Abstract
Protein oxidation has been linked to accelerated aging and is a contributing factor to many diseases. Methionine residues are particularly susceptible to oxidation, but the resulting mixture of methionine R-sulfoxide (Met-RO) and methionine S-sulfoxide (Met-SO) can be repaired by thioredoxin-dependent enzymes MsrB and MsrA, respectively. Here, we describe a knock-out mouse deficient in selenoprotein MsrB1, the main mammalian MsrB located in the cytosol and nucleus. In these mice, in addition to the deletion of 14-kDa MsrB1, a 5-kDa selenoprotein form was specifically removed. Further studies revealed that the 5-kDa protein occurred in both mouse tissues and human HEK 293 cells; was down-regulated by MsrB1 small interfering RNA, selenium deficiency, and selenocysteine tRNA mutations; and was immunoprecipitated and recognized by MsrB1 antibodies. Specific labeling with 75Se and mass spectrometry analyses revealed that the 5-kDa selenoprotein corresponded to the C-terminal sequence of MsrB1. The MsrB1 knock-out mice lacked both 5- and 14-kDa MsrB1 forms and showed reduced MsrB activity, with the strongest effect seen in liver and kidney. In addition, MsrA activity was decreased by MsrB1 deficiency. Liver and kidney of the MsrB1 knock-out mice also showed increased levels of malondialdehyde, protein carbonyls, protein methionine sulfoxide, and oxidized glutathione as well as reduced levels of free and protein thiols, whereas these parameters were little changed in other organs examined. Overall, this study established an important contribution of MsrB1 to the redox control in mouse liver and kidney and identified a novel form of this protein.
Oxidation of proteins has been implicated in a variety of diseases and is a factor that influences aging (1-3). Compared with most other amino acids in proteins, Met is particularly susceptible to oxidation by reactive oxygen species. The product of such oxidation is a diastereomeric mixture of Met sulfoxides: Met R-sulfoxide (Met-RO)2 and Met S-sulfoxide (Met-SO). These oxidized forms, however, can be reduced back to Met by two enzyme families, MsrA (specific for Met-SO) and MsrB (specific for Met-RO) (4, 5).
Mammals contain one MsrA, which is targeted to different cellular compartments by alternative splicing and variable use of signal peptides (6, 7), and three MsrB isozymes, which are individually targeted to different compartments (8-10). MsrB located in the cytosol and nucleus is known as MsrB1 (also designated as SelR or SelX). This protein is interesting in that it is a selenoprotein that contains selenocysteine (Sec) in place of cysteine normally present in the catalytic sites of MsrBs and MsrAs (11). Dietary selenium is known to regulate the expression of selenoproteins and to ultimately control the redox homeostasis in cells (12). Being a selenoprotein, the expression of MsrB1 can also be regulated by dietary selenium (13-15).
Knock-out of the MsrA gene in mice was described several years ago (16). The lack of MsrA results in the accumulation of carbonyl derivatives (indicative of protein oxidation) (17), increased susceptibility to exposure to 100% oxygen, and various neurological abnormalities, such as tip-toe walking, abnormal behavior, and lower locomotor activity (16, 18, 19). In addition, MsrB expression and activity were down-regulated in these mice (13). Most interestingly, MsrA knock-out mice showed a dramatically reduced life span (16), whereas overexpression of this protein in fruit flies increased their life span (20, 21). These studies are consistent with the idea that repair of oxidized proteins is a critical factor in neurological diseases and the aging process (4).
In contrast to MsrA, the consequence of MsrB deficiency in mice or other animal models is not known (although MsrB also can regulate the life span of yeast cells (22)). Selenium deficiency was used to reduce MsrB1 expression (13, 16), which resulted in lower MsrB activity as well as in a reduction in the levels of other selenoproteins. This treatment increased protein oxidation and the amount of Met sulfoxide residues (13). Whereas the knock-out of MsrA removed this protein in all tissues and compartments, the specific removal of individual MsrBs offers an opportunity to examine the effects of MsrB deficiency in different organs, cells, and compartments.
In this work, we describe mice lacking MsrB1. We show that MsrB1 knock-out led to protein oxidation and affected other parameters indicative of oxidative stress. Most effects were observed in the liver and kidney, where MsrB1 levels were particularly high, whereas several other organs were not affected. An additional surprising finding was that MsrB1 knock-out led to the removal of a 5-kDa selenoprotein. We examined this process in more detail and show that this protein corresponds to the C-terminal segment of MsrB1.
EXPERIMENTAL PROCEDURES
Materials—General purpose chemicals were from Sigma, DNA purification kits were from Qiagen, the RNA isolation kit was from Ambion, and mammalian cell culture reagents and protein electrophoresis reagents were from Invitrogen. Restriction enzymes were from Fermentas. KOD XL DNA polymerase (Novagen) was used for genomic DNA amplification and for MsrB1 knock-out allele screening. E. coli NovaBlue cells (Novagen) were used for recombinant DNA transformation and BL21 (DE3) cells (Novagen) for recombinant protein synthesis.
