Summary
The Mre11/Rad50/NBS1 complex (MRN) maintains genomic stability by bridging DNA ends and initiating DNA damage signaling through activation of the ATM kinase. Mre11 possesses DNA nuclease activities that are highly conserved in evolution, but play unknown roles in mammals. To define functions of Mre11 we engineered targeted mouse alleles which either abrogate nuclease activities or inactivate the entire MRN complex. Mre11 nuclease deficiency causes a striking array of phenotypes indistinguishable from absence of MRN, including early embryonic lethality and dramatic genomic instability. We identify a crucial role for the nuclease activities in homology directed double strand break repair, and a contributing role in activating the ATR kinase. However, nuclease activities are not required to activate ATM after DNA damage or telomere deprotection. Therefore, nucleolytic processing by Mre11 is an essential function of fundamental importance in DNA repair distinct from MRN control of ATM signaling.
Introduction
Maintaining genome integrity is essential for the health of an individual and faithful propagation of genetic material to future generations. DNA double strand breaks (DSBs) are a potent cause of genomic instability and when errantly repaired may cause cancer and developmental disorders. Because of the universal threat posed, eukaryotes have evolved an intricate set of responses including cell cycle checkpoints, apoptosis, and pathways of DSB repair. The primary DSB repair pathways are non homologous end joining (NHEJ) which entails direct ligation of broken DNA ends, and homologous recombination (HR) in which sequences homologous to the broken region are used as a template for repair. Proteins exist that function within specific cellular responses or repair pathways, yet certain factors play broad roles (Wyman and Kanaar, 2006).
The MRN complex is a global player in DNA damage responses. It is a sensor of DSBs and physically localizes to sites of damage rapidly after the insult (Berkovich et al., 2007; Lisby et al., 2004; Mirzoeva and Petrini, 2001; Shroff et al., 2004). Once in position MRN secures DNA ends (Williams and Tainer, 2005) and activates DNA damage checkpoint signaling cascades (Lee and Paull, 2007). The complex is comprised of Mre11 and Rad50 which are conserved in all domains of life, and NBS1 (Xrs2 in S. cerevisiae) which is specific to eukaryotes and is less well conserved (Stracker et al., 2004).
Two Mre11 and two Rad50 molecules form a heterotetramer that provides versatile DNA binding capabilities. Rad50 possesses two large flexible coiled arms which foster bridging of DNA ends (Williams and Tainer, 2005). NBS1 interacts with the Rad50/Mre11 tetramer and communicates the presence of DSBs to the cell cycle checkpoint machinery (Difilippantonio and Nussenzweig, 2007). This is accomplished through interaction with the ATM kinase, which is considered a master controller of cellular responses to DSBs (Shiloh, 2003). The interaction, in combination with the presence of DNA ends, causes inactive ATM dimers to dissociate into active monomers which phosphorylate numerous downstream factors (Lee and Paull, 2007).
The Mre11 component of the complex contributes DNA binding capabilities, and as demonstrated in the accompanying manuscript (Williams et al., 2008), functions as a dimer which can bind both sides of a DSB and stabilize them in close proximity. Mre11 possesses DNA nuclease activities (Furuse et al., 1998; Moreau et al., 1999; Paull and Gellert, 1998; Trujillo et al., 1998) which could serve to process ends bound within the dimer. In vitro, Mre11 has exo- and endo-nuclease activities, including double stranded 3' to 5' exonuclease, and single stranded endonuclease that acts on 5' overhangs, 3' flaps, 3' branches, and closed hairpins (D'Amours and Jackson, 2002).
The N-terminus of Mre11 has four highly conserved domains responsible for nuclease activities (Stracker et al., 2004). The third domain (Motif III) contains an invariant histidine residue that in vitro and structural studies have shown acts directly in catalysis by stabilizing the transition state of the sugar-phosphate moiety of the scissile bond during nucleolysis (Arthur et al., 2004; Hopfner et al., 2001; Williams et al., 2008). Surprisingly, mutation of this histidine residue (H125) in S. cerevisiae caused minimal phenotypes compared to Mre11 null. Whereas complete inactivation caused severe radiation hypersensitivity, defective DSB repair, and shortened telomeres, the impact of nuclease deficiency was restricted to mild radiation hypersensitivity (Bressan et al., 1998; Krogh et al., 2005; Lewis et al., 2004; Llorente and Symington, 2004; Moreau et al., 1999). In certain contexts, nuclease deficiencies resembled null, including synthetic lethality in combination with rad27/FEN-1 mutation (Moreau et al., 1999), and deficiency in processing DNA hairpins (Lobachev et al., 2002). Together, these findings suggested that the nuclease activities of Mre11 have specialized roles, but are largely dispensable during DNA damage responses.
Understanding specific in vivo functions of MRN in higher eukaryotes has been hampered by early embryonic lethality conferred by knockout alleles of murine Rad50 and NBS1 (Luo et al., 1999; Zhu et al., 2001). A similar impact of Mre11 inactivation is presumed based on the inability of mouse embryonic stem cells to proliferate when lacking Mre11 (Xiao and Weaver, 1997). Much of what is known comes from studies of inherited hypomorphic alleles of Mre11 or NBS1. The inherited Mre11 mutations caused symptoms very similar to the disease ataxia telangiectasia (A-T), caused by mutation of the ATM (A-T Mutated) gene, including cerebellar degeneration and ionizing radiation (IR) hypersensitivity. Thus this syndrome was named A-T like disorder (ATLD) (Stewart et al., 1999). NBS1 is mutated in Nijmegen breakage syndrome which also shares characteristics with A-T. Studies on NBS1 and ATLD cells demonstrated a clear role for mammalian MRN in controlling checkpoint responses to DNA damage and highlighted the close functional relationship between MRN and ATM (Stracker et al., 2004).
The early embryonic lethality conferred by mouse MRN null alleles is in sharp contrast to the relatively mild impact of ATM inactivation, which permits viability in humans and mice (Stracker et al., 2004). Therefore, MRN has essential roles outside of ATM activation in mitotic cells in mammals. These functions are likely to be operational in the disease associated MRN alleles since they support viability. The known ATLD mutations do not reside in the highly conserved Mre11 nuclease motifs (Stracker et al., 2004). Therefore, despite the minimal impact of nuclease deficiency in S. cerevisiae mitotic cells, we recognized that this function of Mre11 could potentially perform important functions in higher eukaryotes not revealed by studies of existing hypomorphic cell lines. To test this hypothesis we engineered three gene targeted germline alleles of murine Mre11: a nuclease deficient allele with a substitution of the active site histidine within motif III (H129N), a conditionally inactivatable wildtype allele, and a null allele.
We find that nuclease deficient Mre11 maintains interactions within MRN and supports activation of the ATM kinase. Despite these capabilities, abrogation of Mre11 nuclease activities causes a striking array of phenotypes indistinguishable from loss of the entire MRN complex. Therefore in mammals the nuclease activities of Mre11 are not dispensable, but instead represent a paramount function of the MRN complex distinct from ATM activation.
Results
Impact of Mre11 nuclease deficiency in mice
To determine in vivo roles of the nuclease activities of mammalian Mre11 we generated mice deficient specifically in this function. A single amino acid change analogous to the S. cerevisiae Mre11H125N mutation in nuclease motif III (Moreau et al., 1999) was introduced at the endogenous locus. This alteration changes the invariant histidine residue demonstrated to be essential for in vitro nucleolytic activities of Mre11 homologs from species representing three kingdoms of life, including human (Arthur et al., 2004), S. cerevisiae (Moreau et al., 1999), and P. furiosis (Williams et al., 2008). Furthermore, X-ray crystallographic studies have revealed that the invariant motif III histidine is essential for stabilization and positioning of the scissile phosphate in the DNA backbone, but does not have metal ion liganding functions that could influence overall protein structure or interactions (Arthur et al., 2004; Hopfner et al., 2001; Williams et al., 2008).
The mutation in mouse, H129N, was generated by a C to A nucleotide substitution in the first position of the CAT triplet encoding histidine129 in exon 5, resulting in an AAT triplet encoding asparagine (Figure 1A–D). The C to A mutation in total RNA from liver of three Mre11H129N/+ mice was confirmed through sequencing cloned RT-PCR products (not shown).
