Abstract
The hedgehog (Hh) signaling pathway is a key component of cross-talk during vertebrate gut development, involving endodermally secreted Sonic (Shh) and Indian hedgehog (Ihh) proteins that directly signal to adjacent mesoderm. Here we show that the closely linked mesenchymal forkhead transcription factors Foxf1 and Foxl1 are part of this signaling cascade. Analysis of conserved non-coding sequences surrounding Foxf1 and Foxl1 identified seven Gli binding sites, with two sites near Foxl1 being identical among mammalian, bird, fish, and amphibian species. In vitro experiments indicate that Gli2 binds to these Gli sites, several of which are critical for Gli2-mediated activation of a luciferase reporter in 293 cells. In addition, we demonstrate occupancy of one of these elements by Gli proteins in the intestine in vivo using chromatin immunoprecipitation. Furthermore, expression of both Foxf1 and Foxl1 is reduced in the Gli2/Gli3 mutant gut. These results provide compelling evidence that Foxf1 and Foxl1 are mediators of the Hh (endoderm) to mesoderm signaling pathway.
The Hedgehog (Hh)2 signaling pathway is one of the earliest regulatory cascades activated during organogenesis of the gut tube. Shh and Ihh, expressed in the gut endoderm, bind to the receptor Ptch1, which is expressed exclusively in the mesoderm of the vertebrate gut (1-4). Interaction with the Ptch1 receptor causes derepression of Smo, a transmembrane protein, which prevents post-translational processing of Gli2 and Gli3 by inhibiting phosphorylation and proteasomal degradation (5). In the absence of Ptch1-mediated repression of Smo, an N-terminal proteolytic fragment of Gli3, and to a lesser extent Gli2, emerges from the proteasome as a transcriptional repressor, whereas Gli1 appears to be completely degraded.
Both Shh and Ihh are expressed in mouse definitive endoderm from E8.5 onward along the entire gut tube, except in the domain of pancreatic bud endoderm (6). In the intestine, Shh and Ihh are both required for proper specification and development of smooth muscle of the muscularis externa and neurons of the myenteric plexus (2, 4, 7). Severe embryonic inhibition of all Hh signals, through overexpression of an Hhip cDNA or via injection of a neutralizing antibody against Hh proteins, revealed that Shh and Ihh are also indispensable for villus development and later critical for patterning the cryptvillus axis and restricting epithelial proliferation via modulation of adjacent mesenchymal cells (3, 7).
Recent computational approaches have attempted to identify Gli target genes. One investigation used a local alignment algorithm termed “enhancer element locator,” which evaluates the spatial order of transcription factor binding sites from two different species (8). Gli binding sites near Foxf1, Foxc2, and Foxl1, all located within a 50-kb region of mouse chromosome 8, scored very highly using the enhancer element locator method. In addition, expression of Foxf1 is dependent on Shh in the developing oral cavity, lung, and sclerotome, and both Foxf1 and Foxf2 expression can be extinguished in E12.5 intestine explants when treated with the Smo inhibitor cyclopamine (9, 10). These reports suggest that Foxf1 and Foxf2 could be Hh target genes.
Intestinal epithelial phenotypes of Foxf1, Foxf2, and Foxl1 mutants also suggest connections to the Hh signaling pathway. Foxf1+/-, Foxf2-/-, and Foxl1-/- mice all have perturbations in the crypt-villus axis, characterized by increased proliferation and Wnt signaling, reminiscent of epithelial abnormalities caused by Hh signal ablation (3, 7, 11, 12). Foxl1-/- mice also have delayed villus morphogenesis, with very stunted villi evident at E16.5 and E18.5, similar to the severe defects in villus development caused by Hh inhibition (7, 11). The Foxl1 mRNA expression pattern also highly resembles that of the known Hh target genes Ptch1, Gli1, and Bmp4 during intestine development (2, 13-15).
Our studies indicate that Foxfl and Foxl1 are direct target genes of the Hh signaling pathway in the developing stomach and intestine. Their expression is dependent on Gli2 and Gli3 and induced by Shh-N recombinant protein. Several highly conserved Gli binding sites appear crucial for Gli-mediated binding and transcriptional activation of the Foxf1 and Foxl1 promoters. These findings provide a critical link toward understanding the molecular mechanisms that control epithelial proliferation and organization of the developing gastrointestinal (GI) tract.
EXPERIMENTAL PROCEDURES
EMSA Analysis—Stomach and intestine primordia (E14.5) were isolated from ∼36 CD1 embryos and snap-frozen in liquid N2, and nuclear extracts were prepared as described previously (16), with some modifications. Complete (Roche Applied Science) protease inhibitor tablets were used in all buffers, supplemented with 0.1 μm phenylmethylsulfonyl fluoride and 1.0 mm sodium metavanadate. Oligonucleotides (oligos) were labeled by incubating 50 ng of double-stranded DNA with 30 μCi of a [32P]dCTP and 2.5 units of Klenow (New England Biolabs) for 30 min at room temperature followed by removal of unincorporated nucleotides with a MicroSpin G-50 column (Amersham Biosciences). Electrophoretic mobility shift assays (EMSAs) were performed as described previously (17); 3-4 μg of protein extract were incubated with 20 mm Hepes, pH 7.9, with 0.1 mm EDTA, 1 mm MgCl2, 12% glycerol, 60 mm KCl, 0.5 mm dithiothreitol (added fresh), 50 ng/μl sonicated dIdC, and 125 ng/μl bovine serum albumin for 10 min to prebind. Labeled probe (25,000 cpm) was then added and incubated for 30 min at room temperature. For some assays, 1 μg of a polyclonal rabbit antibody to Gli2 (AbCam ab26056) or 1 μg of rabbit IgG (R&D Systems) was added during the prebind. Oligos used in all experiments are listed in supplemental Table 1.