MsrB1 Knock-out Mice—Two strategies were used to prepare MsrB1 knock-out (KO) mice. Two separate targeting constructs were prepared and used to target the MsrB1 gene by homologous recombination (Fig. 1A). The following primer pairs were used to amplify a 4550-bp region, upstream of the first exon of MsrB1 by nested PCR: UP_D1 5′-TTCAGTGTTGGTGTAGGACTGTAGGAC-3′, UP_D2 5′-AGTCAGCGGCCGCCCTTACCAAGGTCATGAGTGACCAG-3′, UP_R1 5′-GCCTCCGAAGAAGCTGCAGAACGACAT-3′, UP_R2 5′-AGTAGCGGCCGCGGGAGCCGCTCCCAGGAAGCTTAG-3′. The amplified fragment was cloned into the NotI site of pPNT targeting vector, and the correct fragment orientation was verified by restriction analysis and sequencing. This region was common to both targeting constructs. The region downstream of the first exon of the MsrB1 gene was amplified by nested PCR with the following primer pairs (the first construct): DWN_D1, 5′-GTTATTGGGACTGGGTCGGGTTTGAGTCC-3′; DWN_D2, 5′-AGTCAGGATCCGGCCTTGAACTTCAAATGTAGATC-3′; DWN_R1_1, 5′-CGAATGCCTGAGGTTCTGACACTAACGTG-3′; DWN_R2_1, 5′-AGTCAGGTACCAGAGAGTGCTGTATGGTAAGTTCCAG-3′. The amplified 1080-bp fragment was cloned into BamHI-KpnI sites of the pPNT targeting vector. For the second construct, the following primer pairs were used: DWN_D1, 5′-GTTATTGGGACTGGGTCGGGTTTGAGTCC-3′; DWN_D2, 5′AGTCAGGATCCGGCCTTGAACTTCAAATGTAGATC-3′; DWN_R1_2, 5′-CTTCGAGTGACTGGAGAACAGCTCATAGC-3′; DWN_R2_2, 5′-AGTCAGGTACCGCCAGTGCTGCAGACACACAATACAG-3′. The amplified 4824-bp fragment was cloned into BamHI-KpnI sites of the pPNT targeting vector.
FIGURE 1.
MsrB1 knock-out mice. A, mouse MsrB1 gene and its flanking regions. The upper part of the panel shows the MsrB1 gene and its flanking genes, and the lower part illustrates MsrB1 exons (in red) and repetitive elements (in blue). The targeting strategy is also shown wherein a Neo cassette (in green) replaces the first exon using the areas of homologous recombination (in gray). Two alternative 3′ arms of the targeting construct are indicated (dashed line). B, the genetrap construct. The trapping cassette was inserted immediately downstream of the first exon. Locations of MsrB1 exons (in orange), Sec (shown as a red U), and the Sec insertion sequence (in blue) are also indicated. C, immunoblot analysis of MsrB1, MsrB2, and MsrA in the liver and heart. Analyses were carried out in wild-type (+/+), heterozygous MsrB1 KO (+/-), and homozygous MsrB1 KO (-/-) mice. Actin was used as a loading control. D, immunoblot analyses of MsrB1 and MsrA in kidney, testis, and lung.
More than 500 ES cell clones were tested for successful MsrB1 targeting with these constructs; however, no positive cells were detected, probably because of the high content of repetitive elements in the vicinity of the MsrB1 gene (predicted with RepeatMasker, available on the World Wide Web).
As an alternative strategy, a mouse (in a 129/Sv genetic background) was generated at Texas Institute for Genomic Medicine by taking advantage of the genetrap cassette inserted immediately downstream of the MsrB1 gene (Fig. 1B). Homozygous MsrB1 KO mice were obtained by mating heterozygous mice and selecting for homozygous MsrB1 KO by genetic screening. Mice were handled following the guidelines and approved protocols at the University of Nebraska-Lincoln.
Preparation of Protein Extracts from Mouse Tissues—Tissues were glass-homogenized and sonicated in PBS, pH 7.6, containing a mixture of protease inhibitors and 20 mm EDTA at 0 °C.
Metabolic Labeling of Mice and Cell Cultures with 75Se—[75Se]Selenite (specific activity 1000 Ci/mmol) was obtained from the Research Reactor Facility, University of Missouri (Columbia, MO). Wild-type and MsrB1 KO mice were injected with 2.5 mCi of 75Se each and maintained for 2 days at a 12-h light/dark cycle on a regular diet. The mice were sacrificed, tissues were extracted, and protein extracts were prepared from various tissues in ice-cold PBS buffer, pH 7.6, supplemented with complete protease inhibitor mixture from Roche Applied Science. HEK 293 cells were grown and labeled with 75Se as described (23). Extracts from the indicated mouse tissues or human cells were subjected to SDS-PAGE, and proteins were transferred onto polyvinylidene difluoride membranes and analyzed with a PhosphorImager.