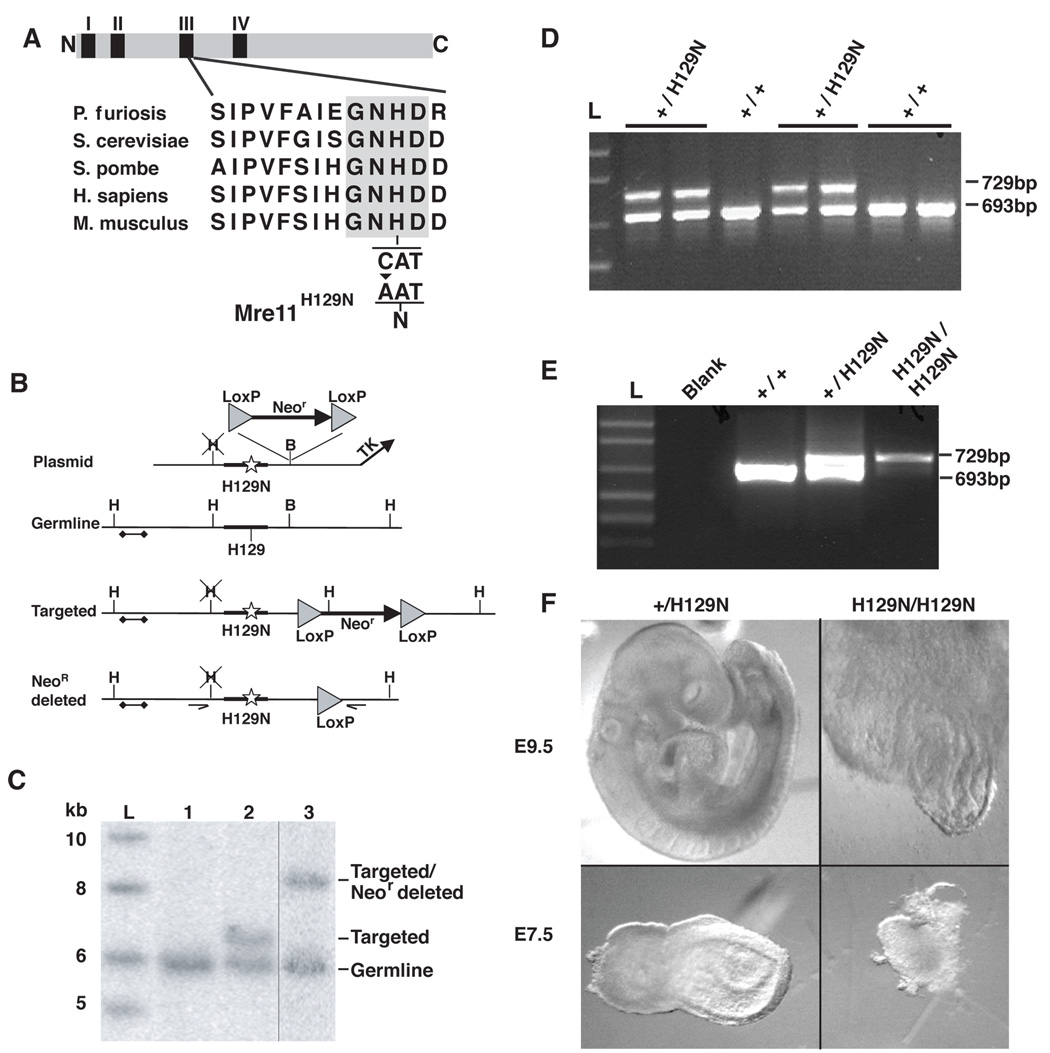
Figure 1. Mre11H129N homozygosity causes early embryonic lethality.
(A) Alignment of Mre11 nuclease motif III. Invariant residues highlighted. The histidine to asparagine change (H129N) is depicted below the active site histidine.
(B) Mre11H129N targeting strategy. Shown are the targeting plasmid with C to A substitution in exon 5 (star in wide line); germline locus; initial targeted configuration; and targeted locus with Neor deleted. Triangles - LoxP sites. Bar with diamonds - Southern blot probe. Arrows - PCR genotyping primers.
(C) Southern blot analysis of targeted embryonic stem cell clone. HindIII digested genomic DNA was probed with 5' probe depicted in (B). Lanes as follows; 1-germline, 2-initial targeting, 3-targeted with Neor gene deleted by Cre. L-ladder.
(D) Genotyping of germline Mre11H129N/+ mice. PCR genotyping of genomic DNA from mouse tails, using primers depicted in (B). Mre11H129N produces larger band due to remaining LoxP.
(E) Genotyping of day e9.5 embryos. Representative Mre11H129N/H129N in right lane.
(F) Light microscopic photographs of day e9.5 (top) and e7.5 (bottom) embryos, comparing Mre11+/H129N (left) to Mre11H129N/H129N (right).
Interbreedings between Mre11H129N/+ mice yielded no Mre11H129N/H129N offspring in more than 300 pups (Table S1). Therefore, homozygosity for the Mre11H129N allele confers embryonic lethality. Dissections at day e13.5 or e10.5 of the 20 day mouse gestation yielded only Mre11+/+ and Mre11H129N/+ embryos (Table S1). We interbred Mre11H129N/+ p53+/− (or p53−/−) mice and isolated embryos from a subset of matings. No Mre11H129N/H129N pups or normal appearing embryos of any p53 genotype were identified (Table S2). Mre11H129N/H129N day e9.5 and 7.5 embryos were recovered (Figure 1E) but were developmentally arrested and highly disordered (Figure 1F and not shown). Whereas wildtype and heterozygous e.9.5 littermates displayed defined fore-, mid-, and hindbrain regions, a beating heart, and forelimb buds, Mre11H129N/H129N embryos displayed a poorly defined body axis and no gross evidence of heart development. Therefore, Mre11H129N/H129N embryos successfully implanted in the uterine wall, but failed to develop normally thereafter, and p53 deficiency provides no apparent rescue.
These observations demonstrate that in mammals Mre11 nuclease activities are not dispensable, and in fact play an essential role among the diverse functions of the MRN complex.
Generation of Mre11 conditional and null alleles
The unanticipated lethality conferred by Mre11H129N homozygosity occurs too early in development to derive cell lines for experimentation. Therefore we generated a wildtype allele of Mre11 that can be conditionally inactivated (Mre11cond) through Cre/LoxP mediated deletion of a region including exon 5 (Figure 2A, B, C). Deletion of a LoxP flanked region (floxed) including exon 5 in a cell line was previously shown to yield no detectable full length or truncated Mre11 (Xiao and Weaver, 1997). Mice homozygous for the Mre11 conditional allele (Mre11cond/cond) appear normal and are born in Mendelian numbers (not shown). We obtained the Mre11 germline null allele (Mre11Δ) by crossing Mre11cond/cond to mice harboring the EIIa-Cre transgene, which expresses Cre in the female germline. Homozygosity for the Mre11 null allele confers embryonic lethality, as no Mre11Δ/Δ pups were identified after more than 200 from crosses with combinations of Mre11cond/Δ and Mre11+/Δ parents (not shown).
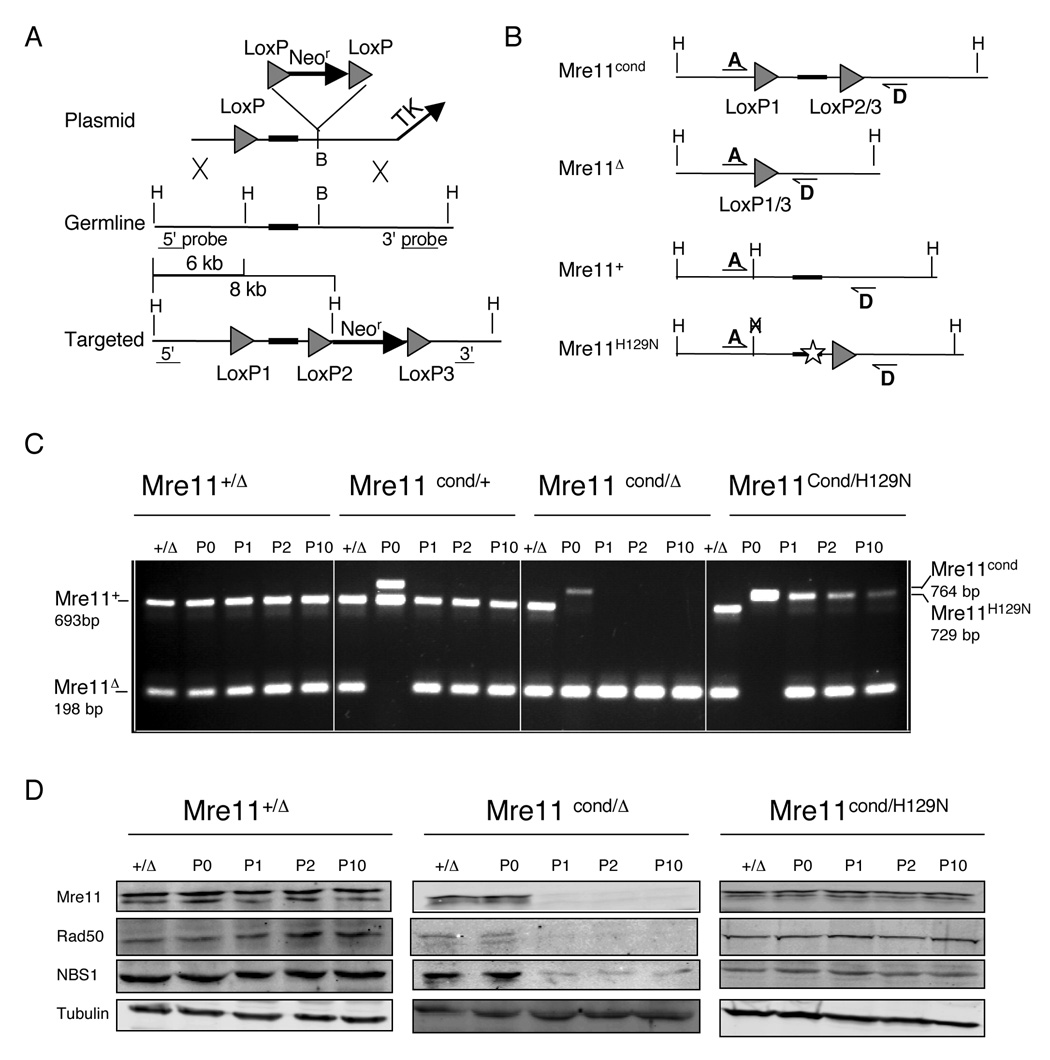
Figure 2. Mre11 conditional allele allows bypass of Mre11H129N embryonic lethality.