Luciferase Assays—HEK293 cells (CRL-1573) were purchased from ATCC. Genomic fragments of CNS1, Foxf1, and Foxl1 were obtained by PCR of mouse genomic tail DNA with PfuUltra (Stratagene). For Foxf1 PCR, a concentration of 1 m betaine was necessary for amplification. PCR products were cloned into pSC-B (Stratagene) and subcloned into pGL3-Basic (Promega) between XhoI and NcoI (Foxf1 promoter). Using PfuUltra (Stratagene), the Foxl1 promoter PCR product was amplified with nested primers and then subcloned into pGL3-Basic. In 24-well plates, 1.5 × 105 293 cells were seeded in each well. About 24 h later, 200 ng of DNA (100 ng of reporter plasmid, 5 ng of pRL-SV40 (Stratagene) and 70 or 45 ng of pBlue-Script II (Stratagene)) were transfected using FuGENE 6 (Roche Applied Science) at a ratio of 3:1 (FuGENE:DNA). For Foxf1 assays, 50 ng of pCS2-Gli2* was co-transfected, whereas 25 ng was co-transfected for Foxl1. Gli2* lacks the 328 N-terminal amino acids of Gli2 and has been shown to be a 30 times more potent transactivator in co-transfection assays than Gli2 (18). Luciferase assays were performed as described previously (17).
Gli sites within the FoxL1 promoter and within CNS1 were mutated according to specifications of the QuikChange site-directed mutagenesis kit (Stratagene). Mutagenesis of the Gli site within the Foxf1 promoter was performed by overlap extension PCR using the PfuUltra polymerase (19).
Quantitative RT-PCR—RNA was isolated using TRIzol (Invitrogen) per the manufacturer's instructions. The Bio-Rad iScript kit was used for cDNA synthesis from 500 ng of total RNA. To prevent amplification of contaminating genomic DNA in RNA samples, 0.5 μl of RQ1 RNase-free DNase (Promega) was added to each reverse transcriptase (RT) reaction, prior to the addition of RT. Reactions were incubated for 15 min at 37 °C, 10 min at 65 °C, and then cooled on ice. Using the Brilliant Sybr Green mix (Stratagene), QPCR amplification was performed in triplicate with 0.5 μl of cDNA per 19-μl reaction on a Stratagene MX3000 thermocycler. Amplification thresholds were normalized to those of HPRT.
Chromatin Immunoprecipitation Assay (ChIP)—Chromatin was prepared from the small intestine of embryos at day 14.5 of gestation (n = 5) as described previously (20). Chromatin was immunoprecipitated with a polyclonal goat anti-Gli1 antibody (Santa Cruz Biotechnology, antibody sc-6153). After reversal of the cross-link, the FoxF1 promoter fragment was PCR-amplified using primers and QPCR as described above. Enrichment between input DNA and immunoprecipitated DNA was calculated relative to that of the promoter of the myelin basic protein gene, which is not expressed in the intestinal mucosa and not a target of Gli regulation.
Isolation of Gli2 and Gli3 Mutant Tissue—Gli2+/- mice were kindly provided by Dr. Alex Joyner (Memorial Sloan-Kettering Cancer Center). Gli3+/- (Gli3Xt-j) mice were obtained from The Jackson Laboratory. Mice were maintained on a mixed CD1-C3H/HeJ background. Embryos were isolated from Gli2+/-Gli3+/- intercrosses and genotyped as described previously (21, 22). Stomach and intestine primordia from E12.5 embryos were removed and homogenized in buffer RLT (Qiagen) in conjunction with QIAshredder columns (Qiagen). RNA was then extracted using the RNeasy micro kit (Qiagen). Quantitative RT-PCR was performed as indicated above.
E14.5 Gut Mesoderm Stimulation with Shh-N—Stomach and intestine primordia (E14.5) were isolated from 12-14 CD1 embryos and placed into cold Ca2+ and Mg2+-free (CMF) HBSS (Invitrogen). Tissues were incubated 10 min at room temperature in CMF-HBSS containing 10 mm EDTA and then cut lengthwise to expose the endoderm and transferred to an Eppendorf tube containing cold CMF-HBSS. Tissue was vortexed until all endoderm was removed, as evaluated under a Leica dissecting microscope. Mesoderm was rinsed in medium (Dulbecco's modified Eagle's medium (catalog number 11995, Invitrogen), 10% fetal calf serum, + penicillin/streptomycin). Mesoderm was then incubated at 37 °C for 30 min, with periodic mixing, in 1 ml of a 50% solution of Dispase (25 caseinolytic units/ml, BD Biosciences) diluted with medium. Medium (9 ml) was added and then centrifuged at 1000 rpm for 5 min at 4 °C. Tissue was washed with 5 ml of medium and then distributed between 6 wells of a 24-well plate in 0.5 ml of medium/well. After 24 h of culture, 0.5% bovine serum albumin or mouse recombinant Shh-N (catalog number 461-SH, R&D Systems) was added to a final concentration of 2 μg/ml. After an additional 72 h of culture, RNA was extracted using TRIzol (Invitrogen) and quantitative RT-PCR was performed as described above.
RESULTS
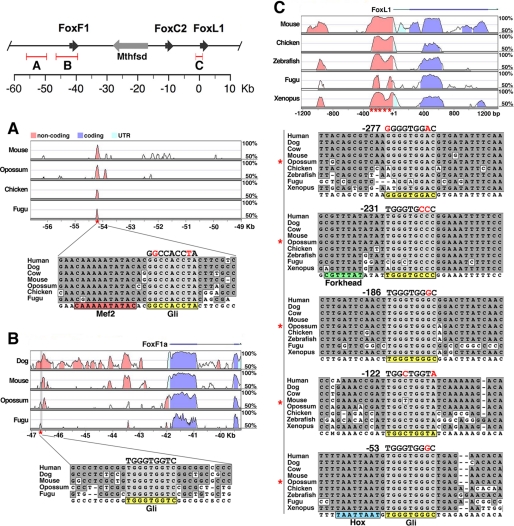
Activation of Hh target genes is mediated by Gli factors binding to the consensus sequence GACCACCCA (23, 24). To identify conserved sequences within a 150-kb region surrounding Foxf1 and Foxl1, we compared mammalian and non-mammalian species using the VISTA browser (25, 26), which provides multispecies global alignment. Using this tool, we discovered two sites near Foxf1 and five sites near Foxl1 that resembled or matched the Gli binding consensus (Fig. 1). One Gli site near Foxf1 was within a small (∼100 bp) conserved region (CNS1), which also contained two forkhead sites (AYAAACA) and one site similar to the consensus of Mef2 transcription factors, known to be involved in striated and smooth muscle development (27) (Fig. 1A). The two Gli sites nearest to Foxl1 were within a region of the proximal promoter that exhibited even greater sequence conservation than the coding region of Foxl1 (Fig. 1C). Gli binding sites often appeared within close proximity to forkhead binding sites, with four out of seven Gli sites located within 16 bp of a conserved forkhead site (data not shown). The Gli site nearest Foxl1, at -53, was also immediately adjacent to a highly conserved Homeobox (Hox) consensus site, TAAT (Fig. 1C).
