Abstract
FIP200 (focal adhesion kinase family interacting protein of 200 kDa) has been shown to interact with other proteins to regulate several intracellular signaling pathways. To study a potential role of FIP200 in tumorigenesis and possibly other disease processes in vivo, we created and analyzed murine mammary tumor virus-Cre-mediated FIP200 conditional knock-out (CKO) mice. We found that deletion of FIP200 in mammary epithelial cells did not result in spontaneous development of breast cancer. Moreover, deletion of FIP200 did not further accelerate or inhibit lymphomagenesis induced by inactivation of p53 in mice. Interestingly, however, FIP200 and p53 double conditional knock-out (dCKO) mice exhibited significant hyperplasia of epidermis (acanthosis), thickening of the cornified layer (hyperkeratosis), and increased vascularity in the dermis. FIP200 CKO mice also showed similar, although less severe, skin defects as dCKO mice. Analyses of primary keratinocytes isolated from dCKO mice did not detect increased proliferation of these cells in vitro, suggesting that epidermis hyperproliferation is not epidermal cell-autonomous but may be a consequence of increased inflammation triggered by immune cells in vivo. Consistent with this possibility, we found infiltration of leukocytes including T cells, macrophages, and granulocytes into the dermis and epidermis, associated with activation of NF-κB and increased expression of several proinflammatory cytokines and chemokines in skin of the dCKO mice. We further found that cultured FIP200 KO keratinocytes showed reduced NF-κB phosphorylation in response to tumor necrosis factor α stimulation, suggesting a paracrine regulation of aberrant NF-κB activation in the skin microenviroment of dCKO and FIP200 CKO mice. Together, these results demonstrate that ablation of FIP200, although not promoting tumorigenesis, can lead to skin inflammatory disorders, suggesting a novel function of FIP200 in the maintenance of normal skin homeostasis in vivo.
Focal adhesion kinase family interacting protein of 200 kDa (FIP200)4 was originally identified by us as an inhibitor of focal adhesion kinase and its related kinase Pyk2 (1, 2). Shortly after these studies, FIP200 was also identified by Chano and colleagues (3) as a potential regulator of the RB1 gene (designated as RB1CC1 for RB1-inducible coiled-coil 1). Chano et al. (4) further showed that 20% of primary breast cancers that they screened contained large deletion mutations in FIP200 that are predicted to generate markedly truncated proteins. In addition, we found that overexpression of FIP200 inhibited cell cycle progression through regulation of both cyclin D1 and cyclin-dependent kinase inhibitor p21 in breast cancer cells (5). Based on these initial studies, FIP200 was suggested to function as a potential tumor suppressor (4).
Recent studies have shown that FIP200 interacts with a number of other proteins besides FAK and Pyk2 and regulates several signaling pathways and cellular processes (6). We have identified an interaction between FIP200 and TSC1 and showed that this interaction negatively regulates TSC1-TSC2 complex function to increase mammalian target of rapamycin activation and cell growth (7). FIP200 was also shown to regulate the size of muscle cells through its activation of mammalian target of rapamycin signaling (8). Martin et al. (9) found recently that association of PIASy with FIP200 increased nuclear localization of FIP200, reducing its cytoplasmic pool and consequently its activity in the inhibition of TSC complex. Conversely, FIP200 enhanced the transcriptional activation of the p21 promoter by PIASy by co-recruitment of both proteins to the promoter as detected by chromatin immunoprecipitation analysis (9). Furthermore, a very recent study showed that FIP200 interacts with ULK1 and -2, mammalian orthologs of the yeast Atg1 protein, which are components of autophagosome essential for its formation (10). FIP200 itself was redistributed to autophagosomes upon induction of autophagy by starvation. Moreover, autophagy induction by various treatments was abolished in FIP200-null mouse embryonic fibroblasts, suggesting that FIP200 may function as a novel regulator of autophagy through its interaction with ULK1 and -2 in mammalian cells.
FIP200 is widely expressed in various human tissues (11) and is an evolutionarily conserved protein present in human, mouse, rat, frog, fly, and worm. The high degree of conservation during evolution suggests potentially important functions of FIP200 in vivo. Consistent with this, we showed recently that total knock-out (KO) of FIP200 in mice resulted in embryonic lethality at mid/late gestation associated with heart failure and liver degeneration (12). As expected based on studies in vitro, FIP200 KO mice showed increased activity of the TSC1-TSC2 complex and reduced cell size in the developing heart, which are associated with the severe defects in the myocardium with thinner ventricular walls. Furthermore, FIP200 KO embryos exhibited significant apoptosis in heart and liver, suggesting a role for FIP200 in the regulation of cell survival and apoptosis. FIP200 KO mouse embryonic fibroblasts and fetal liver cells exhibited an elevated sensitivity to tumor necrosis factor (TNF) α-induced apoptosis due to decreased activation of the JNK signaling pathway. Further analysis showed that FIP200 could interact with ASK1 and TRAF2, thus functioning as a scaffold to orchestrate TRAF2-ASK1 signaling to JNK activation induced by TNFα (12). This may explain the increased sensitivity of FIP200 KO cells to TNFα-induced apoptosis and also possibly for the increased apoptosis in FIP200 KO embryos.
To circumvent the embryonic lethality of the total KO of FIP200 and to examine the potential role of FIP200 in tumorigenesis and other diseases processes in vivo, we generated and analyzed FIP200 conditional KO mice by MMTV-Cre, which mediated FIP200 deletion in mammary epithelial cells and several other cells including keratinocytes (13, 14). We found that inactivation of FIP200 neither promotes mammary tumor development nor accelerates lymphomagenesis induced by loss of p53 in lymphocytes, suggesting that FIP200 may not function as a tumor suppressor as implicated in previous data. Unexpectedly, we found that the FIP200 and p53 double conditional KO (dCKO) mice as well as FIP200 conditional KO (CKO) mice displayed a chronic psoriasis-like inflammatory skin disorder that is characterized by epidermal hyperplasia, accumulation of dermal and epidermal leukocytes, and enlargement and enrichment of blood capillary (15). We also found an activation of the NF-κB pathway and up-regulation of pro-inflammatory cytokines TNFα, interleukin (IL) 1β, IL-6, and IFNγ in the skin of the mutant mice, which are also characteristic of lesions in human psoriasis. We further found that cultured FIP200 KO keratinocytes showed reduced NF-κB phosphorylation in response to TNFα stimulation, suggesting a paracrine regulation of aberrant NF-κB activation in the skin microenviroment of dCKO and FIP200 CKO mice. These results suggest a role for FIP200 in the maintenance of normal skin homeostasis and skin inflammatory diseases in vivo.