(A) Targeting strategy to generate wildtype conditional Mre11 allele. The targeting plasmid was similar to the H129N plasmid depicted in Figure 1, except a LoxP site (triangle) was inserted in the HindIII (H) site upstream of exon 5, which maintained wildtype sequence.
(B) Configurations of four unique Mre11 alleles; Mre11 conditional (Mre11cond), Mre11 null (Mre11), wildtype (Mre11+), nuclease deficient (Mre11H129N). Lines with half arrowheads (A and D) depict PCR primers used to distinguish the four alleles.
(C) PCR analyses of genomic DNA from immortalized MEFs before (passage 0 = P0) and after (passages 1, 2 and 10 = P1, P2, P10) introduction of Cre recombinase. Genotypes indicated were those prior to Cre mediated conversion of Mre11cond to Mre11Δ. Primers are depicted in (B). +/Δ; tail DNA sample used for size comparison of bands.
(D) Western blot analyses of MRN components. Extracts were prepared from the passages shown at top of each panel, and match those in (C). Genotypes indicated were those prior to Cre mediated conversion of Mre11cond to Mre11Δ. Antibodies indicated at left. Tubulin is protein loading control.
Stability of the MH129N RN complex
Mouse embryonic fibroblast (MEF) lines were derived from day e13.5 embryos harvested from matings between Mre11cond/cond and Mre11+/H129N mice, and between Mre11cond/cond and Mre11+/Δ mice. Primary or immortalized Mre11+/Δ (or cond/+), Mre11cond/Δ and Mre11cond/H129N lines allowed for comparative analysis of phenotypes upon conversion of the conditional allele to null.
Cre recombinase was introduced via replication defective adenovirus (Ad-Cre). Deletion of the floxed Mre11 region was evident using a PCR strategy after one passaging of cells (Figure 2B, C, not shown). Western blot analysis comparing Mre11 levels in Mre11+/Δ and Mre11Δ/Δ cells showed a clear deficit of the Mre11 protein which persisted for multiple passages (Figure 2D). Rad50 and NBS1 proteins also displayed reduced levels, likely due to instability in the absence of Mre11. We also observed efficient Cre mediated conversion of Mre11H129N/Cond to Mre11H129N/Δ (Figure 2C). However, the level of Mre11 protein was not reduced after Cre exposure, confirming the Mre11H129N protein is stable in vivo (Figure 2D). Furthermore, levels of Rad50 and NBS1 also remained constant, suggesting that the Mre11H129N protein maintains interactions among MRN components (Figure 2D).
To further address whether Mre11H129N protein maintains interactions, we performed co-immunoprecipitations (co-IP) using an anti-Mre11 antibody. Comparing Mre11Δ/+ and Mre11Δ/H129N lines, equivalent amounts of Rad50 and NBS1 were isolated (Figure 3A, B). We determined if Mre11H129N protein maintains the ability to homodimerize using the yeast two hybrid system. Comparison of murine Mre11H129N or Mre11+ cDNA inserted into bait and prey plasmids demonstrated that the mutant protein retained the capability to homodimerize (Figure 3C). We addressed whether complexes containing Mre11H129N retain the ability to form immunoflourescent foci. After exposure to IR, foci comprised of Mre11H129N or NBS1 are readily detectable (Figure 3D, E).
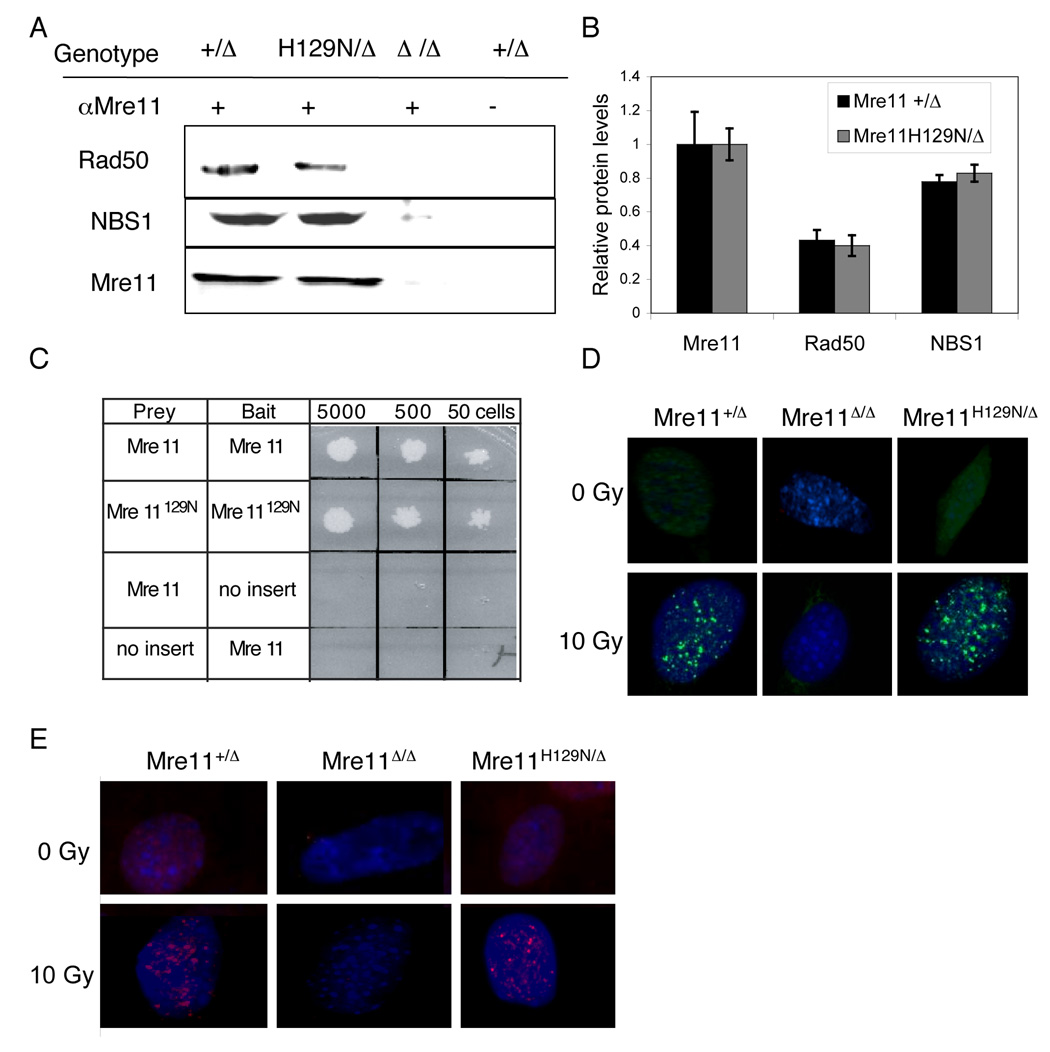
Figure 3. Stability of the MH129N RN complex.
(A) The Mre11+ and Mre11H129N proteins associate with equal quantities of endogenous NBS1 and Rad50. IPs using antibody to Mre11 were performed on MEF extracts. Western blots were performed with antibodies to the MRN components (left).
(B) Quantitation of co-IPs with protein levels expressed relative to Mre11. Error bars represent ± SEM from three independent experiments.
(C) Mre11H129N protein can homodimerize. Yeast two hybrid analyses using the indicated mouse cDNAs as prey or bait. Numbers at top indicate number of cells plated. White coloration represents growth of yeast in selective media, reflecting interaction.
(D) Mre11 (green) and (E) NBS1 (red) immunofluorescent foci in MEFs exposed to 10 Gy IR.
Taken together, our studies indicate that the Mre11H129N nuclease deficient protein is stable and maintains interactions within the MRN complex. These conclusions are supported by prior studies examining mutation of the invariant motif III histidine (Arthur et al., 2004; Bressan et al., 1998; Krogh et al., 2005) and by observations in the accompanying manuscript (Williams et al., 2008).