FIGURE 1.
Identification of phylogenetically conserved Gli binding sites. Foxf1, Foxc2, and Foxll, along with Mthfsd, are located within a 50-kb region of mouse chromosome 8 (top left). Conserved regions identified by VISTA analysis were analyzed for the Gli binding consensus (A, B, and C). About 7.5 kb upstream of the Foxf1 gene, a small, ∼100-bp conserved region (CNS1) contains two forkhead sites (not pictured) and one Gli site adjacent to a Mef2-like consensus (A). One additional Gli site was identified near Foxf1, ∼3.4 kb upstream (B). Five conserved Gli sites were found in the proximal promoter region of Foxl1, at the indicated positions (C). Gli sites are highlighted in yellow, and the consensus binding sites are noted above with mismatches colored red. Physical genomic distances are relative to the Foxl1 gene.
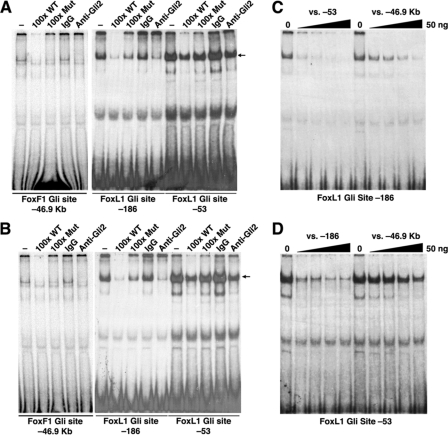
To assess the Gli binding potential of these conserved sequences, we performed EMSAs on sites with the closest match to the Gli consensus. Gli sites near Foxl1 at -186 and -53, with respect to the start of the Foxl1 mRNA at +1, exhibited strong binding, which was diminished upon competition with an unlabeled oligo encompassing a Gli site from the Ptch1 promoter (28) (Fig. 2, A and B). Similarly, an antibody against Gli2 reduced the level of the shifted signal of the -186 and -53 oligos, indicating a disruption of Gli2 binding (Fig. 2, A and B). The same held true in assays performed with the Gli site nearest to Foxf1 (Foxf1 Gli site, -46.9 kb), except that the intensity of the shifted complex was much less (Fig. 2, A and B). To compare the relative binding affinity of these Gli sites, competitions with unlabeled oligos were performed (Fig. 2, C and D). These indicated that sequences at -53 and -186 had higher affinity than the Gli site near Foxf1 at -46.9 kb. These experiments also revealed that the -53 oligo produced two closely migrating complexes, and the top band in this doublet was partially resistant to competition with the -186 oligo (Fig. 2D). This upper band could represent binding of a Hox factor to the highly conserved Hox site immediately adjacent to the Gli site.
FIGURE 2.
Gli2 from E14. 5 gut extracts binds to Gli consensus sequences near Foxf1 and Foxl1. Nuclear extracts from E14.5 stomach (A) and intestine (B) bind Gli sites from the Foxf1 promoter (-46.9 kb) and from the Foxl1 promoter (-186 and -53). Binding is diminished by competition with an unlabeled oligo encompassing a known Gli binding site from the Ptch1 promoter (100x WT) but is not effectively competed by a mutant oligo (100x Mut). Anti-Gli2 disrupts binding with all three oligos, whereas rabbit IgG does not. Using E14.5 intestine extracts, oligos from each site were competed against increasing amounts of unlabeled oligos (0, 5, 10, 25, and 50 ng) to determine relative binding affinity (C and D). The -186 site was effectively competed with 10-25 ng of cold -53 oligo, whereas the -46.9-kb oligo did not completely quench binding, even at 50 ng (C). Similar results were seen with the -53 site, although this site appears to have an upper band not effectively competed by either cold competitor (D). This may represent association of other transcription factors, such as Hox proteins, binding independently of Gli.
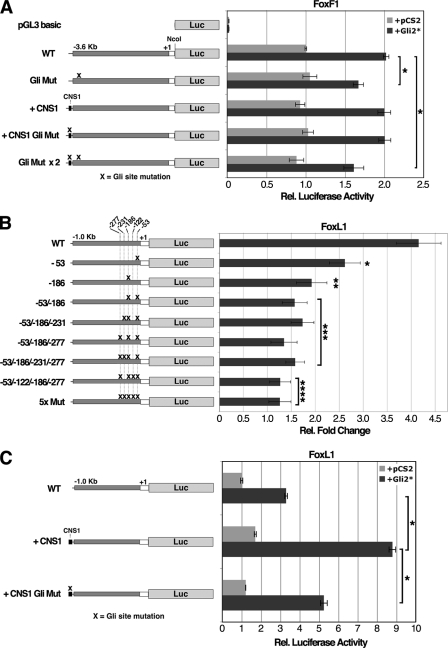
Next, we tested the Gli sites within Foxf1 and Foxl1 promoter fragments for their ability to activate a luciferase reporter in HEK293 cells. Individual Gli sites were mutated, and promoter activity was assayed in the context of a constitutively active Gli2 protein that contains an N-terminal deletion of the repressor domain (Gli2ΔN or Gli2*) (18). The 3.6-kb Foxf1 promoter exhibited very high level reporter activity in 293 cells but was still stimulated ∼2-fold when co-transfected with a Gli2*-expressing plasmid (Fig. 3A). Mutation of the Gli site significantly reduced Gli2*-mediated enhancement but did not affect the basal level of promoter activity (Fig. 3A). The conserved non-coding region (CNS1) near Foxf1, about 7.5 kb distal to the Gli site near Foxf1, was assayed for its effects on reporter activity in the context of the 3.6-kb Foxf1 promoter. CNS1 did not affect Foxf1 promoter activity, and likewise, when the Gli site within CNS1 was mutated, reporter activity was unchanged (Fig. 3A). This may not be so surprising considering that a transgenic mouse with a LacZ cDNA driven by a Foxf1 promoter lacking the CNS1 region largely recapitulates the expression pattern of Foxf1 in the developing and adult gut (29).