EXPERIMENTAL PROCEDURES
Mice and Genotype—MMTV-Cre transgenic mice (line F) (13, 14) were obtained from National Institutes of Health NCI Mouse Models of Human Cancers Consortium. P53 floxed mice (16) and Rosa26 transgenic mice (17) were kind gifts from Dr. Alexander Nikitin (Cornell University). FIP200 floxed mice were described previously (12). Mice were observed daily for signs of poor health. Sick mice were sacrificed and checked for solid tumors. Mice were genotyped by PCR analysis of genomic DNA isolated from tail tips using specific primers as described for the Cre recombinase (18); for the wild type and floxed FIP200 allele or deleted FIP200 allele (12); and for wild type and floxed p53 allele or deleted p53 allele (16). Mice were kept pathogen-free throughout the study. All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Michigan and were performed in compliance with local, state, and federal regulations.
Isolation and Culture of Mouse Keratinocytes—The backskin from newborn mice was incubated in dispase II (Invitrogen) overnight at 4 °C. The epidermis was separated from dermis and briefly treated with trypsin to release keratinocytes. The reaction was terminated by the addition of soybean trypsin inhibitor (Invitrogen). Isolated keratinocytes were cultured on plates coated with collagen I (10 μg/cm2 plate, Sigma) and fibronectin (10 μg/ml, Sigma) in keratinocyte-SFM medium (Invitrogen) supplemented with 0.02 mm CaCl2.
RNA Isolation and Semiquantitative Reverse Transcriptase-PCR—Total RNA was isolated from full skin samples using TRIzol according to the manufacturer's instructions (Invitrogen). Equal RNA amounts were added to reverse transcriptase reaction mixture (Themo Scientific) with oligo(dT) as primer. The resulting templates were subjected to PCR using specific primers. Primer sequences are available upon request.
Histology, Immunofluorescence, Immunohistochemistry, and Antibodies—Skin samples were fixed in 10% neutral buffered formalin (Sigma) overnight and embedded in paraffin, and 4-μm sections were stained with hematoxylin and eosin (H&E). For frozen section, skin samples were embedded in OCT compound (Tissue-Tek) and frozen on dry ice. 10-μm sections were cut. Immunofluorescence staining was performed with the following primary antibodies: Ki67 (Novocastra); phospho-NF-κB p65 (Ser536) and cleaved caspase-3 (Asp175) (both from Cell Signaling); von Willebrand factor (DakoCytomation); CD45 (eBioscience); CD3 (eBioscience); Gr1 (eBioscience); and F4/80 (Serotec). Secondary antibodies were conjugated to fluorescein isothiocyanate or Texas Red (Jackson ImmunoResearch). Sections were counterstained with 4,6-diamidino-2-phenylindole (DAPI) (Sigma) for visualization of nuclei. Mice were injected intraperitoneally with 5-bromo-2′-deoxyuridine (BrdU) (Sigma) (100 μg/g of dam weight) and sacrificed after 3 h. BrdU staining was carried out on paraffin sections using the BrdU staining kit according to the manufacturers protocol (Invitrogen).
Western Blotting Analysis—Dorsal skins or cultured keratinocytes were homogenized in modified RIPA buffer (20 mm Tris, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 10% glycerol, 1 mm CaCl2, 1 mm MgCl2, 10 mm sodium fluoride, and 1 mm NaVO4 supplemented with protease inhibitor (10 μg/ml leupeptin, 10 μg/ml aprotinin, and 1 mm phenylmethylsulfonyl fluoride). Total protein lysates were separated by denaturing SDS-PAGE. Western blotting analysis was performed as described (19).
Transmission Electron Microscopy—Samples were fixed in 2.5% glutaraldehyde in 0.1 m Sorensen buffer, pH 7.4, overnight at 4 °C. After several buffer rinses, samples were fixed in 1% osmium tetroxide in the same buffer. It was then rinsed in double distilled water to remove phosphate salt and then stained by aqueous uranyl acetate (3%) for 1 h. The sample was dehydrated in ascending concentrations of ethanol, rinsed two times in propolynoxide, and embedded in epoxy resin. The samples were ultra-thin sectioned 70 nm in thickness and stained with uranyl acetate and lead citrate. The sections were examined using a Philips CM100 electron microscope at 60 kV. Images were recorded digitally using a Hamamatsu ORCA-HR digital camera system operated using AMT software (Advanced Microscopy Techniques Corp., Danvers, MA).
Statistics—Two-tailed Student's test was used to calculate statistical significance of difference between groups. p ≤ 0.05 was considered statistically significant.
RESULTS
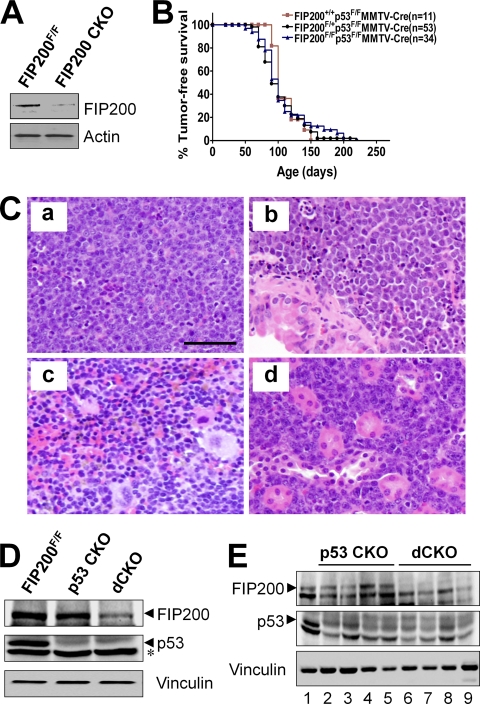
Conditional KO of FIP200 Mediated by MMTV-Cre Does Not Promote Tumorigenesis—Previous studies showing truncation mutations of the FIP200 gene in 20% human breast cancer samples screened suggest that FIP200 may function as a tumor suppressor (4). To study a potential role and mechanisms of FIP200 in breast cancer, we generated FIP200 conditional KO mice (FIP200F/F;MMTV-Cre genotype and designated FIP200 CKO mice) by crossing the floxed FIP200 mice (12) with the MMTV-Cre transgenic mice (13, 14). FIP200 CKO mice were obtained and appeared normal and indistinguishable from control littermates at the perinatal stage. Lysates were then prepared from mammary glands of FIP200 CKO and FIP200F/F mice (as a control) and analyzed by Western blotting using anti-FIP200 antibody. Fig. 1A shows efficient deletion of FIP200 in mammary glands of FIP200 CKO mice, as expected. However, no mammary tumors or other malignancy were observed in female FIP200 CKO mice during 15 months of observation, suggesting that inactivation of FIP200 alone does not predispose mice to breast or other cancers in this mouse model.
FIGURE 1.