Requirement for Mre11 nuclease activities in cellular proliferation and genomic stability
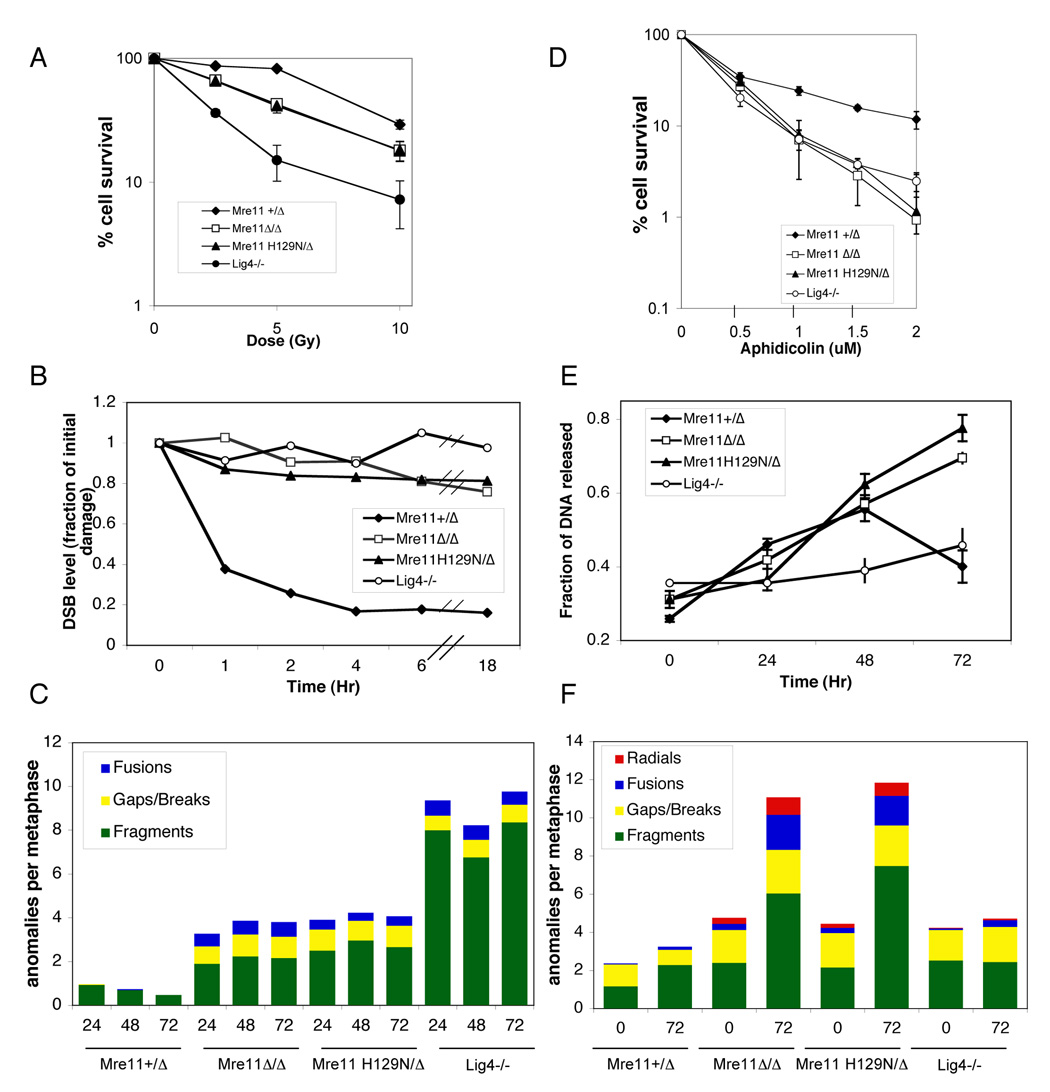
Relative to control (Mre11+/Δ), both Mre11Δ/Δ and Mre11H129N/Δ MEFs exhibited significantly reduced growth rates by the second passage after Cre exposure, and no detectable increase in numbers beyond passage 3 (Figure 4A). Parallel experiments were performed on T antigen immortalized cells, and Mre11Δ/Δ and Mre11H129N/Δ again grew at a slower rate relative to control, but were improved compared to primary MEFs (Figure 4B). Beyond passage 10, growth of these lines progressively slowed, and were occasionally overgrown by cells maintaining the Mre11cond allele. Continued absence of the Mre11 protein in the Mre11Δ/Δ line was confirmed by Western blot (Figure 2D, middle panel) and absence of the Mre11cond allele by PCR in all three lines (Figure 2C). By passage 3, both Mre11Δ/Δ and Mre11H129N/Δ primary cultures showed a dramatic senescence phenotype, with approximately 50% of cells being positive for SA-β-gal, compared to 4.6% for control (Figure 4C).
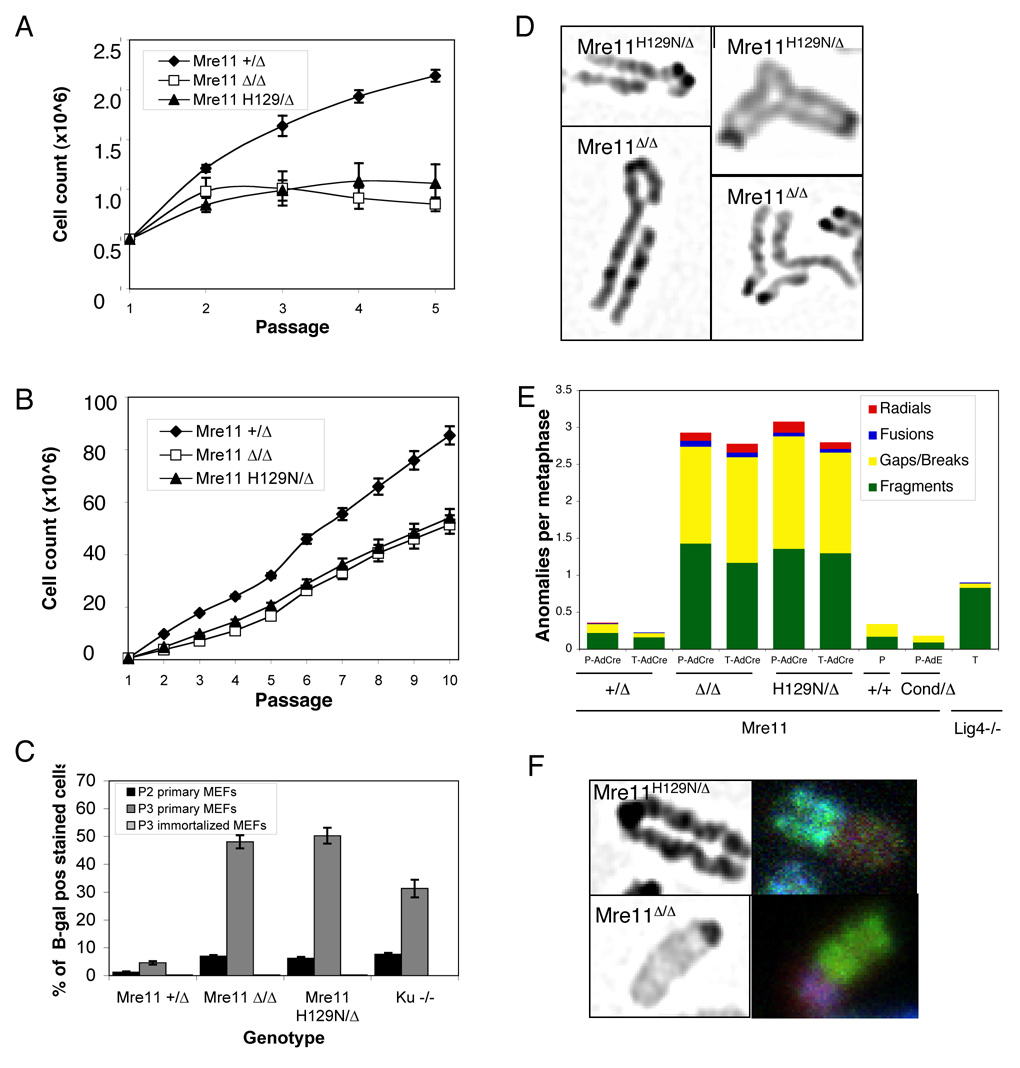
Figure 4. Cellular phenotypes of Mre11H129N phenocopy MRN deficiency.
(A – F) Genotypes indicated are after Cre mediated conversion of Mre11cond to Mre11Δ. (A and B) Primary (A) or immortalized (B) MEFs were passaged and counted every 3 days. Symbols and error bars represent averages ± SEM from three independent experiments, using two independent cell lines for each genotype. Cell numbers plotted as a function of passage number.
(C) Primary and immortalized MEFs were assessed for cellular senescence using SA-β-Gal staining at passages 2 and 3 (P2 and P3) in primary MEFs and P3 in immortalized MEFs. Bars for P3 immortalized MEFs are depicted, but very short. Immortalized Ku−/− not presented. Error bars represent ± SEM from three independent experiments.
(D) Spontaneous genomic instability in Mre11H129N/Δ versus Mre11Δ/Δ MEFs. Metaphase spreads were stained with DAPI. Examples of single chromatid gaps (left) and radial chromosomes (right) are shown.
(E) Spectrum of chromosome anomalies in Mre11Δ/Δ and Mre11H129N/Δ MEFs. Primary (P) or T antigen immortalized (T) cells were exposed to adenovirus expressing Cre recombinase (AdCre), no virus, or a control adenovirus (AdE) without Cre. Lig4−/− MEFs were used for comparison. Metaphase spreads were stained with DAPI. Y-axis indicates anomalies per metaphase. Colors indicate proportions of different anomalies.
(F) Representative chromosome translocations in Mre11Δ/Δ and Mre11H129N/Δ metaphases. DAPI staining (left) and SKY paint mixture (right).
Metaphases from Mre11+/Δ, Mre11Δ/Δ and Mre11H129N/Δ MEFs were stained with DAPI and scored for spontaneous chromosomal anomalies. Deficiency of the MRN complex (Mre11Δ/Δ) caused dramatic chromosome instability. Structural anomalies included chromosome or chromatid fragments, chromosomes with a break or obvious gap in a single chromatid, occasional radial structures and dicentrics (Figure 4D, E, Table S3). We also performed Spectral Karyotyping (SKY), which revealed non clonal translocations (Figure 4F, Table S4).