FIGURE 3.
Gli sites near Foxf1 and Foxl1 are necessary for Gli2-mediated transcriptional activation. A 3.6-kb promoter fragment of Foxf1, plus a 239-bp 5′untranslated region, were assayed for luciferase (Luc) activity in 293 cells (A). The Foxf1 promoter was assayed in the context of the CNS1 element and with the indicated Gli site mutations (Gli Mut) (A). Data represent the average of two experiments, each performed in triplicate. Cells were harvested 24 h after transfection. WT, wild type. Rel., relative. As in A, a 1.0-kb promoter fragment of Foxl1, plus a 190-bp 5′untranslated region, were similarly assayed in B, except that -fold changes represent induction of reporter activity by Gli2* when compared with empty vector. Asterisks indicate a significant decrease in Gli2*-mediated induction (B). Data represent the average of four experiments, each done in triplicate (B). C, Foxl1 promoter activity in the context of the CNS1 element, with or without a mutation in the Gli site. Data are representative of three experiments, each done in triplicate (C). pGL3-Basic basal luciferase activity, not shown (B and C), is greater than the basal activity of the Foxl1 promoter, suggesting that the Foxl1 promoter may have repressive cis-regulatory elements. Data represent the mean ± S.E. For A and C, (*) indicates p value < 0.001; for B, (*) indicates p value < 0.05, (**) indicates p value < 0.01, (***) indicates p value < 0.005, and (****) indicates p value <0.001 according to Student's t test.
In the context of a 1.0-kb Foxl1 promoter fragment, we observed a 4-fold induction of reporter activity upon co-transfection of the Gli2*-expressing plasmid (Fig. 3B). Gli sites at -53 and -186 were assessed first because of their high conservation and ability to bind Gli2 in EMSAs. The Gli site mutation at -186 caused substantial attenuation of Gli2*-mediated induction, whereas mutation of the -53 site had somewhat lesser, although significant, effects. Mutation of both sites decreased Gli2*-mediated induction to less than 1.6-fold, indicating that these two sites provide the large majority of the Gli2 responsiveness of the Foxl1 promoter in this assay (Fig. 3B). However, a small induction was still observed in the -53/-186 double mutant, suggesting that some Gli2 responsiveness may remain. Additional mutations at the -277, -231, and -122 Gli sites, in which all sites except -231 are mutated or in which all five sites are mutated, completely eliminated any significant induction by Gli2* (Fig. 3B). Among all five Gli sites, the -186 site appears to have preeminence because of its comparatively high conservation, strong Gli binding affinity, and key importance in Gli2*-mediated induction in this assay. Although the CNS1 element did not enhance the activity of the Foxf1 promoter, there was a pronounced stimulatory effect on Foxl1 regulatory sequences, more than doubling the Gli2*-mediated induction of this promoter (Fig. 3C). Mutation of the Gli site within CNS1 significantly diminished the effect of CNS1 (Fig. 3C). These data suggest that CNS1 may act as a distal enhancer for the Foxl1 promoter.
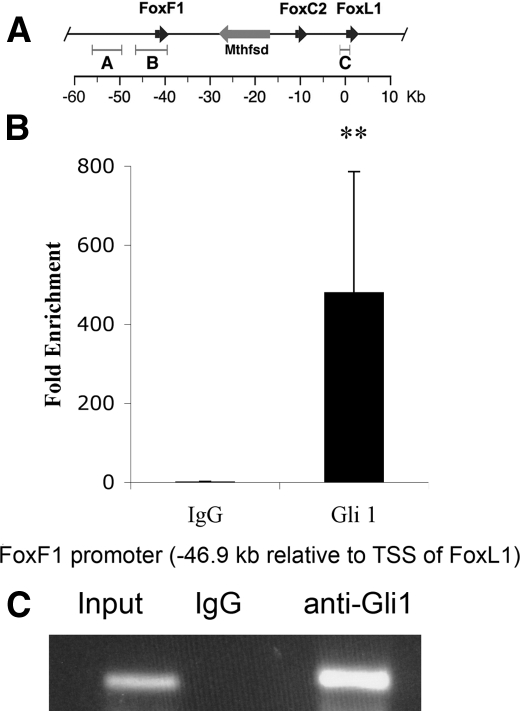
To determine whether Gli proteins occupy any of the potential binding sites identified above in vivo, we performed chromatin immunoprecipitation assays with an antibody specific to Gli1 on chromatin isolated from fetal intestine (see “Experimental Procedures”). As shown in Fig. 4, the Gli binding site in the promoter of the Foxf1 gene, located at position -46.9 kb with respect to the transcriptional start site of Foxl1, is highly enriched in the immunoprecipitated DNA when compared with input DNA, indicating strong binding to this site by Gli1 in vivo. To our knowledge, this is the first description of Gli1 binding to in vivo targets in the GI tract using chromatin immunoprecipitation.
FIGURE 4.
Gli1 occupies its site in the Foxf1 promoter in intestinal chromatin. A, schematic representation of the Foxf1/Foxl1 locus. Physical genomic distances are relative to the Foxl1 gene. The binding site assayed here is located in region B. B, quantification of real-time PCR analysis of the enrichment of the Foxf1 promoter element at position -46.9 relative to the transcriptional start site (TSS) of Foxl1. QPCR was performed on five input samples and five ChIP samples, and the enrichment was calculated relative to a non-target gene (myelin basic protein). (**) p value <0.01 according to Student's t test. Data represent the mean ± S.E. C, representative agarose gel of the ChIP experiment showing the presence of the Foxf1 promoter fragment in the Gli1 ChIP but not the IgG control ChIP.