MsrB1 Knockdown—To down-regulate MsrB1 expression, we used siRNA from Dharmacon. 4 μl of siRNA in RNase-free buffer were mixed with 1 μl of DharmaFECT transfection reagent and antibiotic-free/serum-free medium to a total volume of 40 μl. Samples were incubated for 30 min at room temperature and added to HEK 293 cells cultured in 6-well plates. After 24-48 h of incubation, medium was substituted with that containing 75Se, and cells were incubated for an additional 12 h and then washed with PBS, pH 7.6, and lysed with mammalian cell lysis reagent from Sigma. The lysate was subjected to SDS-PAGE electrophoresis and PhosphorImager analysis.
Dietary Studies—One-month-old BALB/c mice were maintained for 3 months on Torula yeast-based diets supplemented with 0.1 ppm selenium or 0.4 ppm selenium (as sodium selenite) or not supplemented with Se (0 ppm selenium) (Harlan Teklad) as described (15). After 8 weeks, mice were sacrificed, and protein extracts were prepared from the indicated tissues and analyzed by Western blot assays with antibodies specific for MsrB1, MsrA, MsrB2, MsrB3, or β-actin.
Immunoprecipitation of MsrB1 Forms—MsrB1 antibodies were added to protein extracts of mouse livers in PBS, pH 7.6, containing complete protease inhibitor mixture. Samples were further incubated at 4 °C with continuous shaking for 12 h. MsrB1 was pooled using a Protein G immunoprecipitation kit (Sigma) according to the manufacturer's recommendations.
Affinity Enrichment of MsrB1 Forms—MsrB1 antibodies were linked to activated BrCN-Sepharose from Sigma according to the manufacturer's recommendations. Total protein extracts from 30 wild-type mouse livers in PBS, pH 7.6, containing complete protease inhibitor mixture were incubated with the resin containing immobilized antibodies for 12 h at 4 °C. The resin was extensively washed with 50 mm Tris-HCl, pH 8.0, 200 mm NaCl, to remove nonspecifically bound proteins. MsrB1 forms were eluted with 3 ml of 0.1 m glycine, pH 2.5. Proteins were reduced with 20 mm dithiothreitol for 30 min, and cysteines/Sec were modified with 100 mm iodoacetamide for 1 h at room temperature. Proteins were concentrated using 3 kDa cut-off Microcon concentrators (Millipore), and MsrB1 forms were visualized by immunoblot assays with MsrB1 antibodies. The band corresponding to the 5-kDa protein form was cut from an SDS-polyacrylamide gel, and proteins were digested with trypsin or V8 protease and subjected to LC-MS/MS protein sequence analysis at Midwest Bio Services, LLC.
MsrA and MsrB Activity Assays—MsrA and MsrB activities were assayed in homogenates of the indicated mouse tissues. 200 μg of protein were used in each assay. The reaction (100 μl) proceeded at 37 °C for 30 min in the presence of dithiothreitol (20 mm), and either 200 μm dabsyl-Met-SO (MsrA assay) or 200 μm dabsyl-Met-RO (MsrB assay) was added to the reaction mixture. After stopping the reaction by adding 200 μl of acetonitrile, the mixtures were centrifuged at 4 °C for 15 min at 13,000 rpm, and then the supernatant (50 μl) was injected onto a C18 column (ZORBAX Eclipse XDB-C18) to quantify dabsyl-Met in an HPLC system.
Analyses of Oxidative Stress Parameters—Protein extracts from liver, kidney, heart, testis, and brain were prepared in PBS containing complete protease inhibitor mixture. Malondialdehyde was measured using the Oxiselect TBARS assay kit (Cell Biolabs, San Diego, CA) according to the manufacturer's instructions. Protein carbonyl content was assayed using 2,4-dinitrophenyl hydrazine and calculated using a molar extinction coefficient of 22 mm-1 cm-1 (24). Total and protein thiol contents were measured using 5,5′-dithiobis(2-nitrobenzoic acid) as described (25) and expressed as nmol/mg protein. Reduced and oxidized glutathione were assayed using the Bioxytech GSH/GSSG kit (OxisResearch, Portland OR) according to the manufacturer's instructions.
Quantitation of Met-RO and Met-SO Residues in Proteins—Liver samples (2 mg of protein) were hydrolyzed with 3 m p-toluene sulfonic acid at 110 °C for 22 h as described (26), and Met sulfoxides were separated by HPLC after precolumn O-phthalaldehyde derivatization using a Zorbax C18-Rx column as described (27).
MsrB1 Three-dimensional Structure Modeling—The three-dimensional structure of mouse MsrB1 was modeled with Modeler 8.2 based on sequence similarity to MsrB from Bacillus subtilis (Protein Data Bank code 1XM0).