Analysis of Mre11H129N/Δ MEFs revealed a frequency and pattern of genomic instability with striking similarity to Mre11Δ/Δ (Figure 4D, E, F, Table S3, Table S4). To understand whether this phenocopying has mechanistic implications, or is a common manifestation of DNA repair deficiencies, we compared the Mre11 mutant lines to MEFs deficient for DNA Ligase IV (Lig4−/−) (Ferguson et al., 2000; Frank et al., 1998). Unlike MRN, the role of Lig4 in DSB repair is well characterized, performing the final ligation step in NHEJ (Wilson et al., 1997). The overall frequency of chromosomal anomalies in Lig4−/− was less than either Mre11 deficiency (Figure 4E, Table S3). Importantly, the spectrum of anomalies was distinct from the Mre11 deficiencies, in that most were isolated chromosome fragments, with few gaps or breaks within single chromatids, and no radial structures (Figure 4E, Table S3). Therefore the MRN complex plays roles in maintaining genomic stability that are distinct from Lig4, and which require Mre11 nuclease activities.
ATM activation does not require Mre11 nuclease activities
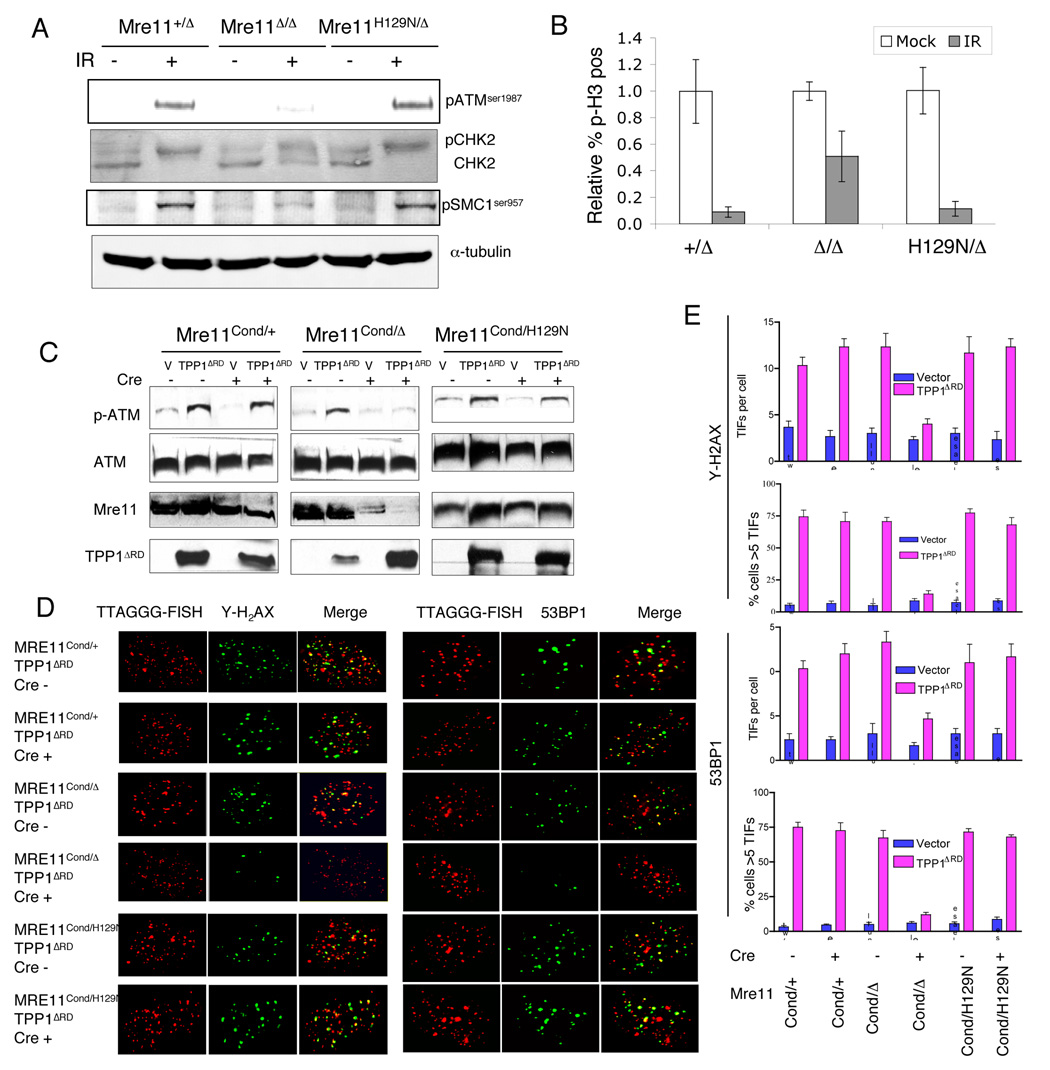
Ectopically expressed nuclease deficient Mre11 was reported to minimally complement defective ATM activation in cells from ATLD patients (Uziel et al., 2003). Because both the nuclease deficient and endogenous ATLD Mre11 proteins likely participated in complexes, and because Mre11 requires post translational modification that might be impacted by transient expression (Boisvert et al., 2005), we determined if Mre11 nuclease activities are required for ATM activation using our engineered mouse alleles. ATM activation can be addressed experimentally by measuring levels of autophosphorylation at serine 1987 (1981 in human)(Bakkenist and Kastan, 2003), and phosphorylation of substrates (Lee and Paull, 2007).
A significant defect ATM activation was clearly present in Mre11Δ/Δ relative to Mre11+/Δ control, in agreement with prior studies examining MRN deficiencies (Lee and Paull, 2007) (Figure 5A). In contrast, ATM autophosphorylation and phosphorylation of the ATM substrates Chk2 and SMC1 were not reduced in Mre11H129N/Δ cells. Reduced efficiency of ATM activation would be expected to manifest as a defective G2/M cell cycle checkpoint. We determined the integrity of this checkpoint after IR by measuring mitotic index. Relative to Mre11+/Δ control, significantly more Mre11Δ/Δ cells entered mitosis after IR, indicating a defective G2/M checkpoint (Figure 5B). In stark contrast, the percentage of Mre11H129N/Δ cells in mitosis was similar to control. Together, these studies demonstrate that while the MRN complex as a whole is required for ATM activation after IR, the nuclease activities of Mre11 are dispensable for this function.
Figure 5. Mre11 nuclease activities are not required for ATM activation.
(A–B) Genotypes indicated reflect those after Cre mediated conversion of MreCond to Mre11Δ. (A) Immunoblotting for ATM substrates (right) was performed on whole cell lysates with the indicated antibodies 1 hour after 10 Gy IR. Tubulin - loading control. Each blot is a representative example of at least 3 independent experiments.
(B) Assessment of the G2/M checkpoint. MEFs were harvested 1 hour after 10 Gy IR. Mock - no IR. Cells committed to M phase were identified by positive staining (flow cytometry) for phosphorylated histone H3 (p-H3) (Y axis). The mitotic index obtained from the mock sample from each genotype was set to 1.0 and after irradiation is expressed as a percentage of mock. Error bars indicate ± SEM from three independent experiments.
(C–E) Mre11 genotypes shown are those prior to Cre mediated conversion of Mre11Cond to Mre11Δ.
(C) ATM1987 autophosphorylation induced by telomere deprotection caused by HA-TPP1ΔRD expression. TPP1ΔRD was detected by anti-HA antibody. Total ATM served as loading control.
(D) γ-H2AX (left panel) and 53BP1 (right panel) foci at telomeres (TIFs) induced by expression of TPP1ΔRD. MEFs expressing TPP1ΔRD were infected either with AdE or AdCre and analyzed by telomere (TTAGGG) PNA-FISH (red) and antibody (green) to γ-H2AX or 53BP1. (E) Quantitation of TIFs from experiments described in (D). The number of TIFs per cell, as well as the percentage of cells containing five or more γ-H2AX or 53BP1 TIFs are shown. Error bars represent ±SEM.
ATM activation is not restricted to radiation responses, and is triggered by DSBs in many contexts (Lee and Paull, 2007). It has recently been shown that ATM is activated by telomeres rendered dysfunctional (Denchi and de Lange, 2007; Guo et al., 2007). Telomeres are maintained by a multiprotein complex termed Shelterin which maintains a protective cap that prevents telomeres from being recognized as DSBs (de Lange, 2005). We rendered telomeres dysfunctional through transient transfection of a dominant negative allele of the Shelterin component TPP1 (TPP1ΔRD) used effectively in the past to uncap telomeres (Guo et al., 2007). We examined ATMser1987 autophosphorylation in Mre11+/Δ, Mre11Δ/Δ and Mre11H129N/Δ MEFs infected with either vector alone or retrovirus expressing TPP1ΔRD. Expression of TPP1ΔRD induced robust ATM autophosphorylation in Mre11+/Δ and Mre11H129N/Δ, but not in Mre11Δ/Δ MEFs (Figure 5C).
At uncapped telomeres, ATM phosphorylates the histone H2AX to form γ-H2AX and causes localization of 53BP1, events that normally occur at DSBs (Denchi and de Lange, 2007; Guo et al., 2007). We monitored telomeric association of γ-H2AX and 53BP1 utilizing the telomere-dysfunction-induced foci (TIF) assay. In Mre11+/Δ and Mre11H129N/Δ MEFs, expression of TPP1ΔRD resulted in the formation of γ-H2AX and 53BP1 foci that colocalized with telomeric DNA, with approximately 75% of cells exhibiting a minimum of five TIFs (Figure 5 D, E). However, only minimal TIF formation was observed in Mre11Δ/Δ MEFs, with less than 10% of cells containing greater than 5 TIFs (Fig. 5D, E). These studies demonstrate a major role for the MRN complex in ATM activation at dysfunctional telomeres, but like IR induced damage, this role does not require the nuclease activities of Mre11.
