Thielebein, J., Cichutek, K., Bravo, I. G., O'Brien, S., Loechelt, M., and Yuhki, N. (2008) Genome Biology 9 R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mbisa, J. L., Barr, R., Thomas, J. A., Vandegraaff, N., Dorweiler, I. J., Svarovskaia, E. S., Brown, W. L., Mansky, L. M., Gorelick, R. J., Harris, R. S., Engelman, A., and Pathak, V. K. (2007) J. Virol. 81 7099-7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo, K., Wang, T., Liu, B., Tian, C., Xiao, Z., Kappes, J., and Yu, X. F. (2007) J. Virol. 81 7238-7248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mangeat, B., Turelli, P., Caron, G., Friedli, M., Perrin, L., and Trono, D. (2003) Nature 424 99-103 [DOI] [PubMed] [Google Scholar]
- 7.Mariani, R., Chen, D., Schröfelbauer, B., Navarro, F., König, R., Bollman, B., Münk, C., Nymark-McMahon, H., and Landau, N. R. (2003) Cell 114 21-31 [DOI] [PubMed] [Google Scholar]
- 8.Holmes, R. K., Malim, M. H., and Bishop, K. N. (2007) Trends Biochem. Sci. 32 118-128 [DOI] [PubMed] [Google Scholar]
- 9.Sheehy, A. M., Gaddis, N. C., Choi, J. D., and Malim, M. H. (2002) Nature 418 646-650 [DOI] [PubMed] [Google Scholar]
- 10.Zheng, Y. H., Irwin, D., Kurosu, T., Tokunaga, K., Sata, T., and Peterlin, B. M. (2004) J. Virol. 78 6073-6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiegand, H. L., Doehle, B. P., Bogerd, H. P., and Cullen, B. R. (2004) EMBO J. 23 2451-2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liddament, M. T., Brown, W. L., Schumacher, A. J., and Harris, R. S. (2004) Curr. Biol. 14 1385-1391 [DOI] [PubMed] [Google Scholar]
- 13.Bishop, K. N., Holmes, R. K., Sheehy, A. M., Davidson, N. O., Cho, S. J., and Malim, M. H. (2004) Curr. Biol. 14 1392-1396 [DOI] [PubMed] [Google Scholar]
- 14.Marin, M., Rose, K. M., Kozak, S. L., and Kabat, D. (2003) Nat. Med. 9 1398-1403 [DOI] [PubMed] [Google Scholar]
- 15.Mehle, A., Strack, B., Ancuta, P., Zhang, C., McPike, M., and Gabuzda, D. (2004) J. Biol. Chem. 279 7792-7798 [DOI] [PubMed] [Google Scholar]
- 16.Stopak, K., de Noronha, C., Yonemoto, W., and Greene, W. C. (2003) Mol. Cell 12 591-601 [DOI] [PubMed] [Google Scholar]
- 17.Yu, X., Yu, Y., Liu, B., Luo, K., Kong, W., Mao, P., and Yu, X. F. (2003) Science 302 1056-1060 [DOI] [PubMed] [Google Scholar]
- 18.Sheehy, A. M., Gaddis, N. C., and Malim, M. H. (2003) Nat. Med. 9 1404-1407 [DOI] [PubMed] [Google Scholar]
- 19.Mehle, A., Goncalves, J., Santa-Marta, M., McPike, M., and Gabuzda, D. (2004) Genes Dev. 18 2861-2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu, Y., Xiao, Z., Ehrlich, E. S., Yu, X., and Yu, X. F. (2004) Genes Dev. 18 2867-2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo, K., Xiao, Z., Ehrlich, E., Yu, Y., Liu, B., Zheng, S., and Yu, X. F. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 11444-11449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian, C., Yu, X., Zhang, W., Wang, T., Xu, R., and Yu, X. F. (2006) J. Virol. 80 3112-3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russell, R. A., and Pathak, V. K. (2007) J. Virol. 81 8201-8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi, M., Takaori-Kondo, A., Shindo, K., Abudu, A., Fukunaga, K., and Uchiyama, T. (2004) J. Virol. 78 8238-8244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doehle, B. P., Schafer, A., Wiegand, H. L., Bogerd, H. P., and Cullen, B. R. (2005) J. Virol. 79 8201-8207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abudu, A., Takaori-Kondo, A., Izumi, T., Shirakawa, K., Kobayashi, M., Sasada, A., Fukunaga, K., and Uchiyama, T. (2006) Curr. Biol. 16 1565-1570 [DOI] [PubMed] [Google Scholar]