RESULTS AND DISCUSSION
Mice Lacking MsrB1 Are Viable—To generate the MsrB1 knock-out mouse, we initially utilized a strategy wherein a Neo cassette was inserted by homologous recombination. The mouse MsrB1 gene is located on chromosome 17 and flanked by Rpl3l and Hstst6 genes, the former being particularly close to the MsrB1 gene (Fig. 1A). In addition, the MsrB1 gene is flanked by a series of repetitive elements, especially abundant at the 3′-end as well as in some MsrB1 introns. We employed the targeting vector as shown in Fig. 1A, with the 3′-arm either 1 or 4 kb (to avoid repetitive elements). However, no ES cells positive for homologous recombination at the MsrB1 gene were found following repeated attempts.
We then took advantage of the genetrap clone with the disrupting cassette inserted immediately after the first exon of MsrB1 (Fig. 1B). Mice were prepared that contained this cassette in both gene copies (i.e. homozygous mice) and were found to be viable. These mice had undetectable levels of MsrB1 mRNA (data not shown), and MsrB1 protein was also not detected in liver, heart, kidney, testis, and lung (Fig. 1, C and D). In addition, heterozygous mice containing the genetrap cassette had lower MsrB1 levels than wild-type mice. We further refer to the mice lacking the MsrB1 protein and mRNA as MsrB1 knock-out mice (MsrB1 KO). We also examined the expression of MsrA in MsrB1 KO mice and found that it was unchanged compared with wild-type mice. However, MsrB2 (mitochondrial MsrB) expression was slightly elevated in the liver and heart (this protein was not detected in other tissues examined). In contrast, MsrB3 levels in heart were not changed, but it showed slightly increased levels in skeletal muscle (both tissues are among those with highest MsrB3 expression) (Fig. S1).
A 5-kDa Selenoprotein Is an Alternative Form of MsrB1—Since MsrB1 is a selenoprotein, we examined how its levels, as well as the expression of other selenoproteins, were affected in the knock-out mice. MsrB1 KO and wild-type mice were metabolically labeled with 75Se, and the selenoprotein expression was examined in liver, kidney, and testis (Fig. 2A). We found no differences in the levels of ∼14-kDa 75Se-labeled proteins (which correspond in size to MsrB1). Apparently, MsrB1 over-lapped on SDS-polyacrylamide gels with other selenoproteins of similar size, including SelM, Sep15, SelH, and SelK. Some of these selenoproteins were more abundant than MsrB1, which precluded MsrB1 detection with 75Se. The overall expression of selenoproteins was not changed by MsrB1 KO when assayed by 75Se labeling (Fig. 2A) or immunoblot assays (Fig. S2).
FIGURE 2.
Identification of the short form of mouse MsrB1. A, metabolic labeling of MsrB1 KO mice with 75Se. Wild-type (+/+) and MsrB1 KO (-/-) mice were injected with 75Se, and, after 2 days, tissues were extracted and proteins were analyzed by SDS-PAGE, followed by PhosphorImager analysis. Migration of full-length and short forms of MsrB1 is shown by arrows on the left. B, immunoprecipitation of 75Se-labeled MsrB1 from mouse liver. MsrB1 was immunoprecipitated from liver extracts of wild-type (+/+, left panel) and MsrB1 KO (-/-, right panel) mice with MsrB1 antibodies. Initial, unbound (flow-through), wash, and eluted fractions were collected and separated by SDS-PAGE, and the labeled protein bands were visualized with a PhosphorImager. Migration of full-length and short forms of MsrB1 is shown by arrows on the left. C, MsrB1 was affinity-isolated from mouse liver extracts on the column containing MsrB1 antibodies and analyzed by immunoblot assays. Migration of full-length and short forms of MsrB1 is shown by the arrows on the left. Molecular weight markers are shown on the right in each panel.
Interesting, however, was an observation of specific removal of a 5-kDa selenoprotein band in liver and kidney of MsrB1 KO mice, the two tissues with highest MsrB1 expression (Fig. 2A). To determine if the 5-kDa selenoprotein has a relation to MsrB1, we immunoprecipitated MsrB1 from 75Se-labeled livers of wild-type and MsrB1 KO mice with antibodies specific for mouse MsrB1 (Fig. 2B). The 75Se-labeled 5 kDa band was specifically removed by the MsrB1 antibodies from samples derived from wild-type mice, and the pooled fraction contained both the 14- and 5-kDa protein bands, the former corresponding to the full-length MsrB1 and the latter to the trapped 5-kDa selenoprotein. These bands were not immunoprecipitated by the MsrB1 antibodies from the MsrB1 KO samples.
We prepared an affinity column containing the MsrB1 antibodies and used it to affinity-isolate MsrB1 forms from liver extracts of wild-type mice. The samples were then analyzed by immunoblot assays, and both bands were trapped on the MsrB1 affinity column (Fig. 2C).
Next, regulation of the 5-kDa form was examined in mice with altered Sec tRNA status (Fig. S3). Sec tRNA is essential in mammals, but its knock-out can be rescued with wild-type or mutant Sec tRNA transgenes. We found that expression of wild-type Sec tRNA transgene in mice lacking the natural Sec tRNA gene rescued expression of both 5 and 14 kDa bands as detected by immunoblot assays with MsrB1 antibodies. However, when the Sec tRNA transgene containing a mutation in position 37 was expressed in the Sec tRNA knock-out mice, neither 14 nor 5 kDa bands were expressed. Thus, both MsrB1 forms were similarly regulated by Sec tRNA status.