Mre11 nuclease activities function in DNA repair
Our findings demonstrate that Mre11 focus formation and ATM activation occur in the context of Mre11 nuclease deficiency. Therefore we considered the possibility that Mre11 nuclease activities are dispensable for all DSB related functions of MRN. To this end we determined relative sensitivities to IR. Mre11Δ/Δ and Mre11H129N/Δ lines both displayed hypersensitivity to IR, and remarkably, yielded nearly identical survival curves (Figure 6A). Lig4−/− MEFs displayed a more severe hypersensitivity compared to either Mre11 deficiency. Therefore, the nuclease activities of Mre11 do perform important functions in the context of DSBs that are independent of control over ATM activation.
Figure 6. DSB repair defects in Mre11 nuclease and MRN deficiencies.
(A) Comparison of ionizing radiation sensitivities. The Y axis indicates percent of cells relative to unirradiated control populations, and the X axis indicates increasing doses of IR. Error bars represent ±SEM of 3 independent experiments.
(B) Quantitation of DNA damage levels after IR. MEFs of the indicated genotypes were exposed to 80 Gy IR and allowed to recover for various times (X axis). The DSB level (Y axis) based on quantitation of CHEF gels was calculated as a ratio of the amount of damage at each recovery time to that at time 0. Shown is representative of at least three experiments for each line.
(C) Radiation induced chromosome instability persists in Mre11 nuclease and MRN deficiencies. MEFs were exposed to 2 Gy IR, followed by recovery times of 24, 48 and 72 hours (X axis). Metaphases were stained with DAPI. Y-axis - anomalies per metaphase.
(D) Comparison of sensitivities to the DNA polymerase inhibitor aphidicolin. Y axis indicates percent of cells relative to untreated control populations, and X axis indicates increasing doses of aphidicolin. Error bars represent ±SEM of 3 independent experiments.
(E) Quantitation of DNA damage levels after aphidicolin exposure. MEFs of the indicated genotypes were exposed to aphidicolin for 24 hours and allowed to recover for various times (X axis, 0 represents 24 hour exposure with no recovery). The fraction of DNA released (Y axis) at each recovery time was calculated from the ratio of DNA entering the gel (damaged) to total. Error bars represent ±SEM.
(F) Aphidicolin induced chromosome instability persists in Mre11 nuclease and MRN deficiencies. MEFs were exposed to 0.4µM aphidicolin for 24 hours followed by 0 or 72 hours recovery. Metaphase spreads were stained with DAPI. Y-axis indicates anomalies per metaphase.
The requirement for Mre11 nuclease activities after IR, in the context of proficient ATM activation, led us to hypothesize that in mammals nucleolytic processing by Mre11 plays a vital role directly in DNA repair. Therefore, we performed a pulsed field gel electrophoresis (PFGE)-based DSB assay after IR to directly determine DNA repair capacities. The fragmentation of chromosomes can be detected as DNA entering the gel, and is quantitated as a ratio of DNA within the lane relative to the total DNA loaded. We observed that control (Mre11+/Δ) cells displayed progressively less DNA damage in a time course from 0 to 18 hours after IR, whereas Lig4−/− displayed no capability to repair (Figure 6B). Mre11Δ/Δ and Mre11H129N/Δ lines both displayed a striking defect in DNA repair. Therefore the hypersensitivity to IR conferred by absence of Mre11 nuclease activities likely reflects a defect in the repair process.
As an independent means to identify a DNA repair defect we determined the impact of IR on the degree and persistence of genomic instability. Mre11+/Δ, Mre11Δ/Δ, Mre11H129N/Δ, and Lig4−/− lines were irradiated with 2 Gy and allowed recover for 24, 48 or 72 hours. In contrast to control, all three mutant lines showed no indication of a downward trend in frequency of chromosome anomalies during the 72 hour timeframe (Figure 6C, Table S5). Therefore, absence of Mre11 nuclease activities or of MRN confer similar increases in IR induced genomic instability, which like Lig4 deficiency, persists for at least several days.
Stalled DNA replication forks can be processed into DSBs to facilitate replication re-start (Lambert et al., 2007). The DSBs that arise during replication stress are therefore distinct from those induced by IR, since they are generated by endogenous processes. We determined if the nuclease activities of Mre11 are required during replication stress by examining the impact of the DNA polymerase α/δ inhibitor aphidicolin on survival, DSB repair and genomic stability. Hypersensitivity to aphidicolin as well as persistent DNA damage and genomic instability was observed for the Mre11Δ/Δ line relative to Mre11+/Δ control (Figure 6D, E, F, Table S6). Once again, the Mre11H129N/Δ nuclease deficient line phenocopied the Mre11Δ/Δ line. To confirm that DSBs arise under these conditions we also analyzed the Lig4−/− line, and found that it is hypersensitive to aphidicolin, although DNA damage does not persist to the same degree as in the Mre11 mutants. Therefore, DSBs do arise under these conditions. Taken together, these studies reveal that the nuclease activities of Mre11 are required for repair of DSBs that are induced by IR or that arise due to replication stress.
Mre11 nuclease activities in resection and recombination
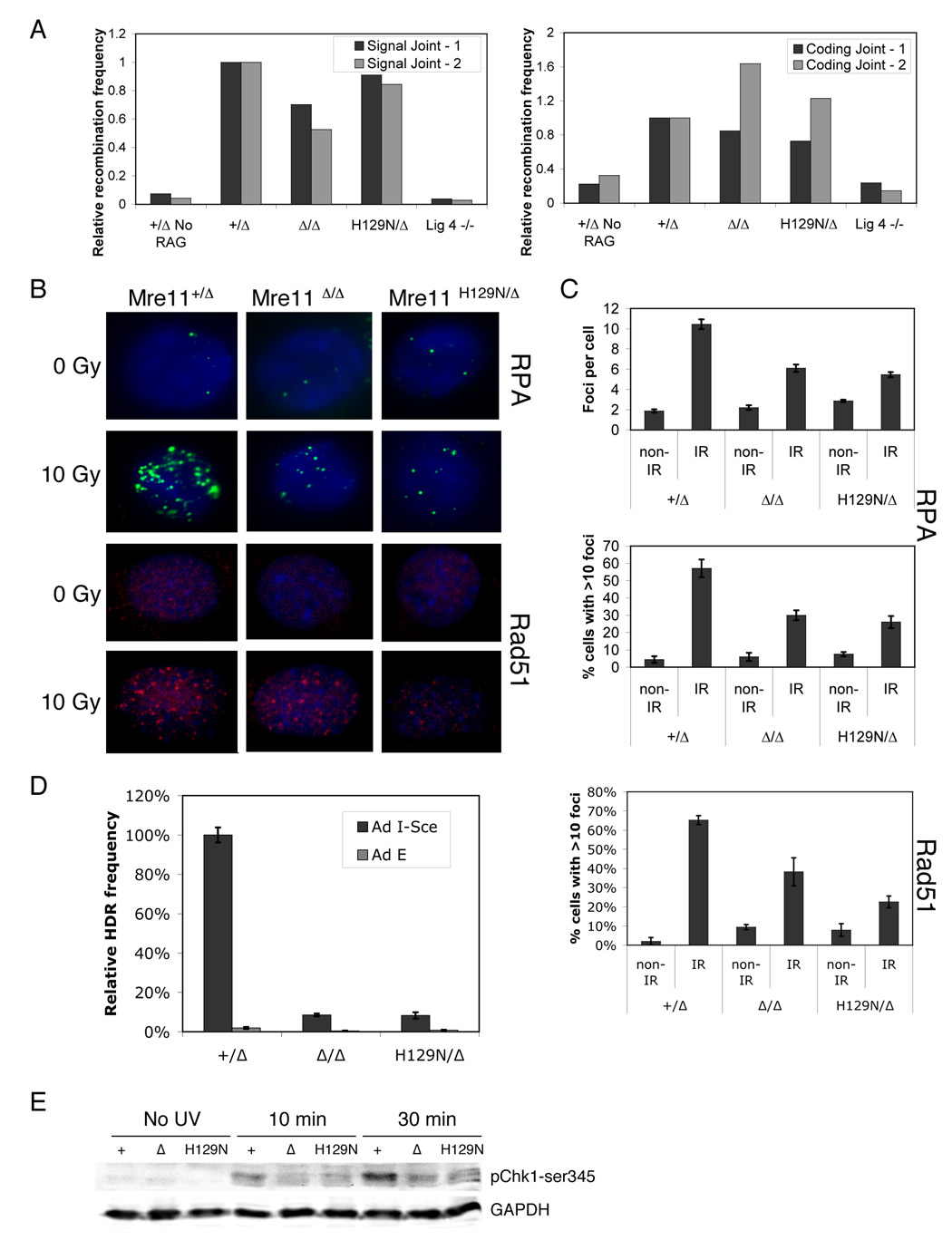
We wished to determine which DSB repair pathway(s) require the nuclease activities of Mre11. NHEJ is required for V(D)J recombination, which generates antibody diversity through a process initiated by DSBs generated by the RAG1/2 endonuclease (Fugmann et al., 2000). Co-transfection of RAG1/2 expression vectors with a signal end (blunt) or coding end (hairpin end requiring processing) reporter construct allows comparison of recombination frequencies, as well as analysis of the joining process by examining sequences of products (Rooney et al., 2003). Comparing Mre11Δ/+ to Mre11Δ/Δ and Mre11Δ/H129N lines, no significant differences in recombination frequencies (Figure 7A) or the nature of sequenced joints were detected (Figure S4, not shown). As previously reported for Lig4−/− the frequency of signal and coding join formation was reduced to levels similar to controls without RAG1/2 (Frank et al., 1998; Grawunder et al., 1998). Thus, the MRN complex does not appear to play a significant role in NHEJ in mammals. However, a function in NHEJ not required for V(D)J recombination cannot be entirely ruled out.