MMTV-Cre-mediated conditional KO of FIP200 does not affect lymphomagenesis induced by p53 inactivation. A, lysates were prepared from mammary glands of the first lactation day of FIP200 CKO and FIP200F/F mice and analyzed by Western blotting using anti-FIP200 (top) or anti-actin (bottom). B, Kaplan-Meier analysis of tumor development in mice with the indicated genotypes. n represents the number of animals analyzed. C, representative micrographs of lymphoma in thymus (a), lung (b), spleen (c), and kidney (d) in dCKO mice. Scale bar = 50 μm. D, lysates were prepared from thymus of 4-week-old mice and analyzed by Western blotting using anti-FIP200 (upper), anti-p53 (middle), or anti-vinculin (lower) antibodies. The asterisk marks a background band. E, lysates prepared from thymic lymphoma samples in p53 CKO (lanes 2-5) and dCKO (lanes 6-9) mice or thymus of wild type mice (lane 1) were analyzed by Western blotting using anti-FIP200 (upper), anti-p53 (middle), or anti-vinculin (lower) antibodies.
We next explored the potential synergistic effect of loss of FIP200 with mutations in other tumor suppressor genes. p53 is a major tumor suppressor and more than 50% of human tumors contain mutations or deletion of the p53 gene. To address whether the loss of FIP200 could accelerate p53-mediated tumorigenesis, we introduced the floxed p53 allele into FIP200 CKO mice and generated mice with the FIP200F/F;p53F/F; MMTV-Cre genotype (designated as dCKO mice) as well as control mice with FIP200+/+;p53F/F;MMTV-Cre and FIP200F/+;p53F/F;MMTV-Cre genotypes (both designated as p53 CKO mice). Characterization of all three mice cohorts revealed similar cancer predisposition phenotypes (Fig. 1B). About half of the mice in all three groups became moribund with tumors by 90 days of age, whereas other littermates without p53 deletion (e.g. p53F/F mice) did not develop any tumor at this stage (data not shown). Pathological examination showed that the most frequently observed tumors were thymic lymphoma. Extranodular lymphoma was also found in the lung, spleen, kidney (Fig. 1C), liver, and intestine (data not shown) in all three cohorts of mice. No breast tumors were detected in these mice before their death due to lymphomas. Although there is no report on similar analysis of conditional KO of p53 using MMTV-Cre transgenic mice, total KO of p53 is known primarily to induce lymphomas in mice (20), raising the possibility that mice in our studies developed lymphomas due to leaking expression of MMTV-Cre in other cells such as T lymphocytes. To test this possibility, lysates were prepared from thymuses of dCKO, p53 CKO, or wild type mice and analyzed by Western blot. Fig. 1D shows a significantly reduced expression of p53 in samples from both p53 CKO and dCKO mice and the diminished level of FIP200 in a sample from dCKO mice, which are consistent with leaking activity of MMTV-Cre in the thymus leading to deletion of the floxed p53 and FIP200 alleles in these mice. To further validate this possibility, protein extracts were prepared from thymic lymphomas and subjected to Western blotting analysis. Fig. 1E shows expression of FIP200 in the tumors from p53 CKO mice (lanes 2-5), but not those from dCKO mice (lanes 6-9). As expected, p53 is absent in all tumors (middle panel). Taken together, these results demonstrated that loss of FIP200 does not accelerate or inhibit tumorigenesis induced by p53 inactivation in mouse models.
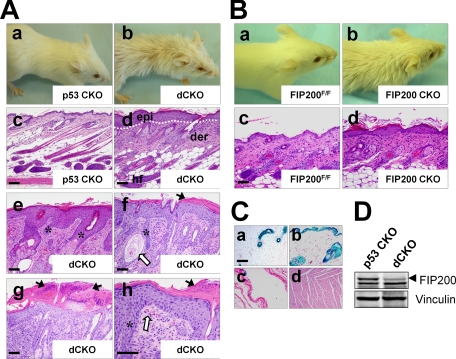
Development of an Acanthosis Skin Disorder in dCKO and FIP200 CKO Mice—During the course of generation of dCKO and p53 CKO mice, we noticed that by 2 weeks after birth, the dCKO (but not p53 CKO) mice began to develop systemically ruffled and sparse fur with scurfy and itchy areas (Fig. 2A, panels a and b), although the skin of these mice appeared no different from the controls at birth (data not shown). The dCKO mice frequently scratched their skin, reflecting a skin pruritus. Around 20% of these mice showed more severe phenotypes at 4 weeks or later time points with skin ulceration often at symmetrical positions. Histological examination of dorsal skin sections from 2-week-old dCKO mice revealed mild epidermal thickening (hyperplasia) when compared with p53 CKO mice (data not shown). After 4 weeks of age, dCKO mice showed a much more thickened epidermis (acanthosis) accompanied by interepidermal edema (spongiosis), and thickened keratinized upper layers (hyperkeratasis) (Fig. 2A, panels c-h, supplementary Fig. S1A and S1B). In severe lesions, there were epidermal microabscesses consisting of neutrophil accumulation in the superficial layer of the epidermis (Fig. 2A, panel g). In addition, more severely affected animals exhibited uneven elongation structures resembling rete ridges (fingerlike epidermal projections into dermis) that extend deeply downward into the dermis (Fig. 2A, panels e, f, and h). Aside from these skin defects, we did not find any apparent abnormality in the spleen, liver, kidney, lung, heart, and intestine in dCKO mice by 8 weeks of age (data not shown). Because the skin defects appeared before any overt signs of lymphoma in dCKO mice and both dCKO and p53 CKO mice developed lymphomas to a similar extent (see Fig. 1B), these results suggest that skin defects in dCKO mice is likely a direct consequence of FIP200 deletion in the skin, rather than caused by secondary effects due to tumorigenesis in these mice.
FIGURE 2.
Double conditional KO of FIP200 and p53 and single conditional KO of FIP200 result in multiple skin abnormalities in mice. A, panels a and b show representative 6-week-old p53 CKO and dCKO mice, respectively. Note the ruffled fur and hair loss phenotype in dCKO mice. Panels c and d show histological analysis of backskin sections of 4-week-old p53 CKO and dCKO mice. Note the thickening of the epidermis in dCKO mice (d) compared with p53 CKO mice (c). Panels e-h show examples of dCKO mice with more severe skin defects. Dotted lines mark the boundary between epidermis and dermis (panels d, e, and f). Black arrows in g indicate subcorneal microabscesses with neutrophil exudates. Open arrows in f and h indicate dilated hair follicles filled with keratinized cell masses and clustered erythrocytes in dermal papillae, respectively. Asterisks (e, f, and h) indicate uneven elongation structures resembling rete ridges in the dermis. epi, epidermis; der, dermis; hf, hair follicle. Scale bars = 200 μm. B, panels a and b show representative 6-week-old FIP200F/F and FIP200 CKO mice, respectively. Note the ruffled fur and hair loss phenotype in FIP200 CKO mice. Panels c and d show histological analysis of backskin sections of 4-week-old FIP200F/F and FIP200 CKO mice, respectively. Note the thickening of the epidermis in FIP200 CKO mice compared with FIP200F/F mice. Scale bars = 200 μm. C, LacZ staining of sections from mammary gland (a), skin (b), lung (c), and heart (d) of MMTV-Cre mice crossed to Rosa26-lacZ reporter mice. Scale bar = 200 μm. D, lysates were prepared from skin of 4-week-old mice and analyzed by Western blotting using anti-FIP200 (upper) or anti-vinculin (lower) antibodies.