- 27.Browne, E. P., and Littman, D. R. (2008) J. Virol. 82 1305-1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rulli, S. J., Jr., Mirro, J., Hill, S. A., Lloyd, P., Gorelick, R. J., Coffin, J. M., Derse, D., and Rein, A. (2008) J. Virol. 82 6566-6575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Derse, D., Hill, S. A., Princler, G., Lloyd, P., and Heidecker, G. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 2915-2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolfe, N. D., Switzer, W. M., Carr, J. K., Bhullar, V. B., Shanmugam, V., Tamoufe, U., Prosser, A. T., Torimiro, J. N., Wright, A., Mpoudi-Ngole, E., McCutchan, F. E., Birx, D. L., Folks, T. M., Burke, D. S., and Heneine, W. (2004) Lancet 363 932-937 [DOI] [PubMed] [Google Scholar]
- 31.Calattini, S., Betsem, E. B., Froment, A., Mauclere, P., Tortevoye, P., Schmitt, C., Njouom, R., Saib, A., and Gessain, A. (2007) Emerg. Infect. Dis. 13 1314-1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Achong, B. G., Mansell, P. W., Epstein, M. A., and Clifford, P. (1971) J. Natl. Cancer Inst. 46 299-307 [PubMed] [Google Scholar]
- 33.Herchenröder, O., Turek, R., Neumann-Haefelin, D., Rethwilm, A., and Schneider, J. (1995) Virology 214 685-689 [DOI] [PubMed] [Google Scholar]
- 34.Switzer, W. M., Salemi, M., Shanmugam, V., Gao, F., Cong, M. E., Kuiken, C., Bhullar, V., Beer, B. E., Vallet, D., Gautier-Hion, A., Tooze, Z., Villinger, F., Holmes, E. C., and Heneine, W. (2005) Nature 434 376-380 [DOI] [PubMed] [Google Scholar]
- 35.Löchelt, M., Romen, F., Bastone, P., Muckenfuss, H., Kirchner, N., Kim, Y. B., Truyen, U., Rosler, U., Battenberg, M., Saib, A., Flory, E., Cichutek, K., and Münk, C. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 7982-7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell, R. A., Wiegand, H. L., Moore, M. D., Schafer, A., McClure, M. O., and Cullen, B. R. (2005) J. Virol. 79 8724-8731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delebecque, F., Suspene, R., Calattini, S., Casartelli, N., Saib, A., Froment, A., Wain-Hobson, S., Gessain, A., Vartanian, J. P., and Schwartz, O. (2006) J. Virol. 80 605-614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duda, A., Stange, A., Luftenegger, D., Stanke, N., Westphal, D., Pietschmann, T., Eastman, S. W., Linial, M. L., Rethwilm, A., and Lindemann, D. (2004) J. Virol. 78 13865-13870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pietschmann, T., Heinkelein, M., Heldmann, M., Zentgraf, H., Rethwilm, A., and Lindemann, D. (1999) J. Virol. 73 2613-2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dang, Y., Wang, X., Esselman, W. J., and Zheng, Y. H. (2006) J. Virol. 80 10522-10533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muckenfuss, H., Hamdorf, M., Held, U., Perkovic, M., Löwer, J., Cichutek, K., Flory, E., Schumann, G. G., and Münk, C. (2006) J. Biol. Chem. 281 22161-22172 [DOI] [PubMed] [Google Scholar]