We also found that 5- and 14-kDa forms were detected in other mouse tissues, including lung, muscle, and spleen (Fig. S3). When Sec tRNA knock-out was rescued with a transgene lacking the activator region (STAF-binding site),3 expression of both 14- and 5-kDa forms was reduced.
Overall, the data suggested that the 5-kDa selenoprotein is a novel form of mouse MsrB1. Since Sec in MsrB1 is located in the C-terminal portion of the protein, and the 5-kDa protein was recognized by MsrB1 antibodies, the data suggested that this small selenoprotein corresponded to the C-terminal portion of MsrB1. The fact that the immunoblot signal was weaker than the 75Se signal can be explained by unequal immunogenicity of the C-terminal portion and the full-length MsrB1. In fact, comparison of 75Se signals for 14- and 5-kDa MsrB1 forms suggests that the 5-kDa form is more abundant than the 14-kDa form.
Mass Spectrometry Analysis of the 5-kDa MsrB1 Form—We isolated the 5-kDa selenoprotein from mouse liver on an affinity column containing MsrB1 antibodies, resolved the protein by SDS-PAGE, and subjected the 5 kDa band to LC-MS/MS sequence analysis. Standard trypsin digestion did not yield MsrB1 peptides. We next used V8 protease from Staphylococcus aureus, which preferentially cleaves after aspartic and glutamic acid residues. This procedure detected a peptide derived from mouse MsrB1, NGLGHEFLND (Fig. 3A). The C-terminal part of this peptide corresponded to the expected V8 protease cleavage site, but the N-terminal part did not. This observation and the predicted size of the resulting MsrB1 form (∼5 kDa) suggested that the identified peptide corresponded to the N-terminal part of the short form of MsrB1.
FIGURE 3.
The 5-kDa selenoprotein is derived from full-length mouse MsrB1. A, protein sequence of mouse MsrB1. The identified cleavage site between Gly75 and Asn76 in MsrB1 is shown by a caret. The detected peptide derived from V8 protease cleavage is shown in blue. B, structural model of mouse MsrB1. The zinc atom was modeled based on the location of known ligands (4 cysteine residues from two CXXC motifs) of this metal ion. Predicted protease cleavage site is marked by a red dot and highlighted by scissors.
Structural Base for Formation of the Short Form of MsrB1—We modeled the three-dimensional structure of mouse MsrB1 based on the B. subtilis MsrB structure (Protein Data Bank code 1XM0) (Fig. 3B). The predicted cleavage site in MsrB1 was located on the protein surface. Sequences located 4 amino acids up and down the predicted cleavage site showed decreased solvent exposure. Taken together, the observed size of the MsrB1 short form, the identified N-terminal peptide, and the known occurrence of Sec in the C-terminal portion of MsrB1 all suggested that the 5-kDa form was generated from the full-length MsrB1 by proteolytic cleavage between Gly75 and Asn76.
Expression of 14- and 5-kDa MsrB1 Forms Is Regulated by Dietary Selenium—MsrB1 is a selenoprotein, whose expression is regulated by dietary selenium. To examine whether dietary selenium also regulates the expression of the 5-kDa MsrB1, we subjected wild-type mice to three Torula yeast-based selenium diets, including selenium-deficient (i.e. no selenium added), 0.1-ppm selenium, and 0.4-ppm selenium diets. As expected, expression of the full-length MsrB1 in the liver was dramatically reduced by selenium deficiency in these mice (Fig. 4A). Interestingly, the 5-kDa MsrB1 was undetectable under these conditions. Thus, both 14- and 5-kDa MsrB1 forms were regulated by dietary selenium.
FIGURE 4.
Regulation of full-length and short forms of mouse MsrB1. A, regulation by dietary selenium. Wild-type mice were fed selenium-deficient (0 ppm selenium) diet and diets containing 0.1 or 0.4 ppm selenium. Liver extracts from these mice were analyzed by Western blot assays with antibodies against MsrB1 (upper panel). Migration of full-length and short forms of MsrB1 is shown by arrows on the left. Each lane corresponds to liver extract from a different mouse. The lower panel shows protein loading (Coomassie Blue staining). B, stability of full-length and short forms of mouse MsrB1. Freshly prepared liver extract from wild-type mice was incubated with or without protease inhibitor mixture for 0, 3, 6, or 12 h at room temperature, and the two MsrB1 forms were detected by immunoblot assays.