Because p53 CKO mice did not develop any of the skin defects observed in the dCKO mice, our results suggest strongly that conditional KO of FIP200 is responsible for the phenotypes. Therefore, we further examined the potential defects in the skin of FIP200 CKO mice, although none of the mice exhibited the severe skin phenotype such as ulceration in some of dCKO mice that prompted our initial investigations on their skin defects. FIP200 CKO mice also showed ruffled and sparse hairs compared with the floxed FIP200 mice as a control (Fig. 2B, panels a and b). Histological analysis of skin sections showed epidermal hyperplasia (acanthosis) in FIP200 CKO mice compared with the control mice (Fig. 2B, panels c and d). These data suggest that deletion of FIP200 alone also caused similar, albeit milder, skin defects as the dCKO mice.
MMTV-Cre has been observed to be active in the skin in previous studies (13, 14), thus could cause deletion of the floxed FIP200 allele in the skin. To further validate such a possibility in dCKO mice, we crossed MMTV-Cre mice used in our study with the ROSA26-LacZ reporter mice (strain R26R), in which expression of LacZ requires Cre-mediated excision of a stop cassette (17). As shown in Fig. 2C, LacZ activity was detected in mammary epithelial cells (panel a) and several other tissues including skin (panel b), salivary glands, and thymus (data not shown), but little or none in lung (panel c), heart (panel d), kidney, liver, and brain (data not shown). Consistent with these results, Western blotting analysis of lysates from skin showed significantly decreased expression of FIP200 in dCKO mice compared with p53 CKO mice (Fig. 2D). These results provide further support that skin lesions in dCKO and FIP200 CKO mice are caused by FIP200 deletion in keratinocytes mediated by MMTV-Cre.
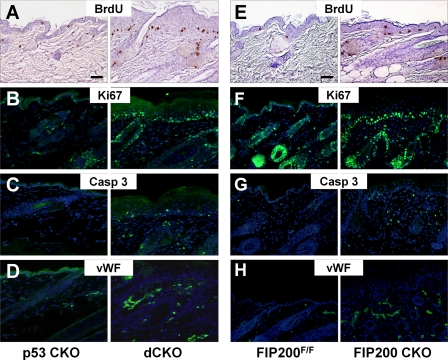
Increased Proliferation, Apoptosis, and Vascularity in the Skin of dCKO and FIP200 CKO Mice in Vivo—To explore the mechanisms underlying the skin defects in dCKO mice, we first analyzed proliferation of keratinocytes because of hyperplasia in the epidermis. Skin sections were prepared from dCKO mice and p53 CKO mice that had been injected intraperitoneally with BrdUrd 3 h prior to euthanasia, and then examined by immunohistochemical staining with anti-BrdU antibody to determine BrdU incorporation. Fig. 3A shows a markedly increased BrdU incorporation at the basal layer of the epidermis in dCKO mice compared with p53 CKO mice. Similarly, staining of the sections by immunofluorescence using antibody against Ki67 also indicated increased proliferation of keratinocytes in dCKO mice (Fig. 3B). Interestingly, increased apoptosis was also found throughout the skin of dCKO mice as measured by immunofluorescent staining for cleaved caspase-3 (Fig. 3C). In addition, we found an increase in the number of blood capillaries with dilated and tortuous morphology as detected by staining with anti-von Willebrand factor in the dermis of the dCKO mice (Fig. 3D). Immunofluorescent staining for proliferation marker BrdU or Ki67 also showed increased proliferation of basal keratinocytes in FIP200 CKO mice (Fig. 3, E and F, respectively). Staining with antibodies against cleaved caspase 3 and von Willebrand factor, respectively, also revealed increased apoptosis and dermal blood vessels in these mice (Fig. 3, G and H, respectively). Together with data from histological analyses, these results suggest that deletion of FIP200 leads to a psoriasis-like phenotype.
FIGURE 3.
Increased epidermal hyperproliferation, dermal apoptosis, and vascularity of dCKO and FIP200 CKO mice. A and E, BrdU incorporation assay; and B and F, anti-Ki67 immunofluorescence showed hyperproliferation in the basal layer of the epidermis in dCKO and FIP200 CKO mice (right panels) compared with p53 CKO and FIP200F/F mice (left panels). C and G, anti-cleaved caspase 3 stainings indicate increased apoptosis in the dermis and epidermis of dCKO and FIP200 CKO mice compared with p53 CKO and FIP200F/F mice. D and H, anti-von Willebrand (vWF) factor stainings show enriched and enlarged blood vessels in the dermis of dCKO and FIP200 CKO mice compared with p53 CKO and FIP200F/F mice. Nuclei were counterstained with DAPI (blue) in panels B-D and F-H. Scale bars = 100 μm.
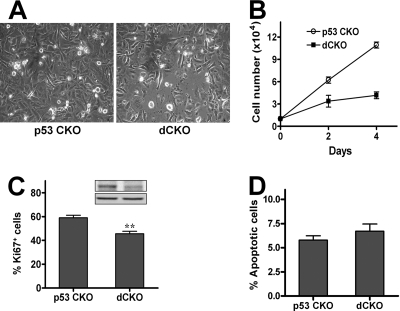
Epidermal hyperproliferation may be epidermal cell autonomous or occur as a secondary response to the influx of inflammatory leukocytes (21, 22). To determine whether deletion of FIP200 led to epidermal hyperproliferation through increased intrinsic proliferative potential of keratinocytes in dCKO mice, primary epidermal keratinocytes were isolated from newborn dCKO and p53 CKO control mice and analyzed in vitro. Surprisingly, however, we found that keratinocytes from dCKO mice showed a reduced growth compared with that from p53 CKO mice under these conditions (Fig. 4A). Quantitative analysis indicated an ∼3-fold decrease in the number of cells over a period of 4 days in culture (Fig. 4B). Consistent with this, staining of the cells with Ki67 also indicated a decrease in the number of mitotic keratinocytes isolated from dCKO mice compared with that from p53 CKO mice (Fig. 4C). However, keratinocytes from both dCKO and p53 CKO control mice did not show a significant difference in apoptosis when cultured on plates coated with collagen I and fibronectin in growth medium (Fig. 4D). These results suggested that deletion of FIP200 did not lead to increased proliferation of epidermal keratinocytes in vitro and that epidermal hyperproliferation observed in vivo may not be keratinocyte autonomous.