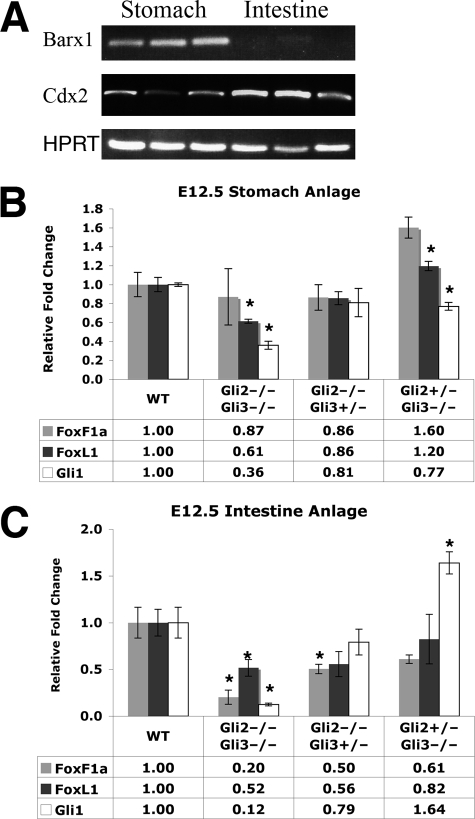
The in vitro and in vivo data described above indicate that Gli transcription factors directly activate the Foxf1 and Foxl1 genes, predicting that genetic inactivation of Gli2 and/or Gli3 would cause reductions of Foxf1 and Foxl1 mRNA levels in vivo. Gli1 is not expressed in the absence of Gli2 and Gli3 and thus cannot compensate for their inactivation (30). Therefore, we evaluated Foxf1 and Foxl1 mRNA levels in E12.5 Gli2 and Gli3 null stomach and intestine (midgut) primordia (Fig. 5, B and C). In the stomach primordium of Gli2-/-Gli3-/- double mutants, Foxl1 expression is significantly decreased, whereas Foxf1 expression is not altered (Fig. 5B). The stomach primordium of Gli2+/-Gli3-/- embryos exhibited a significant increase in expression of Foxf1 and a small increase of Foxl1 mRNA. This may reflect strong repressive effects of Gli3 that normally function to limit expression of Foxf1 (and perhaps Foxl1) in the stomach. However, this did not hold true in the intestine, where Foxf1 and Foxl1 expression was lower when both alleles of either Gli2 or Gli3 are inactivated, although Foxl1 mRNA levels were not significantly reduced in Gli2+/-Gli3-/- embryos (Fig. 5C). In terms of Foxf1 and Foxl1 regulation, expression of these genes appears more dependent on levels of both Gli2 and Gli3 in the developing intestine than in the stomach. Reduction of Foxf1 and Fox1l mRNA levels was not secondary to a deficiency of mesodermal tissue caused by Gli2 and/or Gli3 mutations because QPCR did not reveal significant reductions of the mRNA for vimentin, an intermediate filament broadly expressed in mesenchymal cells (data not shown). As expected, expression of Gli1 was dramatically reduced in Gli2-/-Gli3-/- embryos (Fig. 5B).
FIGURE 5.
Foxf1 and Foxl1 expression are dependent on Gli2 and Gli3, in vivo. A, prospective stomach and intestine were dissected from day 12.5 days postcoital embryos. RT-PCR for Barx1 confirms that our stomach anlage is expressing this gastric marker. Cdx2 expression is strong in the intestine, as expected. Some Cdx2 expression is also present in the stomach fraction, indicating that these samples also contained some prospective duodenal tissue. B and C, Foxf1, Foxl1, and Gli1 mRNA levels in E12.5 stomach (B) and intestine (C) anlage in Gli2 and Gli3 compound mutants, as assessed by quantitative RT-PCR. Tissues from 3-5 embryos were assayed for each genotype. Wild type (WT) levels are set equal to 1.0. (*) p value <0.05 according to Student's independent t test. Data represent the mean ± S.E.
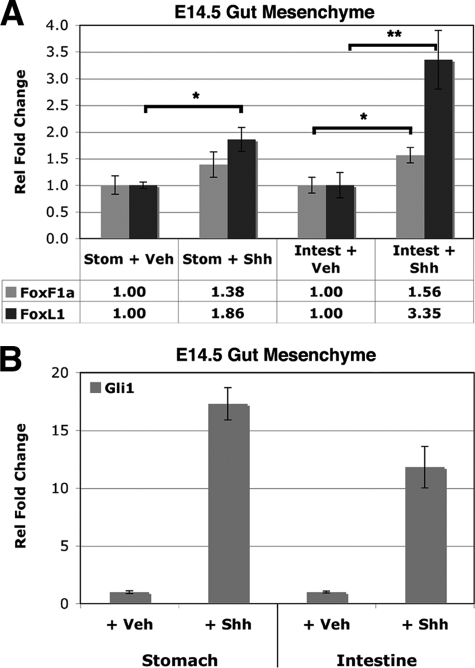
Next we investigated whether activation of the Hh pathway was sufficient to induce the expression of these genes in a relevant tissue. Gut mesoderm from E14.5 embryos was isolated and treated with a recombinant mouse N-terminal fragment of Shh (Shh-N) and assayed for induction of Foxf1 and Foxl1 mRNA. Foxl1 was significantly induced in both stomach and intestine mesoderm, whereas Foxf1 was significantly induced only in the intestine (Fig. 6A). In this assay, the Hh signaling pathway is clearly activated, as indicated by robust induction of the Gli2/Gli3 target gene Gli1 (Fig. 6B). Collectively, these data demonstrate that Foxf1 and Foxl1 are dependent on Gli2 and Gli3 and induced upon Hh pathway activation in the mesoderm of developing stomach and intestine.
FIGURE 6.
Foxf1 and Foxl1 are induced in gut mesenchyme upon administration of Shh. E14.5 mesenchyme from stomach (Stom) and intestine (Intest) were cultured with 0.5% bovine serum albumin (vehicle (Veh), set equal to 1.0) or recombinant Shh-N for 72 h, and Foxf1 and Foxl1 mRNA levels were assessed by quantitative RT-PCR (A). Rel, relative. Gli1 induction by Shh-N is also shown (B). Data are representative of two experiments, each performed in triplicate. (*) p value <0.05 or <0.01 (**) according to Student's independent t test. Data represent the mean ± S.E.