The 5-kDa Form Is Not Due to Sample Preparation—We examined the possibility that the 5-kDa MsrB1 is the result of sample preparation wherein the full-length MsrB1 is degraded by nonspecific proteolytic cleavage. Extracts from mouse liver were incubated with or without protease inhibitors for 0, 3, 6, and 12 h and analyzed for the occurrence of the 14- and 5-kDa MsrB1 forms. In the presence of protease inhibitors along with 20 mm EDTA, both forms were stable for 12 h (Fig. 4B). In the absence of inhibitors, the proteins were stable for 6 h, and then the levels of both protein forms were reduced (rather than converted from the 14- to 5-kDa form). Thus, both MsrB1 forms were stable and resistant to nonspecific proteolytic cleavage. This finding argues against degradation of full-length MsrB1 during sample preparation. It should also be noted that abundant, partially degraded forms are not known for any selenoproteins. All major bands in the 75Se labeling pattern can be accounted for and correspond to known selenoproteins.4 In addition, the short form of MsrB1 is the only selenoprotein that did not match the size of any predicted mouse selenoprotein.
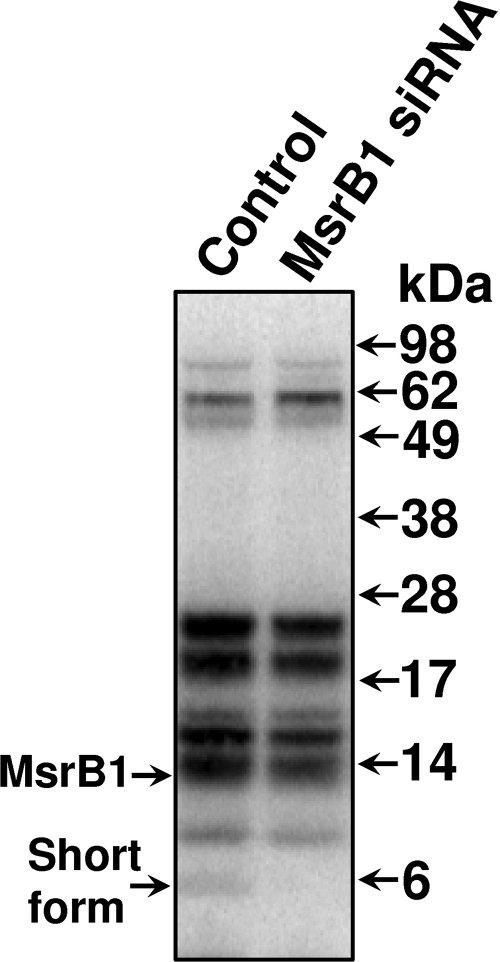
The 5-kDa MsrB1 Form Occurs in Human Cells—To determine if the 5-kDa MsrB1 form is specific to mice or also occurs in other species, we subjected human HEK 293 cells to metabolic 75Se labeling. A 5-kDa selenoprotein band was observed (Fig. 5, left lane). Knockdown of MsrB1 by siRNA completely removed this band without affecting the expression of other selenoproteins. Thus, the short form of MsrB1 also occurs in humans. As in mice, the full-length MsrB1 overlapped with several other selenoproteins on SDS-PAGE gel; therefore, the effect of knockdown on this form was difficult to assess.
FIGURE 5.
The short form of human MsrB1. Human embryonic kidney HEK 293 cells and the HEK 293 cells subjected to MsrB1 siRNA were labeled with 75Se, protein extracts were resolved by SDS-PAGE, and the labeled proteins were visualized with a PhosphorImager. Migration of full-length and short forms of MsrB1 is shown by arrows on the left and molecular weight markers on the right.
The molecular basis for generation of the 5-kDa MsrB1 form is not known. It is also not clear what the function of the MsrB1 short form could be. Being part of the overall fold, the C-terminal fragment could not catalyze the reduction of Met-RO on its own. Further studies are required to ascertain the mechanism by which the 5-kDa selenoprotein form is generated and the biological function of this protein form.
Decreased MsrA Activity in MsrB1 Knock-out Mice—We examined MsrB and MsrA activities in the liver, kidney, heart, testis, lung, and brain of MsrB KO mice (Fig. 6). Compared with wild-type mice, total MsrB activity was most dramatically decreased in liver (p value 0.004) and kidneys (p value 0.012) of MsrB1 KO mice, which are the organs with highest MsrB1 expression. Activity was also partially decreased in the testis (p value 0.001), lung (p value 0.002), and brain (p value 0.0004). The least change in MsrB activity was observed in the heart, an organ where MsrB2 is highly expressed.
FIGURE 6.
MsrA and MsrB activities in MsrB1 KO mice. Total MsrB (shown in orange) and MsrA (shown in green) activities (expressed in pmol/min/mg protein) were analyzed in extracts from the indicated tissues of wild-type mice (+/+), and heterozygous (+/-) and homozygous (-/-) MsrB1 KO mice. Dabsylated Met-RO and Met-SO substrates were used in the reaction as described under “Experimental Procedures.”
Interestingly, we also observed a decreased MsrA activity in each of the organs tested, with liver (p value 0.009) and lung (p value 0.002) showing the largest drop and heart the least change in activity. A previous study revealed that MsrB activity is decreased in MsrA KO mice (13). Thus, MsrA and MsrB1 may regulate each other with respect to activity in the cells. It should be noted that although MsrA activity was decreased (Fig. 6), its expression was not (Fig. 1, C and D).