FIGURE 4.
Reduced proliferation of keratinocytes isolated from dCKO in vitro. Primary mouse keratinocytes were isolated from p53 CKO and dCKO mice. A and B, the same number of cells were plated in keratinocyte-SFM and photographed after 4 days of culture. Cells from p53 CKO mice, but not those from dCKO mice, reached confluence (A). Panel B shows growth curve of these cells. Data represent triplicate independent experiments (mean ± S.D.). C, cells were stained with anti-Ki67 antibody at 20 h in keratinocyte-SFM. The percentage of Ki67 positive cells were determined and data represent triplicate independent experiments (mean ± S.D.). **, p < 0.01. Inset shows Western blotting of samples from p53 CKO (left) or dCKO (right) cultured keratinocytes with anti-FIP200 (top) or anti-vinculin (bottom). D, cultured keratinocytes were stained with Hoechst to determine the fraction of apoptotic cells and data represent triplicate independent experiments (mean ± S.D.). p = 0.15.
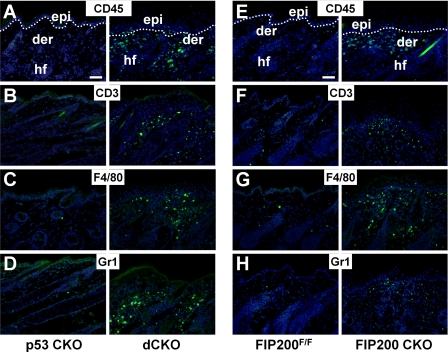
Increased Infiltration of Leukocytes into Dermis and Epidermis of Mutant Mice—To investigate whether epidermal hyperplasia could be caused by infiltration of inflammatory cells, we examined the presence of leukocytes in the skin of 4-week-old dCKO and p53 CKO mice by immunofluorescent labeling using various markers for these cells. Staining with anti-CD45 antibody, which is a common leukocyte marker, confirmed the presence of leukocytes in the dermis and epidermis of dCKO mice, but not in p53 CKO mice (Fig. 5A). Staining with anti-CD3 antibody, a T cell marker, showed a significant increase of T cells in the dermis of dCKO mice compared with that of p53 CKO mice (Fig. 5B). Staining with antibodies against F4/80 and Gr1, markers for macrophages and granulocytes, respectively, also revealed the infiltration of these immune cells in the skin of dCKO mice (Fig. 5, C and D). Ultrastructural analyses also revealed that the dermis of dCKO mice was infiltrated with immune cells compared with controls (supplementary Fig. S1C and S1D). The majority of the inflammatory cells were detected in the superficial dermis, and scattered positive stainings were also found in the epidermis of dCKO mice. Furthermore, leukocyte infiltration can be detected in the skin of dCKO mice at 2 weeks of age when skin acanthosis has not fully developed in these mice (data not shown). Infiltrated leukocytes including T cells, macrophages, and granulocytes were also detected in the skin of FIP200 CKO mice (Fig. 5, E-H). Together, these results suggest that deletion of FIP200 leads to chronic inflammation in the skin of dCKO and FIP200 CKO mice, which is responsible for epidermal hyperplasia in these mice.
FIGURE 5.
Infiltration of the inflammatory cells in the skin of dCKO and FIP200 CKO mice. Cryosections of backskin from 4-week-old p53 CKO (A-D, left), dCKO (A-D, right), FIP200F/F (E-H, left), and FIP200 CKO (E-H, right) mice were immunostained with the indicated antibodies (shown by green fluorescein isothiocyanate fluorescence): A and E, leukocytes (CD45); B and F, T cells (CD3); C and G, macrophages (F4/80); or D and H, granulocytes (Gr1). Nuclei were counterstained with DAPI (blue). Dotted lines mark the boundary between the epidermis and dermis (A and E). epi, epidermis; der, dermis; hf, hair follicle. Scale bars = 100 μm.
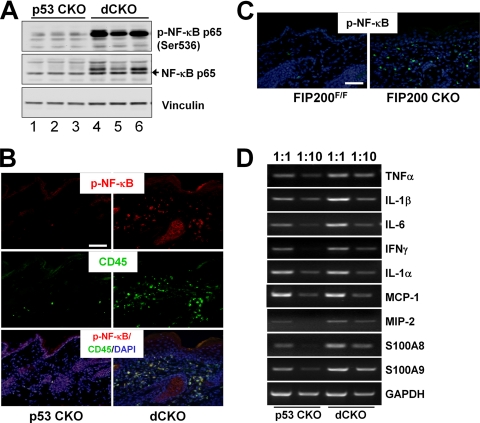
Activation of NF-κB and Increased Expression of Proinflammatory Cytokines and Chemokines in Mutant Mice—Because the NF-κB pathway plays a central role in many inflammation diseases (23, 24), we examined the activation of NF-κB by measuring phosphorylation of the p65 subunit of NF-κB in the skin of dCKO mice. As shown in Fig. 6A, Western blotting analysis of lysates from the skin revealed a significant increase of p65 phosphorylation in the skin of dCKO mice (lanes 4-6) compared with that from p53 CKO mice (lanes 1-3). Immunofluorescent staining of skin sections using anti-phospho-p65 also identified activated NF-κB predominantly in the dermis of dCKO mice, but not in p53 CKO mice (Fig. 6B, top panel). Consistent with the primarily dermal localization of active NF-κB, co-staining of the sections with anti-CD45 showed overlapping patterns with phospho-NF-κB staining (Fig. 6B, middle and lower panels), indicating activation of NF-κB in the infiltrated leukocytes. Activated NF-κB was also found in the dermis of FIP200 CKO mice but not in control mice (Fig. 6C). These results suggest that activation of the NF-κB signaling pathway in infiltrating leukocytes may play a role in inflammation and acanthosis of dCKO and FIP200 CKO mice.
FIGURE 6.