DISCUSSION
In summary, this study provides clear evidence that Foxf1 and Foxl1 are target genes of the Hh signaling pathway via Gli transcription factors. Although a total absence of Gli factors (in Gli2-/-Gli3-/- double mutants) does not completely extinguish Foxf1 and Foxl1 expression in the developing gut, this is not surprising. The remaining expression of Foxf1 and Foxl1 in Gli2-/-Gli3-/- gut tissue is likely due to other signals and transcription factors acting on their respective promoters. Indeed, as shown in Fig. 1, conserved Hox sites, forkhead sites, and other binding sites are located in these promoters and could provide Hh-independent positive influences on Foxf1 and Foxl1 expression. In the developing mouse, expression of Foxf1 in the lateral mesoderm and yolk sac is absent in Shh-/-Ihh-/- and Smo-/- embryos but maintained in the allantois and primitive streak (31), indicating that Hh-independent signals maintain some level of Foxf1 expression in certain tissues.
Although complete inactivation of Gli activity in Gli2-/- Gli3-/- embryos did not entirely eliminate expression of Foxf1 and Foxl1 in the stomach and intestine, an 80% reduction of Foxf1 and 50% reduction of Foxl1 in the intestine could account for a portion of the comprehensive defects seen upon Hh inhibition in the developing gut. In addition to Foxf1 and Foxl1, there are likely to be multiple other Hh target genes activated via Gli transcription factors in the developing gut mesoderm. These target genes may individually execute small roles in larger developmental processes. One remarkable example of this phenomenon involves the platelet-derived growth factor (PDGF) signaling pathway, in which each of 12 identified platelet-derived growth factor target genes individually contributes small phenotypic aspects of the much more serious phenotypes of the Pdgfra and Pdgfrb mutants (32). Indeed, genetic inactivation of Foxl1 results in a developmental phenotype of the gut considerably less severe than that caused by potent inhibition of the Hh signal (7, 11).
Foxf1 and Foxl1 dependence on the Hh signal also differs between different tissues. It is interesting that both Foxf1 and Foxl1 appear less responsive to Shh-N stimulation in stomach mesoderm and somewhat less dependent on Gli2 and Gli3 in the developing stomach when compared with similar experiments with intestinal tissue. This could reflect a difference of Gli2 and Gli3 expression levels in the stomach, the degree to which the Hh pathway is activated there, and/or differential dependence on other transcription factors expressed in the stomach.
Where possible, primary tissue was used to evaluate the connection between the Hh signal and Foxf1 and Foxl1 in this study. Luciferase assays of Foxf1 and Foxl1 Gli-dependent promoter activity were the only instances where primary tissue could not be used due to technical limitations. However, it is compelling that Shh induction of Foxf1 and Foxl1 mRNAs in E14.5 intestinal mesenchyme closely mirrors the induction seen by Gli2* in HEK293 cells; note a 3.4-fold induction of Foxl1 in E14.5 mesenchyme (versus a 4-fold induction in HEK293 cells) and a 1.6-fold induction of Foxf1 in E14.5 mesenchyme (versus a 2-fold induction in HEK293 cells) in Figs. 3 and 6. This similarity may suggest that testing these promoter fragments in HEK293 cells represents an appropriate model for assaying Gli-mediated activation of these mesenchymally expressed forkhead genes.
When fetal intestinal mesenchyme was cultured in the presence of Shh, Foxl1 was more responsive than Foxf1. In contrast, when we analyzed gene expression in the intestine anlage of Gli2-/-Gli3-/-embryos, Foxf1 was the more affected gene, whereas in the stomach, Foxl1 is more sensitive to Gli2/Gli3 ablation. At present, we do not know what explains these slight discrepancies. It is possible that during the relatively long culture with Shh (72 h), there is a partial fate change in the cultured mesenchyme. Alternatively, differences could be due to alternate signaling molecules affecting gene expression in the culture experiment or non-physiological dosage of Shh. Nevertheless, our genetic data strongly support the notion that both Foxf1 and Foxl1 are regulated by hedgehog signaling and Gli factors.
To evaluate the in vivo binding of Gli factors to the Foxf1 and Foxl1 promoters, we developed ChIP from embryonic gut tissue for Gli1, which to our knowledge has not been reported previously. We were thus able to confirm strong Gli binding to the promoter of the Foxf1 gene in vivo. To truly dissect the in vivo relevance of the Hh responsiveness of a target gene, it would be necessary to mutate all Gli binding sites within endogenous regulatory elements. This would be a costly and extremely complicated undertaking for linked genes such as Foxl1 or Foxf1, which have multiple Gli binding sites that could additionally regulate the expression of neighboring loci. The surmounting evidence provided here (phylogenetic conservation of Gli sites, confirmed binding by EMSA, promoter responsiveness to Gli2 in luciferase assays, occupancy of target sites by Gli in vivo, genetic dependence on Gli2 and Gli3, and Shh induction in primary tissue explants) strongly indicates that Foxf1 and Foxl1 are target genes of the Hh pathway.
Because Foxf1 and Foxl1 appear to be controlled in parallel by the same signaling pathway (the Hh signal) and are both expressed in the same tissue (the developing mesoderm of the gut), it is compelling to speculate whether, and to what extent, Foxf1 and Foxl1 are functionally or biochemically redundant. Foxl1 and Foxf2 mutants both exhibit reduced expression levels of Bmp4 in the gut (10, 11), and recent characterizations indicate that Foxf1 mutant embryos have a severe depletion of Bmp4 expression in the lateral and extraembryonic mesoderm (31). If Foxf1 is involved in Bmp4 expression in the developing intestine, then the effects of Foxf1 and Foxl1 could be routed, in part, through Bmp4, a known repressor of epithelial proliferation (33-35), perhaps via direct effects on the Wnt signaling pathway (Fig. 7B). Foxf1 was recently described as having a critical role in mediating fibroblast migration, and the integrin Itgb3 was identified as a direct target gene (36). Foxf1 deficiency in the gut could thus cause defects in mesodermal migration during villus development. This could affect the distribution of mesenchymal cells along the crypt-villus axis, and in turn, affect epithelial organization. Still, there are few known target genes of Foxf1 and Foxl1, and no described targets that are candidate repressors of the Wnt signaling pathway, except that Foxl1 inactivation results in higher expression of the Wnt-enhancing proteoglycan, Syndecan-1 (12). Elucidation of the Foxl1 in vivo DNA binding consensus will aid bioinformatic identification of Foxl1 target genes and enable the evaluation of whether Foxf1 and Foxl1 bind to the same cis-regulatory elements and cooperatively or redundantly regulate identical target genes.