Figure 7. Mre11 nuclease activities in resection and recombination.
(A) Transient transfection based V(D)J recombination assay. (Left) Signal joining (blunt ends). (Right) coding joins (hairpin ends). Two representative experiments shown for each. For relative recombination frequency (Y axis), results from control (Mre11Δ/+) were set to 1.0.
(B) IR induced RPA1 (top) and Rad51 (bottom) foci.
(C) Quantitation of foci described in (B). Error bars represent ±SEM of 3 independent experiments.
(D) Relative frequencies of HDR using DR-GFP substrate. HDR events were scored as the percentage of GFP-positive populations after I-SceI expression. The percentage of GFP-positive cells in Mre11+/Δ was set to 100%. Error bars represent ±SEM from three independent DR-GFP clones of each genotype.
(E) Immunoblotting for phospho-Chk1ser345. Cells exposed to 5 J/m2 UV recovered for the indicated times. GAPDH - loading control. Blot is representative of 3 experiments.
HR is initiated at DSBs by the generation of single stranded DNA, which is bound by RPA and ultimately by filaments of Rad51 family members which carry out the search for homology and strand exchange (Wyman and Kanaar, 2006). On a cellular level, these events can be tracked by examining immunofluorescent foci. We quantitated RPA and Rad51 foci after IR in Mre11+/Δ, Mre11Δ/Δ and Mre11H129N/Δ cells and found that deficiency of the MRN complex, or of Mre11 nuclease activities, reduces foci formation of both proteins (Figure 7B, C). In each case, foci formation was reduced by approximately 50 % relative to control. This suggests that Mre11 nuclease activities play a role in the early steps of HR (Sartori et al., 2007). To more definitively identify a role in HR, we employed the DR-GFP assay, which measures homology directed repair of an I-SceI endonuclease induced DSB in an integrated GFP reporter gene (Pierce et al., 1999). For each genotype, three independent lines containing single copy genomic integrants of the intact DR-GFP plasmid were analyzed. I-SceI was introduced via replication defective adenovirus, with virus containing no I-SceI insert as control. Relative to Mre11Δ/+, Mre11Δ/Δ and Mre11Δ/H129N lines showed a striking reduction in the percentage of GFP+ cells, to approximately 10% of control (Figures 7D and S5). We confirmed using propidium iodide staining that there is no significant difference in the percentage of cells in S/G2 phase among the lines, indicating that differences in recombination efficiencies do not reflect cell cycle differences (data not shown). Therefore the nuclease activities of Mre11 function in HR in mammals, likely at an early step in the pathway.
In addition to initiating HR, the generation of single stranded DNA serves to activate the ATM- and rad3-related (ATR) kinase (Petermann and Caldecott, 2006). ATR is activated by agents that inhibit DNA replication, such as low dose ultraviolet light (UV). Certain deficiencies of MRN reduced ATR activation, leading to the notion that nuclease activities of Mre11 could contribute to resection in this context (Stiff et al., 2005; Zhong et al., 2005). We tested this by examining UV induced phosphorylation of the Chk1 kinase. Phospho-Chk1 levels were reduced in Mre11Δ/Δ and Mre11Δ/H129N, indicating that Mre11 is involved in this process (Figure 7E).
Discussion
A direct DNA repair function of MRN distinct from ATM activation
Roles of the ATM kinase and the MRN complex are intimately linked in mammals. This relationship was highlighted by defective activation of ATM in cells harboring hypomorphic alleles of Mre11 (Stracker et al., 2004). However, the observation that mouse knockouts of MRN components confer early embryonic lethality while ATM inactivation permits viability suggested there are essential mitotic functions of MRN unrelated to ATM control. Our studies demonstrate that Mre11 nuclease activities provide such a function. These activities play a major role in HR, and contribute to ATR activation (Sartori et al., 2007). Thus, demise in early embryogenesis caused by deficient Mre11 nuclease activities likely results from combined impact of deficiencies in these responses to DNA damage.
We have provided evidence supporting the notion that in mammals the MRN complex operates at DSBs during HR, but not during repair by the classic Lig4 dependent NHEJ pathway (Yan et al., 2007). The differing contexts in which these essential factors operate are illustrated by the distinct impact on genomic stability when either is absent. The Mre11 deficiencies conferred higher overall levels of spontaneous chromosome anomalies, correlating with the earlier embryonic lethality relative to Lig4−/−. The spectrum of types of spontaneous anomalies observed in the Mre11 deficiencies resembled that induced by aphidicolin, with notable levels of radial structures and dicentric chromosomes. These are not prevalent in Lig4−/− or in any cells exposed to IR. Taken together, these findings suggest that replication stress is a major contributor to early embryonic demise in the absence of Mre11 nuclease activities or MRN, while more straightforward forms of DSBs lead to late embryonic demise without Lig4.
Why the nucleolytic functions of Mre11 have such a striking impact when deficient in mammals as compared to S. cerevisiae is an intriguing evolutionary question. A possible explanation has been raised by two recent reports. In cooperation with another DNA repair factor Sae2, Mre11 cleaves in single stranded DNA adjacent to hairpins in vitro (Lengsfeld et al., 2007). In this study, Sae2 itself was shown to possess endonuclease activity. In a separate study the proposed mammalian Sae2 homolog, CTIP, was shown to interact with Mre11 and stimulate its endonuclease activity, but did not display nuclease activities of its own (Sartori et al., 2007). Therefore it is possible that in higher eukaryotes situations requiring MRN employ Mre11 as the only nuclease, whereas in S. cerevisiae Mre11 and Sae2 act redundantly.
It is interesting to note that in S. cerevisiae there are Rad50 separation of function alleles (RAD50S) which confer phenotypes resembling Mre11 nuclease deficiency. These mutations block processing of DSBs that initiate meiosis, but have minimal impact on mitotic cells (Cao et al., 1990). One Rad50S allele generated in mice yielded phenotypes distinct from Mre11 nuclease deficiency, permitting viability with minimal genomic instability and DNA damage hypersensitivity, while causing constitutive ATM activation (Bender et al., 2002; Morales et al., 2005). However, two other Rad50S alleles proved to be embryonic lethal (Bender et al., 2002). It will be interesting to determine if the severe impact of these alleles reflects abnormal regulation of Mre11 nuclease activity.
The MRN complex at dysfunctional telomeres
Our studies demonstrate a critical role for the MRN complex in activating the ATM kinase at sites of deprotected telomeres, but as observed for activation after IR, this role does not require the nuclease activities of Mre11. MRN is found at telomeres and interacts with the Shelterin component TRF2, which binds double stranded telomeric DNA (Zhu et al., 2000). We therefore postulate that MRN is an important sensor of dysfunctional telomeres, and is pre-positioned to rapidly detect uncapped telomeric sequences. While this is seemingly the same role proposed for IR responses, there is an important distinction. Telomere deprotection in the context of the TPP1ΔRD allele does not result in significant numbers of chromosome fusions (Guo et al., 2007). Thus, the uncapped telomeres in our study are unlike general DSBs in that most are one-sided. Typical DSBs have two sides which ultimately undergo ligation during repair. This distinction suggests that functions of MRN involving two sides, such as long range bridging by Rad50 or short range bridging functions by the Mre11 dimer (Williams et al., 2008) are not required for ATM activation.
Our work in combination with the accompanying manuscript (Williams et al., 2008) now provide a more detailed mechanistic understanding of early events at DSBs. In addition to long range bridging functions of Rad50, the Mre11 dimer can position two sides of a DSB in close proximity. Within the dimer interface the nuclease activities of Mre11 process ends prior to, or in association with, final repair. We have shown that this processing is not dispensable in mammals, but instead represents a paramount function of the MRN complex and an activity of fundamental importance to DSB repair. This essential function, provided by a portion of MRN of ancient evolutionary origin, operates independently of the more recently evolved and non essential interplay between MRN and ATM.
Experimental Procedures
Generation of germline Mre11 alleles
Targeting constructs used plasmid PLN-TK as backbone with arms generated by PCR using genomic DNA from TC1 mouse cells. Positive/negative selection used G418 and gancyclovir. (details in supplementary methods).
Growth and analysis of MEFs
MEFs were isolated from day e13.5 embryos and grown in standard culture conditions as described (Gu et al., 1997). (See Supplemental Procedures). Primary MEFs were immortalized by transfection with pBsSVD2005 (SV40 large T antigen expression vector). Adeno-Cre at MOIs from 100 to 500:1 gave comparable results. MEFs were grown 3 days post infection and split once prior to experiments.
Metaphases on glass slides were stained with DAPI (Invitrogen), SKY probe (ASI) or telomere PNA FISH-FITC (DakoCytomation) according to the manufacturers' instructions. Images acquired on an Olympus BX-61 microscope and viewed using SKYview software (ASI).