- 42.Cullen, B. R. (1986) Cell 46 973-982 [DOI] [PubMed] [Google Scholar]
- 43.Muckenfuss, H., Kaiser, J. K., Krebil, E., Battenberg, M., Schwer, C., Cichutek, K., Münk, C., and Flory, E. (2007) Nucleic Acids Res. 35 3784-3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hatziioannou, T., Perez-Caballero, D., Yang, A., Cowan, S., and Bieniasz, P. D. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10774-10779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Löchelt, M., Zentgraf, H., and Flügel, R. M. (1991) Virology 184 43-54 [DOI] [PubMed] [Google Scholar]
- 46.Simm, M., Shahabuddin, M., Chao, W., Allan, J. S., and Volsky, D. J. (1995) J. Virol. 69 4582-4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jarmuz, A., Chester, A., Bayliss, J., Gisbourne, J., Dunham, I., Scott, J., and Navaratnam, N. (2002) Genomics 79 285-296 [DOI] [PubMed] [Google Scholar]
- 48.Su, A. I., Cooke, M. P., Ching, K. A., Hakak, Y., Walker, J. R., Wiltshire, T., Orth, A. P., Vega, R. G., Sapinoso, L. M., Moqrich, A., Patapoutian, A., Hampton, G. M., Schultz, P. G., and Hogenesch, J. B. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 4465-4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larue, R. S., Jonsson, S. R., Silverstein, K. A., Lajoie, M., Bertrand, D., El-Mabrouk, N., Hotzel, I., Andresdottir, V., Smith, T. P., and Harris, R. S. (2008) BMC Mol. Biol. 9 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu, Q., Chen, D., Konig, R., Mariani, R., Unutmaz, D., and Landau, N. R. (2004) J. Biol. Chem. 279 53379-53386 [DOI] [PubMed] [Google Scholar]
- 51.Hill, C. L., Bieniasz, P. D., and McClure, M. O. (1999) J. Gen. Virol. 80 2003-2009 [DOI] [PubMed] [Google Scholar]
- 52.Meiering, C. D., and Linial, M. L. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 15130-15135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alke, A., Schwantes, A., Kido, K., Flotenmeyer, M., Flügel, R. M., and Löchelt, M. (2001) Virology 287 310-320 [DOI] [PubMed] [Google Scholar]
- 54.Bock, M., Heinkelein, M., Lindemann, D., and Rethwilm, A. (1998) Virology 250 194-204 [DOI] [PubMed] [Google Scholar]
- 55.Diaz-Griffero, F., Kar, A., Perron, M., Xiang, S. H., Javanbakht, H., Li, X., and Sodroski, J. (2007) J. Virol. 81 10362-10378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deleted in proof
- 57.Chiu, Y. L., and Greene, W. C. (2008) Annu. Rev. Immunol. 26 317-353 [DOI] [PubMed] [Google Scholar]
- 58.Kao, S., Goila-Gaur, R., Miyagi, E., Khan, M. A., Opi, S., Takeuchi, H., and Strebel, K. (2007) Virology 369 329-339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heneine, W., Schweizer, M., Sandstrom, P., and Folks, T. (2003) Curr. Top. Microbiol. Immunol. 277 181-196 [DOI] [PubMed] [Google Scholar]
- 60.Linial, M. L. (1999) J. Virol. 73 1747-1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hahn, H., Baunach, G., Brautigam, S., Mergia, A., Neumann-Haefelin, D., Daniel, M. D., McClure, M. O., and Rethwilm, A. (1994) J. Gen. Virol. 75 2635-2644 [DOI] [PubMed] [Google Scholar]
- 62.Weikel, J., Löchelt, M., and Truyen, U. (2003) J. Vet. Med. A Physiol. Pathol. Clin. Med. 50 415-417 [DOI] [PubMed] [Google Scholar]
- 63.Alke, A., Schwantes, A., Zemba, M., Flügel, R. M., and Löchelt, M. (2000) Virology 275 170-176 [DOI] [PubMed] [Google Scholar]