FIGURE 7.
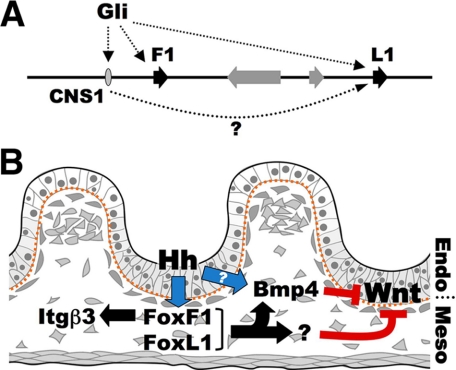
Reciprocal signaling in the gastrointestinal tract. A, summary of Gli-mediated regulation of Foxf1 and Foxl1 chromosomal region, and possible enhancer function of CNS1, which may impact Foxl1 activity from a distance. B, model of Hh regulatory cascade in E14-E15 developing gut, indicating possible roles for Foxf1 and Foxl1. Endo, endoderm; Meso, mesoderm.
Although mutations in either Foxl1 or Foxf1 affect mesenchymal signaling, their combined role in GI development cannot be easily addressed by genetic means due to their genomic proximity. Construction of a double mutant is further hampered by the presence of at least two genes, Foxc2 and Mthfsd, between Foxf1 and Foxl1. Nevertheless, our analysis of the regulatory circuits operative in the developing GI tract provides firm evidence placing Foxf1 and Foxl1 at a central node in the reciprocal signaling mediating organogenesis of the mammalian GI tract (Fig. 7B).
Although multiple genes are involved in the morphological processes of gut development, the mutation of single genes can have profound effects on the homeostasis of the adult gastrointestinal epithelium. Mutations of the Wnt signaling pathway represent the most salient causes of sporadic and familial colorectal cancer in humans. However, very few non-cell-autonomous regulators of intestinal epithelial proliferation and/or Wnt signaling have been described. One non-autonomous regulator, Foxl1, is a modifier of the mouse ApcMin mutation, acting as a suppressor of tumors formed from unchecked Wnt signaling, probably through the regulation of secreted proteins (37). Together, Foxl1 with Foxf1 could provide indispensable control of proliferation and homeostasis of the adult intestinal epithelium. As more mesenchymal regulators of epithelial proliferation are discovered in mice and humans, genetic risk factors associated with colorectal cancer could include an array of mutations relevant to this indirect tumor suppression, thus expanding the power of genetic testing and pharmacological intervention for preventative and therapeutic measures.
Supplementary Material
Acknowledgments
We thank Elizabeth Helmbrecht for the care of our animals and the morphology core of the Penn Center for Molecular Studies in Digestive and Kidney Diseases (DK-050306).
This work was supported, in whole or in part, by National Institutes of Health Grant DK-053839 through the NIDDK. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains a supplemental table.
Footnotes
The abbreviations used are: Hh, hedgehog; Shh, Sonic hedgehog; Ihh, Indian hedgehog; Hox, Homeobox; E, embryonic day; GI, gastrointestinal; EMSA, electrophoretic mobility shift assay; RT-PCR, reverse transcription-PCR; QPCR, quantitative PCR; ChIP, chromatin immunoprecipitation; CMF, Ca2+ and Mg2+-free; HBSS, Hanks' balanced salt solution; oligo, oligonucleotide.
References
- 1.Motoyama, J., Heng, H., Crackower, M. A., Takabatake, T., Takeshima, K., Tsui, L. C., and Hui, C. (1998) Mech. Dev. 78 81-84 [DOI] [PubMed] [Google Scholar]
- 2.Ramalho-Santos, M., Melton, D. A., and McMahon, A. P. (2000) Development (Camb.) 127 2763-2772 [DOI] [PubMed] [Google Scholar]
- 3.Wang, L. C., Nassir, F., Liu, Z. Y., Ling, L., Kuo, F., Crowell, T., Olson, D., Davidson, N. O., and Burkly, L. C. (2002) Gastroenterology 122 469-482 [DOI] [PubMed] [Google Scholar]
- 4.Sukegawa, A., Narita, T., Kameda, T., Saitoh, K., Nohno, T., Iba, H., Yasugi, S., and Fukuda, K. (2000) Development (Camb.) 127 1971-1980 [DOI] [PubMed] [Google Scholar]
- 5.Wang, Y., McMahon, A. P., and Allen, B. L. (2007) Curr. Opin. Cell Biol. 19 159-165 [DOI] [PubMed] [Google Scholar]
- 6.Apelqvist, A., Ahlgren, U., and Edlund, H. (1997) Curr. Biol. 7 801-804 [DOI] [PubMed] [Google Scholar]
- 7.Madison, B. B., Braunstein, K., Kuizon, E., Portman, K., Qiao, X. T., and Gumucio, D. L. (2005) Development (Camb.) 132 279-289 [DOI] [PubMed] [Google Scholar]
- 8.Hallikas, O., Palin, K., Sinjushina, N., Rautiainen, R., Partanen, J., Ukkonen, E., and Taipale, J. (2006) Cell 124 47-59 [DOI] [PubMed] [Google Scholar]
- 9.Mahlapuu, M., Enerback, S., and Carlsson, P. (2001) Development (Camb.) 128 2397-2406 [DOI] [PubMed] [Google Scholar]
- 10.Ormestad, M., Astorga, J., Landgren, H., Wang, T., Johansson, B. R., Miura, N., and Carlsson, P. (2006) Development (Camb.) 133 833-843 [DOI] [PubMed] [Google Scholar]