Oxidative Stress in MsrB1 KO Mice—We examined the parameters of oxidative stress in MsrB1 KO mice and found increased levels of malondialdehyde and protein carbonyls in liver (p value 0.01), kidney (p value 0.001), and brain (p value 0.01) samples of MsrB1 KO mice, whereas these parameters did not change significantly in heart and testis (Fig. 7). We also found reduced protein and total thiol levels in liver (p value 0.001) and kidneys (p value 0.001) of MsrB1 KO mice, but other organs were not significantly affected (Fig. 8). Liver and kidneys were further analyzed with respect to glutathione status. We found that the level of reduced glutathione was decreased (p value 0.04), and GSSG increased in these organs (p value 0.01) (Fig. 9). Accordingly, the GSH/GSSG ratio was reduced in the MsrB1 KO samples. Finally, we assayed for protein-based methionine sulfoxides in liver and kidneys and found elevated levels of methionine sulfoxide residues in MsrB1 KO mice (Fig. 10). The last finding is fully consistent with the idea that MsrB1 is the main MsrB in liver and kidneys. Overall, the data suggested an important role of MsrB1 in the redox control in liver and kidney, organs with maximal levels of MsrB1, but a lower impact of this protein in other organs.
FIGURE 7.
Analyses of malondialdehyde and protein carbonyl levels in MsrB1 KO mice. Levels of malondialdehyde (A) and protein carbonyls (B) in liver, kidney, heart, testes, and brain of wild-type mice (+/+), and heterozygous (+/-) and homozygous (-/-) MsrB1 KO mice are shown. The assays are described under “Experimental Procedures.”
FIGURE 8.
Analyses of total and protein thiols in MsrB1 KO mice. The total (A) and protein-based (B) thiols in the liver, kidney, heart, testes, and brain of wild-type mice (+/+) and heterozygous (+/-) and homozygous (-/-) MsrB1 KO mice are shown. The assays are described under “Experimental Procedures.”
FIGURE 9.
Analyses of reduced and oxidized glutathione in MsrB1 KO mice. A, GSH, reduced glutathione. B, GSSG, oxidized glutathione. C, the ratio of reduced/oxidized glutathione. Analyses were carried out in livers and kidneys of wild-type mice (+/+) and heterozygous (+/-) and homozygous (-/-) MsrB1 KO mice. The assays are described under “Experimental Procedures.”
FIGURE 10.
Analyses of methionine sulfoxide in proteins. Levels of Met-SO and Met-RO residues in proteins were determined in liver and kidney samples from wild-type (+/+), and heterozygous (+/-) and homozygous (-/-) MsrB1 KO mice. The assays are described under “Experimental Procedures.”
MsrB1 KO Mice Do Not Show Phenotypes Seen in MsrA KO Mice—Previous studies revealed a dramatically reduced life span (the mice lived only for 1 year) and a tip-toe walking behavior of MsrA KO mice (16). Our MsrB1 KO mice appeared to be normal (without strong phenotypes). The oldest animals, currently 20 months old, are still alive, suggesting that the same dramatic effect on the life span found in MsrA KO mice will not be seen with the MsrB1 KO mice. However, considering the apparent oxidative stress seen in these mice, some reduction in the life span may be expected. We also did not observe a tip-toe walking behavior in young and middle-aged MsrB1 KO mice. Thus, there are important differences between MsrA KO and MsrB1 KO mice.
In summary, knock-out of the MsrB1 gene in mice is viable and leads to the conditions of oxidative stress; however, the phenotypes are milder than in the MsrA knock-out mice. In addition, our analyses revealed that liver and kidney are severely affected, whereas the redox status of other organs examined was not changed significantly. Since MsrB1 is also highly expressed in liver and kidney, these organs appear to be the prime sites of MsrB1 function. Another interesting observation made in this study is the discovery of a novel selenoprotein form of MsrB1, which is apparently even more abundant than the full-length form. The short, 5-kDa form occurs in both mice and humans and corresponds to the C-terminal segment of the protein. Expression of this protein is regulated by dietary selenium, similar to that of the 14-kDa full-length MsrB1, and both forms appear to be stable proteins. Further studies will be needed to determine the mechanism of formation and function of the 5-kDa MsrB1 form.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant AG021518 (to V. N. G.) and the Intramural Research Program of the Center for Cancer Research, NCI, National Institutes of Health (to D. L. H.). The Nebraska Redox Biology Center is supported by National Institutes of Health Grant RR017675. This work was also supported by Korea Science and Engineering Foundation Grant R13-2005-005-01004-0 (to H. Y. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S3.
Footnotes
The abbreviations used are: Met-RO, methionine R-sulfoxide; Met-SO, methionine S-sulfoxide; KO, knock-out; Sec, selenocysteine; PBS, phosphate-buffered saline; siRNA, small interfering RNA; LC, liquid chromatography; MS, mass spectrometry; Se, selenium.