Activated NF-κB pathway and production of proinflammatory cytokines and chemokines in the skin of dCKO and FIP200 CKO mice. A, lysates were prepared from the skin of p53 CKO (lanes 1-3) or dCKO (lanes 4-6) mice and analyzed by Western blotting using anti-phospho-NF-κB p65 (upper), anti-NF-κB p65 (middle), or anti-vinculin (lower) antibodies. B, two-color immunofluorescence of skin sections from p53 CKO (left) or dCKO (right) mice using fluorescein isothiocyanate-conjugated anti-CD45 (green) and Texas Red-conjugated anti-phospho-NF-κB antibodies. Note the CD45+ phospho-NF-κB+ double-positive leukocytes in the skin of dCKO mice. Sections were counterstained with DAPI (blue). Scale bar = 100 μm. C, skin sections from FIP200F/F (left) and FIP200 CKO (right) mice were immunostained by fluorescein isothiocyanate-conjugated anti-phospho-NF-κB antibody (green). Sections were counterstained with DAPI (blue). Scale bar = 100 μm. D, semi-quantitative reverse transcriptase-PCR analysis of cytokines and chemokines gene expression for mRNA isolated from the dorsal skin of p53 CKO and dCKO mice. cDNA were used directly (1:1) or diluted 10-fold (1:10) before PCR. Data are representative of more than five independent mouse pairs. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Previous studies have shown that NF-κB signaling promotes inflammation by transcriptional activation of a number of proinflammatory cytokines including TNFα, IL-1β, and IL-6 (25). Therefore, we further analyzed the expression of various cytokines and chemokines implicated in skin inflammation by using semi-quantitative reverse transcriptase-PCR. As shown in Fig. 6D, skin from dCKO mice expressed higher levels of proinflammatory cytokines including TNFα, IL-1β, IL-6, and IFNγ, which have been implicated in the pathogenesis of psoriasis (26-28). The dCKO skin also showed elevated expression of macrophage inflammatory protein 2 and antimicrobial peptide transcript S100A8 (calgranulin A) and S100A9 (calgranulin B). In contrast, IL-1α and monocyte chemotactic protein-1 were expressed in comparable levels in the skin from dCKO and p53 CKO mice. These results suggest that activation of NF-κB in infiltrated leukocytes may stimulate production of multiple proinflammatory cytokines and chemokines. These cytokines released within the dermis may promote proliferation of keratinocytes within the epidermal compartment and further induce the activation of NF-κB possibly as a feedback loop, which could account for increasing inflammatory response and epidermal proliferation in the skin of mutant mice over time.
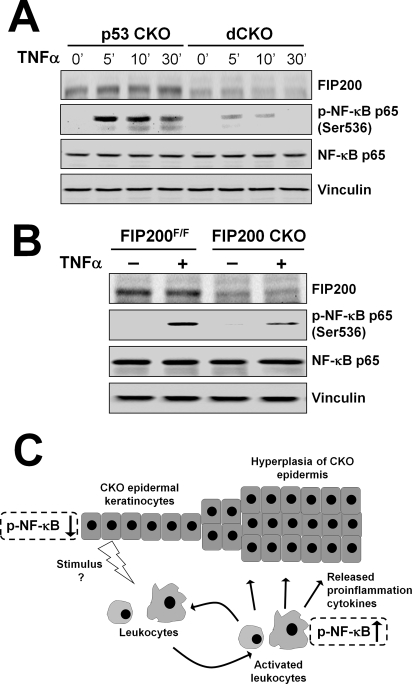
FIP200 Deletion Leads to Defective TNFα-induced NF-κB Signaling in Keratinocytes—Previous studies suggested that perturbation of TNFα-NF-κB signaling pathways in the epidermis causes a psoriasis-like skin inflammatory phenotype (21, 29). We therefore examined NF-κB activation by TNFα in isolated keratinocytes. As seen in Fig. 7A, stimulation of NF-κB activation by TNFα is significantly reduced in dCKO keratinocytes compared with p53 CKO cells, as measured by phosphorylation of its p65 subunit. Furthermore, activation of NF-κB upon TNFα stimulation is also decreased in FIP200 CKO keratinocytes compared with the FIP200F/F keratinocytes (Fig. 7B). Together, these results suggest that deletion of FIP200 diminished TNFα-induced NF-κB activation in keratinocytes, which may be responsible for triggering increased recruitment of leukocytes into the dermis leading to the skin inflammatory phenotype in the FIP200 CKO and dCKO mice.
FIGURE 7.
Reduced NF-κB activation in response to TNFα stimulation in FIP200-null keratinocytes. A, keratinocytes isolated from p53 CKO and dCKO mice were serum starved overnight. They were then left untreated or treated with 50 ng/ml TNFα for the different periods of time as indicated. Cell lysates were then analyzed by Western blotting using various antibodies as indicated. B, keratinocytes were isolated from FIP200F/F and FIP200 CKO mice and analyzed as in A. C, a working model summarizing the possible mechanisms of the skin inflammatory phenotype of mutant mice with FIP200 conditional deletion in keratinocytes.
DISCUSSION
Using a conditional KO approach, we present data here suggesting that FIP200 is involved in skin inflammatory diseases resembling aspects of human psoriasis. Deletion of FIP200 either alone or together with p53 led to skin abnormalities including acanthosis, hyperkeratosis, as well as infiltration of inflammatory leukocytes into the dermis and epidermis accompanied by increased vascularity in the mutant mice. Furthermore, the mutant mice showed activation of NF-κB and up-regulation of proinflammatory cytokines TNFα, IL-1β, IL-6, and IFNγ in the skin, which are also seen in lesions of human psoriasis. Psoriasis is among the most common autoimmune chronic inflammatory diseases of unsolved pathogenesis affecting skin and joints in ∼2% of the general population. It is characterized by a marked epidermal hyperplasia (acanthosis) accompanied by parakeratosis (retention of keratinocyte nuclei in the stratum corneum), epidermal rete ridge formation (downward papillary projections of the epidermis), and skin infiltration of inflammatory cells such as T-cells, dendritic cells, macrophages, and mast cells (15).
Interactions between the immune system and various cells in the skin have been shown to play critical roles in the development of inflammatory skin diseases (30, 31). A tightly regulated microenvironment of cytokines and growth factors produced by keratinocytes, dermal fibroblasts, and immune cells is crucial for the maintenance of physiological skin homeostasis (32, 33). In particular, the NF-κB signaling pathway has been shown to play an essential role in many types of inflammation diseases such as psoriasis (23, 24). Several previous studies have generated mouse models that mimic aspects of psoriasis (34, 35), including transgenic mice expressing IFNγ (36), integrin (β1, α2β1, α5β1) (37), or STAT3 (38) in the epidermis as well as conditional KO of IκBα (39, 40) and IKK2 (21, 29). All of these genes have been implicated in the regulation of NF-κB signaling pathways either directly or indirectly, and changes in NF-κB activities have been demonstrated in all of the mouse models. Interestingly, these studies suggest that either the inhibition or activation of NF-κB signaling can lead to skin inflammation phenotypes similar to what we observed in the dCKO and FIP200 CKO mice. This suggests that skin homeostasis requires a balanced NF-κB activity. Although previous studies suggested that inactivation of FIP200 did not affect NF-κB signaling in mouse embryonic fibroblasts (12), our analysis showed deficient TNFα-induced activation of NF-κB in keratinocytes isolated from either dCKO or FIP200 CKO mice, suggesting a cell-type specific function of FIP200 in the regulation of NF-κB signaling in these cells. Therefore, ablation of FIP200 from keratinocytes could interfere with the balance of NF-κB activity, as discussed above, thus triggering an inflammatory response that is marked by infiltration of immune cells, activation of NF-κB in these cells, and induction of inflammatory cytokines such as TNFα, IL-1β, IFNγ, and IL-6 in skin microenvironment, resulting in the development of skin inflammation disease in the mutant mice.