- 11.Kaestner, K. H., Silberg, D. G., Traber, P. G., and Schutz, G. (1997) Genes Dev. 11 1583-1595 [DOI] [PubMed] [Google Scholar]
- 12.Perreault, N., Katz, J. P., Sackett, S. D., and Kaestner, K. H. (2001) J. Biol. Chem. 276 43328-43333 [DOI] [PubMed] [Google Scholar]
- 13.Kaestner, K. H., Bleckmann, S. C., Monaghan, A. P., Schlondorff, J., Mincheva, A., Lichter, P., and Schutz, G. (1996) Development (Camb.) 122 1751-1758 [DOI] [PubMed] [Google Scholar]
- 14.Karlsson, L., Lindahl, P., Heath, J. K., and Betsholtz, C. (2000) Development (Camb.) 127 3457-3466 [DOI] [PubMed] [Google Scholar]
- 15.Fukuda, K., Yoshida, H., Sato, T., Furumoto, T. A., Mizutani-Koseki, Y., Suzuki, Y., Saito, Y., Takemori, T., Kimura, M., Sato, H., Taniguchi, M., Nishikawa, S., Nakayama, T., and Koseki, H. (2003) Dev. Biol. 255 278-289 [DOI] [PubMed] [Google Scholar]
- 16.Greenbaum, L. E., Li, W., Cressman, D. E., Peng, Y., Ciliberto, G., Poli, V., and Taub, R. (1998) J. Clin. Investig. 102 996-1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta, R. K., Gao, N., Gorski, R. K., White, P., Hardy, O. T., Rafiq, K., Brestelli, J. E., Chen, G., Stoeckert, C. J., Jr., and Kaestner, K. H. (2007) Genes Dev. 21 756-769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roessler, E., Ermilov, A. N., Grange, D. K., Wang, A., Grachtchouk, M., Dlugosz, A. A., and Muenke, M. (2005) Hum. Mol. Genet. 14 2181-2188 [DOI] [PubMed] [Google Scholar]
- 19.Horton, R. M., Ho, S. N., Pullen, J. K., Hunt, H. D., Cai, Z., and Pease, L. R. (1993) Methods Enzymol. 217 270-279 [DOI] [PubMed] [Google Scholar]
- 20.Rubins, N. E., Friedman, J. R., Le, P. P., Zhang, L., Brestelli, J., and Kaestner, K. H. (2005) Mol. Cell. Biol. 25 7069-7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hui, C. C., and Joyner, A. L. (1993) Nat. Genet. 3 241-246 [DOI] [PubMed] [Google Scholar]
- 22.Mo, R., Freer, A. M., Zinyk, D. L., Crackower, M. A., Michaud, J., Heng, H. H., Chik, K. W., Shi, X. M., Tsui, L. C., Cheng, S. H., Joyner, A. L., and Hui, C. (1997) Development (Camb.) 124 113-123 [DOI] [PubMed] [Google Scholar]
- 23.Kinzler, K. W., and Vogelstein, B. (1990) Mol. Cell. Biol. 10 634-642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vokes, S. A., Ji, H., McCuine, S., Tenzen, T., Giles, S., Zhong, S., Longabaugh, W. J., Davidson, E. H., Wong, W. H., and McMahon, A. P. (2007) Development (Camb.) 134 1977-1989 [DOI] [PubMed] [Google Scholar]
- 25.Dubchak, I., Brudno, M., Loots, G. G., Pachter, L., Mayor, C., Rubin, E. M., and Frazer, K. A. (2000) Genome Res. 10 1304-1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., and Dubchak, I. (2004) Nucleic Acids Res. 32 (Web Server issue) W273-W279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Black, B. L., and Olson, E. N. (1998) Annu. Rev. Cell Dev. Biol. 14 167-196 [DOI] [PubMed] [Google Scholar]
- 28.Agren, M., Kogerman, P., Kleman, M. I., Wessling, M., and Toftgard, R. (2004) Gene (Amst.) 330 101-114 [DOI] [PubMed] [Google Scholar]
- 29.Kim, I. M., Zhou, Y., Ramakrishna, S., Hughes, D. E., Solway, J., Costa, R. H., and Kalinichenko, V. V. (2005) J. Biol. Chem. 280 37908-37916 [DOI] [PubMed] [Google Scholar]
- 30.Bai, C. B., Stephen, D., and Joyner, A. L. (2004) Dev. Cell 6 103-115 [DOI] [PubMed] [Google Scholar]
- 31.Astorga, J., and Carlsson, P. (2007) Development (Camb.) 134 3753-3761 [DOI] [PubMed] [Google Scholar]
- 32.Schmahl, J., Raymond, C. S., and Soriano, P. (2007) Nat. Genet. 39 52-60 [DOI] [PubMed] [Google Scholar]
- 33.Haramis, A. P., Begthel, H., van den Born, M., van Es, J., Jonkheer, S., Offerhaus, G. J., and Clevers, H. (2004) Science 303 1684-1686 [DOI] [PubMed] [Google Scholar]
- 34.Batts, L. E., Polk, D. B., Dubois, R. N., and Kulessa, H. (2006) Dev. Dyn. 235 1563-1570 [DOI] [PubMed] [Google Scholar]
- 35.He, X. C., Zhang, J., Tong, W. G., Tawfik, O., Ross, J., Scoville, D. H., Tian, Q., Zeng, X., He, X., Wiedemann, L. M., Mishina, Y., and Li, L. (2004) Nat. Genet. 36 1117-1121 [DOI] [PubMed] [Google Scholar]
- 36.Malin, D., Kim, I. M., Boetticher, E., Kalin, T. V., Ramakrishna, S., Meliton, L., Ustiyan, V., Zhu, X., and Kalinichenko, V. V. (2007) Mol. Cell. Biol. 27 2486-2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perreault, N., Sackett, S. D., Katz, J. P., Furth, E. E., and Kaestner, K. H. (2005) Genes Dev. 19 311-315 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.