Western Blots and Immunoprecipitation
Cell extracts were prepared in Laemmli buffer (4% SDS, 20% glycerol, 120 mM Tris-HCl, pH 6.8). resolved by SDS-PAGE and transferred using standard procedures. Primary antibodies were: Mre11 (Novus or Cell Signaling); NBS1 (Novus); Rad50 (Bethyl); p-ATM Ser 1981(Rockland); ATM (Santa Cruz), γ-tubulin (AbCam); p-CHK2, clone 7 (Upstate); and SMC1-ser957 (Cell Signaling), γ-H2AX (Upstate), 53BP1 (kind gift from Phil Carpenter, UTHMB), Chk1-ser345(Rockland). Secondary antibodies for western blots were IRDye800CW conjugated goat anti-rabbit or anti-mouse (Li-Cor Biosciences).
Immunoprecipitations were performed using protein G sepharose beads (GE healthcare) with 2 mg protein extract incubated with anti-Mre11 antibody (Cell Signaling) for 16 hours at 4° C. Western blots performed as above.
TIF (Telomere dysfunction Induced Foci) assay
Performed as described (Guo, et al. 2007), and Supplemental Procedures. Cells were infected and selected for 7–10 days for stable expression of TPP1ΔRD. Secondary antibodies against mouse or rabbit IgG were labeled with Alexa 488 (Molecular Probes). Tamra-(TTAGGG)3 PNA telomere probe (Applied Biosystems) and DAPI DNA counterstain were used. Only cells with ≥ 5 γ-H2AX signals or 53BP1 foci (green) co-localized with telomere signal (TTAGGG)3 (red) were scored.
Pulsed field gel electrophoresis
MEFs were exposed to 80 Gy IR or 0.4 µM aphidicolin. DNA analyses were performed using Bio-Rad CHEF DRII optimized to the 1 – 4 Mb range. Gels were stained with Sybr green 1 (Invitrogen), imaged on a Typhoon 9400 imager (GE HealthCare) and quantified with ImageQuant software (GE Healthcare).
Recombination assays
MEF lines containing a single integrated intact copy of reporter substrate DR-GFP were constructed as described (Pierce et al., 1999). I-SceI was transiently expressed via infection with I-SceI containing adenovirus (AdNGUS24i), and recombination frequency determined by % GFP+ cells.
V(D)J transient assays were performed as previously described (Rooney et al., 2003). Full-length RAG1/2 expression constructs and pJH290 coding joining or pJH200 RS joining substrate plasmids were transfected using Superfect (Qiagen). 48 h after transfection plasmids were isolated by alkaline lysis and electroporated into Escherichia coli MC1061. Recombination efficiencies calculated by ratio of ampicillin- and chloramphenicol-resistant colonies to ampicillin-resistant colonies.
Presentation of images
Images were cropped, and when appropriate placed in composites, using Adobe Photoshop Software (Adobe).
Supplementary Material
Acknowledgments
Support for this work to DOF. provided by NIH (HL079118), Sidney Kimmel Cancer Research Foundation, University of Michigan Cancer Center Support Grant 5-P30-CA46592 and Munn Endowment, and by K08 HL067580 under initial mentorship of Dr. F.W. Alt. DOF. thanks Drs. John Moran, Tom Wilson, Mats Ljungman, Chris Canman and Tobias Else for advice on the manuscript and experiments. SC acknowledges M.D. Anderson Cancer Center's Molecular Cytogenetics Core (CA016672) and support from the Welch, Elsa U. Pardee, Abraham and Phyllis Katz Foundations, Michael Kadoorie Cancer Genetics Research Program, NIH (AG028888, CA129037). Y.D. supported by 1K01CA124461-01. JMS supported by NIH AI063058 and Pew Foundation Scholars Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arthur LM, Gustausson K, Hopfner KP, Carson CT, Stracker TH, Karcher A, Felton D, Weitzman MD, Tainer J, Carney JP. Structural and functional analysis of Mre11-3. Nucleic Acids Res. 2004;32:1886–1893. doi: 10.1093/nar/gkh343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Bender CF, Sikes ML, Sullivan R, Huye LE, Le Beau MM, Roth DB, Mirzoeva OK, Oltz EM, Petrini JH. Cancer predisposition and hematopoietic failure in Rad50(S/S) mice. Genes Dev. 2002;16:2237–2251. doi: 10.1101/gad.1007902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- Boisvert FM, Dery U, Masson JY, Richard S. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 2005;19:671–676. doi: 10.1101/gad.1279805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan DA, Olivares HA, Nelms BE, Petrini JH. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics. 1998;150:591–600. doi: 10.1093/genetics/150.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S cerevisiae. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- D'Amours D, Jackson SP. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat Rev Mol Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- Difilippantonio S, Nussenzweig A. The NBS1-ATM connection revisited. Cell cycle (Georgetown, Tex. 2007;6:2366–2370. doi: 10.4161/cc.6.19.4758. [DOI] [PubMed] [Google Scholar]
- Ferguson DO, Sekiguchi JM, Chang S, Frank KM, Gao Y, DePinho RA, Alt FW. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc Natl Acad Sci U S A. 2000;97:6630–6633. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, Cheng HL, Davidson L, Kangaloo L, Alt FW. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annual review of immunology. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. Embo J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grawunder U, Zimmer D, Fugmann S, Schwarz K, Lieber MR. DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Mol Cell. 1998;2:477–484. doi: 10.1016/s1097-2765(00)80147-1. [DOI] [PubMed] [Google Scholar]
- Gu Y, Seidl KJ, Rathbun GA, Zhu C, Manis JP, van der Stoep N, Davidson L, Cheng HL, Sekiguchi JM, Frank K, et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 1997;7:653–665. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- Guo X, Deng Y, Lin Y, Cosme-Blanco W, Chan S, He H, Yuan G, Brown EJ, Chang S. Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. Embo J. 2007;26:4709–4719. doi: 10.1038/sj.emboj.7601893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- Krogh BO, Llorente B, Lam A, Symington LS. Mutations in Mre11 phosphoesterase motif I that impair Saccharomyces cerevisiae Mre11-Rad50-Xrs2 complex stability in addition to nuclease activity. Genetics. 2005;171:1561–1570. doi: 10.1534/genetics.105.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S, Froget B, Carr AM. Arrested replication fork processing: interplay between checkpoints and recombination. DNA Repair (Amst) 2007;6:1042–1061. doi: 10.1016/j.dnarep.2007.02.024. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LK, Storici F, Van Komen S, Calero S, Sung P, Resnick MA. Role of the nuclease activity of Saccharomyces cerevisiae Mre11 in repair of DNA double-strand breaks in mitotic cells. Genetics. 2004;166:1701–1713. doi: 10.1534/genetics.166.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Llorente B, Symington LS. The Mre11 nuclease is not required for 5' to 3' resection at multiple HO-induced double-strand breaks. Mol Cell Biol. 2004;24:9682–9694. doi: 10.1128/MCB.24.21.9682-9694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- Luo G, Yao MS, Bender CF, Mills M, Bladl AR, Bradley A, Petrini JH. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc Natl Acad Sci U S A. 1999;96:7376–7381. doi: 10.1073/pnas.96.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzoeva OK, Petrini JH. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol Cell Biol. 2001;21:281–288. doi: 10.1128/MCB.21.1.281-288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales M, Theunissen JW, Kim CF, Kitagawa R, Kastan MB, Petrini JH. The Rad50S allele promotes ATM-dependent DNA damage responses and suppresses ATM deficiency: implications for the Mre11 complex as a DNA damage sensor. Genes Dev. 2005;19:3043–3054. doi: 10.1101/gad.1373705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M. The 3' to 5' exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- Petermann E, Caldecott KW. Evidence that the ATR/Chk1 pathway maintains normal replication fork progression during unperturbed S phase. Cell cycle (Georgetown, Tex. 2006;5:2203–2209. doi: 10.4161/cc.5.19.3256. [DOI] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney S, Alt FW, Lombard D, Whitlow S, Eckersdorff M, Fleming J, Fugmann S, Ferguson DO, Schatz DG, Sekiguchi J. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. J Exp Med. 2003;197:553–565. doi: 10.1084/jem.20021891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- Stiff T, Reis C, Alderton GK, Woodbine L, O'Driscoll M, Jeggo PA. Nbs1 is required for ATR-dependent phosphorylation events. Embo J. 2005;24:199–208. doi: 10.1038/sj.emboj.7600504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Theunissen JW, Morales M, Petrini JH. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair (Amst) 2004;3:845–854. doi: 10.1016/j.dnarep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Trujillo KM, Yuan SS, Lee EY, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. Embo J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, et al. Mre11-DNA Structures and Mutants Reveal Key Mre11 Roles in MRN Complex DNA End Synapsis and Nuclease Processing. Cell. 2008 doi: 10.1016/j.cell.2008.08.017. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Tainer JA. A nanomachine for making ends meet: MRN is a flexing scaffold for the repair of DNA double-strand breaks. Mol Cell. 2005;19:724–726. doi: 10.1016/j.molcel.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Wilson TE, Grawunder U, Lieber MR. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature. 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- Wyman C, Kanaar R. DNA double-strand break repair: all's well that ends well. Annual review of genetics. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Weaver DT. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997;25:2985–2991. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- Zhong H, Bryson A, Eckersdorff M, Ferguson DO. Rad50 depletion impacts upon ATR-dependent DNA damage responses. Hum Mol Genet. 2005;14:2685–2693. doi: 10.1093/hmg/ddi302. [DOI] [PubMed] [Google Scholar]
- Zhu J, Petersen S, Tessarollo L, Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol. 2001;11:105–109. doi: 10.1016/s0960-9822(01)00019-7. [DOI] [PubMed] [Google Scholar]
- Zhu XD, Kuster B, Mann M, Petrini JH, de Lang T. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat Genet. 2000;25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.