Besides mammary gland and skin cells, MMTV-Cre also exhibited Cre activity in several other cell types including some lineages of hematopoietic cells such as T- and B-cells, megakaryocytes of spleen, and erythroid cells (41, 42). Thus increased infiltration of inflammatory cells could be the consequence of immune cell activation upon FIP200 deletion in some of these cells. However, this is not likely because we did not observe any significant inflammation in other tissues in these mice. Consistent with this, we did not find any differences in the spontaneous activation of splenocytes from FIP200 CKO mice compared with those from FIP200F/F mice, as measured by CD44 expression (supplementary Fig. S2). Furthermore, no differences in the activation of NF-κB were observed in the thymus and spleen of FIP200 CKO mice compared with FIP200F/F mice (supplementary Fig. S3A and S3B). Besides T cells, macrophages and granulocytes also play important roles in skin inflammation, although MMTV-Cre-mediated Cre recombination has not been characterized in these cells. We found that FIP200 was not deleted in primary macrophages isolated from FIP200 CKO mice, presumably due to the absence of Cre activity in these cells (supplementary Fig. S3C). Therefore, our results suggest that the primary cause of the inflammatory phenotype of the FIP200 CKO and dCKO mice is due to FIP200 deletion in keratinocytes, but not due to abnormality of various immune cells.
It should be noted that although they are not the primary culprit for the phenotype, macrophages and/or other leukocytes could still play a crucial role in skin inflammation in FIP200 CKO mice. Previous studies using epidermis-specific IKK2 conditional KO mice showed that depletion of macrophages, but not granulocytes or T cells, from these mice abolished the skin phenotype, suggesting an essential contribution of macrophages in the psoriasis-like skin lesions caused by inhibition of NF-κB signaling upon deletion of IKK2 in the skin (29). In an analogous manner, FIP200 deletion in keratinocytes and consequent inhibition of the NF-κB pathway could cause recruitment into the dermis of immune cells such as macrophages and their activation by mechanisms as yet to be characterized. Activated inflammatory cells with increased NF-κB signaling may produce and secrete cytokines to which epidermal keratinocytes respond. Thus, the sustained activation of NF-κB in the skin microenvironment of dCKO and FIP200 CKO mice could trigger a paracrine NF-κB signaling loop between epidermal keratinocytes and various inflammatory cells that may increase with aging (43), which is consistent with our observation of more severe skin lesions in 4 weeks or older mice than younger ones. Such a working hypothesis is summarized in Fig. 7C.
FIP200 has been proposed as a putative tumor suppressor based on initial studies on breast cancer cells and analysis of mutations of FIP200 in human breast cancer samples (4). Nevertheless, direct evidence for a tumor suppressor role of FIP200 in vivo is lacking and the embryonic lethality of FIP200 total KO mice precludes its use as a model to assess the function of FIP200 in tumorigenesis. In this study, deletion of FIP200 did not accelerate or inhibit lymphomagenesis upon inactivation of p53 in mice. Therefore, FIP200 is unlikely to function as a tumor suppressor in vivo, despite previous data suggesting such a possibility (4). However, these data could not exclude the possibility that FIP200 may act as a tumor suppressor in combination with other gene mutations.
In summary, we find that the ablation of FIP200 is dispensable for tumorigenesis induced by p53 deficiency, but causes skin epidermal hyperplasia and chronic inflammation in our mouse models. These studies provide novel insight into the pathogenesis of inflammatory skin disease and could also be of relevance to other inflammatory diseases. The dCKO and FIP200 CKO mice may provide useful mouse models for new and promising therapeutic approaches for the treatment of inflammatory skin disorder in humans.
Supplementary Material
Acknowledgments
We are grateful to our colleagues Chunchi Liang, Fei Liu, Ming Luo, Chenrang Wang. and Huei Jin Ho for critical reading of the manuscript and helpful suggestions, and Chenrang Wang for help with statistical analysis. We thank Dr. Alexander Nikitin, Cornell University, for providing p53 floxed mice and Rosa26 transgenic mice and Dr. Yang Liu, University of Michigan, for providing anti-CD3 and anti-Gr1 antibodies.
This work was supported, in whole or in part, by National Institutes of Health Grant GM52890 (to J.-L. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S3.
Footnotes
The abbreviations used are: FIP200, focal adhesion kinase family interacting protein of 200 kDa; KO, knock-out; TNF, tumor necrosis factor; dCKO, double conditional knock-out; CKO, conditional knock-out; DAPI, 4,6-diamidino-2-phenylindole; BrdU, 5-bromo-2′-deoxyuridine; JNK, c-Jun NH2-terminal kinase; MMTV, murine mammary tumor virus; IL, interleukin; IFN, interferon.