B. A. Carlson and D. L. Hatfield, unpublished results.
S. V. Novoselov and V. N. Gladyshev, unpublished results.
References
- 1.Stadtman, E. R. (1992) Science 257 1220-1224 [DOI] [PubMed] [Google Scholar]
- 2.Stadtman, E. R., Van Remmen, H., Richardson, A., Wehr, N. B., and Levine, R. L. (2005) Biochim. Biophys. Acta 1703 135-140 [DOI] [PubMed] [Google Scholar]
- 3.Picot, C. R., Perichon, M., Lundberg, K. C., Friguet, B., Szweda, L. I., and Petropoulos, I. (2006) Exp. Gerontol. 41 663-667 [DOI] [PubMed] [Google Scholar]
- 4.Weissbach, H., Resnick, L., and Brot, N. (2005) Biochim. Biophys. Acta 1703 203-212 [DOI] [PubMed] [Google Scholar]
- 5.Hansel, A., Heinemann, S. H., and Hoshi, T. (2005) Biochim. Biophys. Acta 1703 239-247 [DOI] [PubMed] [Google Scholar]
- 6.Kim, H. Y., and Gladyshev, V. N. (2005) Biochemistry 44 8059-8067 [DOI] [PubMed] [Google Scholar]
- 7.Vougier, S., Mary, J., and Friguet, B. (2003) Biochem. J. 373 531-537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim, H. Y., and Gladyshev, V. N. (2004) Biochem. Biophys. Res. Commun. 320 1277-1283 [DOI] [PubMed] [Google Scholar]
- 9.Kim, H. Y., and Gladyshev, V. N. (2004) Mol. Biol. Cell 15 1055-1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marchetti, M. A., Pizarro, G. O., Sagher, D., Deamicis, C., Brot, N., Hejtmancik, J. F., Weissbach, H., and Kantorow, M. (2005) Invest. Ophthalmol. Vis. Sci. 46 2107-2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kryukov, G. V., Kryukov, V. M., and Gladyshev, V. N. (1999) J. Biol. Chem. 274 33888-33897 [DOI] [PubMed] [Google Scholar]
- 12.Burk, R. F. (2002) Nutr. Clin. Care 5 75-79 [DOI] [PubMed] [Google Scholar]
- 13.Moskovitz, J., and Stadtman, E. R. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 7486-7490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uthus, E. O., and Moskovitz, J. (2007) Biol. Trace Elem. Res. 115 265-276 [DOI] [PubMed] [Google Scholar]
- 15.Novoselov, S. V., Calvisi, D. V., Labunskyy, V. M., Factor, V. M., Carlson, B. A., Fomenko, D. E., Moustafa, M. E., Hatfield, D. L., and Gladyshev V. N. (2005) Oncogene 24 8003-8011 [DOI] [PubMed] [Google Scholar]
- 16.Moskovitz, J., Bar-Noy, S., Williams, W. M., Requena, J., Berlett, B. S., and Stadtman, E. R. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 12920-12925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oien, D., and Moskovitz, J. (2007) Amino Acids 32 603-606 [DOI] [PubMed] [Google Scholar]
- 18.Pal, R., Oien, D. B., Ersen, F. Y., and Moskovitz, J. (2007) Exp. Brain Res. 180 765-774 [DOI] [PubMed] [Google Scholar]
- 19.Oien, D. B., Osterhaus, G. L., Latif, S. A., Pinkston, J. W., Fulks, J., Johnson, M., Fowler, S. C., and Moskovitz, J. (2008) Free Radic. Biol. Med. 45 193-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruan, H., Tang, X. D., Chen, M. L., Joiner, M. L., Sun, G., Brot, N., Weissbach, H., Heinemann, S. H., Iverson, L., Wu, C. F., and Hoshi, T. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 2748-2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wassef, R., Haenold, R., Hansel, A., Brot, N., Heinemann, S. H., and Hoshi, T. (2007) J. Neurosci. 27 12808-12816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koc, A., Gasch, A. P., Rutherford, J. C., Kim, H. Y., and Gladyshev, V. N. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 7999-8004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novoselov, S. V., Lobanov, A. V., Hua, D., Kasaikina, M. V., Hatfield, D. L., and Gladyshev, V. N. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7857-7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sohal, R. S., Agarwal, S., Dubey, A., and Orr, W. C. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 7255-7259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Natarajan, S. K., Thomas, S., Ramamoorthy, P., Basivireddy, J., Pulimood, A. B., Ramachandran, A., and Balasubramanian, K. A. (2006) J. Gastroenterol. Hepatol. 21 947-957 [DOI] [PubMed] [Google Scholar]
- 26.Hayashi, R., and Suzuki, F. (1985) Anal. Biochem. 149 521-528 [DOI] [PubMed] [Google Scholar]
- 27.Sharov, V. S., Ferrington, D. A., Squier, T. C., and Schöneich C. (1999) FEBS Lett. 455 247-250 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.