References
- 1.Abbi, S., Ueda, H., Zheng, C., Cooper, L. A., Zhao, J., Christopher, R., and Guan, J. L. (2002) Mol. Biol. Cell 13 3178-3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ueda, H., Abbi, S., Zheng, C., and Guan, J. L. (2000) J. Cell Biol. 149 423-430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chano, T., Ikegawa, S., Kontani, K., Okabe, H., Baldini, N., and Saeki, Y. (2002) Oncogene 21 1295-1298 [DOI] [PubMed] [Google Scholar]
- 4.Chano, T., Kontani, K., Teramoto, K., Okabe, H., and Ikegawa, S. (2002) Nat. Genet. 31 285-288 [DOI] [PubMed] [Google Scholar]
- 5.Melkoumian, Z. K., Peng, X., Gan, B., Wu, X., and Guan, J. L. (2005) Cancer Res. 65 6676-6684 [DOI] [PubMed] [Google Scholar]
- 6.Gan, B., and Guan, J. L. (2008) Cell. Signal. 20 787-794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gan, B., Melkoumian, Z. K., Wu, X., Guan, K. L., and Guan, J. L. (2005) J. Cell Biol. 170 379-389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chano, T., Saji, M., Inoue, H., Minami, K., Kobayashi, T., Hino, O., and Okabe, H. (2006) Int. J. Mol. Med. 18 425-432 [PubMed] [Google Scholar]
- 9.Martin, N., Schwamborn, K., Urlaub, H., Gan, B., Guan, J. L., and Dejean, A. (2008) Mol. Cell. Biol. 28 2771-2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hara, T., Takamura, A., Kishi, C., Iemura, S., Natsume, T., Guan, J. L., and Mizushima, N. (2008) J. Cell Biol. 181 497-510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bamba, N., Chano, T., Taga, T., Ohta, S., Takeuchi, Y., and Okabe, H. (2004) Int. J. Mol. Med. 14 583-587 [PubMed] [Google Scholar]
- 12.Gan, B., Peng, X., Nagy, T., Alcaraz, A., Gu, H., and Guan, J. L. (2006) J. Cell Biol. 175 121-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner, K. U., Wall, R. J., St-Onge, L., Gruss, P., Wynshaw-Boris, A., Garrett, L., Li, M., Furth, P. A., and Hennighausen, L. (1997) Nucleic Acids Res. 25 4323-4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagner, K. U., McAllister, K., Ward, T., Davis, B., Wiseman, R., and Hennighausen, L. (2001) Transgenic Res. 10 545-553 [DOI] [PubMed] [Google Scholar]
- 15.Lowes, M. A., Bowcock, A. M., and Krueger, J. G. (2007) Nature 445 866-873 [DOI] [PubMed] [Google Scholar]
- 16.Jonkers, J., Meuwissen, R., van der Gulden, H., Peterse, H., van der Valk, M., and Berns, A. (2001) Nat. Genet. 29 418-425 [DOI] [PubMed] [Google Scholar]
- 17.Soriano, P. (1999) Nat. Genet. 21 70-71 [DOI] [PubMed] [Google Scholar]
- 18.Peng, X., Kraus, M. S., Wei, H., Shen, T. L., Pariaut, R., Alcaraz, A., Ji, G., Cheng, L., Yang, Q., Kotlikoff, M. I., Chen, J., Chien, K., Gu, H., and Guan, J. L. (2006) J. Clin. Investig. 116 217-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei, H., Wang, X., Gan, B., Urvalek, A. M., Melkoumian, Z. K., Guan, J. L., and Zhao, J. (2006) J. Biol. Chem. 281 16664-16671 [DOI] [PubMed] [Google Scholar]
- 20.Jacks, T., Remington, L., Williams, B. O., Schmitt, E. M., Halachmi, S., Bronson, R. T., and Weinberg, R. A. (1994) Curr. Biol. 4 1-7 [DOI] [PubMed] [Google Scholar]
- 21.Pasparakis, M., Courtois, G., Hafner, M., Schmidt-Supprian, M., Nenci, A., Toksoy, A., Krampert, M., Goebeler, M., Gillitzer, R., Israel, A., Krieg, T., Rajewsky, K., and Haase, I. (2002) Nature 417 861-866 [DOI] [PubMed] [Google Scholar]
- 22.Zhang, J. Y., Green, C. L., Tao, S., and Khavari, P. A. (2004) Genes Dev. 18 17-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, Q., and Verma, I. M. (2002) Nat. Rev. Immunol. 2 725-734 [DOI] [PubMed] [Google Scholar]
- 24.Silverman, N., and Maniatis, T. (2001) Genes Dev. 15 2321-2342 [DOI] [PubMed] [Google Scholar]
- 25.Karin, M., and Greten, F. R. (2005) Nat. Rev. Immunol. 5 749-759 [DOI] [PubMed] [Google Scholar]
- 26.Austin, L. M., Ozawa, M., Kikuchi, T., Walters, I. B., and Krueger, J. G. (1999) J. Investig. Dermatol. 113 752-759 [DOI] [PubMed] [Google Scholar]
- 27.Robert, C., and Kupper, T. S. (1999) N. Engl. J. Med. 341 1817-1828 [DOI] [PubMed] [Google Scholar]
- 28.Arican, O., Aral, M., Sasmaz, S., and Ciragil, P. (2005) Mediators Inflamm. 2005 273-279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stratis, A., Pasparakis, M., Rupec, R. A., Markur, D., Hartmann, K., Scharffetter-Kochanek, K., Peters, T., van Rooijen, N., Krieg, T., and Haase, I. (2006) J. Clin. Investig. 116 2094-2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leung, D. Y. (1999) J. Allergy Clin. Immunol. 104, S99-108 [DOI] [PubMed] [Google Scholar]
- 31.Nickoloff, B. J. (1999) J. Clin. Investig. 104 1161-1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szabowski, A., Maas-Szabowski, N., Andrecht, S., Kolbus, A., Schorpp-Kistner, M., Fusenig, N. E., and Angel, P. (2000) Cell 103 745-755 [DOI] [PubMed] [Google Scholar]
- 33.Perez-Moreno, M., Davis, M. A., Wong, E., Pasolli, H. A., Reynolds, A. B., and Fuchs, E. (2006) Cell 124 631-644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nickoloff, B. J., and Wrone-Smith, T. (1997) Nat. Med. 3 475-476 [DOI] [PubMed] [Google Scholar]
- 35.Gudjonsson, J. E., Johnston, A., Dyson, M., Valdimarsson, H., and Elder, J. T. (2007) J. Investig. Dermatol. 127 1292-1308 [DOI] [PubMed] [Google Scholar]
- 36.Carroll, J. M., Crompton, T., Seery, J. P., and Watt, F. M. (1997) J. Investig. Dermatol. 108 412-422 [DOI] [PubMed] [Google Scholar]
- 37.Carroll, J. M., Romero, M. R., and Watt, F. M. (1995) Cell 83 957-968 [DOI] [PubMed] [Google Scholar]
- 38.Sano, S., Chan, K. S., Carbajal, S., Clifford, J., Peavey, M., Kiguchi, K., Itami, S., Nickoloff, B. J., and DiGiovanni, J. (2005) Nat. Med. 11 43-49 [DOI] [PubMed] [Google Scholar]
- 39.Klement, J. F., Rice, N. R., Car, B. D., Abbondanzo, S. J., Powers, G. D., Bhatt, P. H., Chen, C. H., Rosen, C. A., and Stewart, C. L. (1996) Mol. Cell. Biol. 16 2341-2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rebholz, B., Haase, I., Eckelt, B., Paxian, S., Flaig, M. J., Ghoreschi, K., Nedospasov, S. A., Mailhammer, R., Debey-Pascher, S., Schultze, J. L., Weindl, G., Forster, I., Huss, R., Stratis, A., Ruzicka, T., Rocken, M., Pfeffer, K., Schmid, R. M., and Rupec, R. A. (2007) Immunity 27 296-307 [DOI] [PubMed] [Google Scholar]
- 41.Thompson, J. E., Phillips, R. J., Erdjument-Bromage, H., Tempst, P., and Ghosh, S. (1995) Cell 80 573-582 [DOI] [PubMed] [Google Scholar]
- 42.Whiteside, S. T., Epinat, J. C., Rice, N. R., and Israel, A. (1997) EMBO J. 16 1413-1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Werner, S., and Smola, H. (2001) Trends Cell Biol. 11 143-146 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.