Abstract
Resistance mutations to HCV NS3 protease inhibitors in <1% of the viral quasispecies may still allow >1000-fold viral load reductions upon treatment, consistent with their reported reduced replicative fitness in vitro. Recently, however, an R155K protease mutation was reported as the dominant quasispecies in a treatment-naïve individual, raising concerns about possible full drug resistance. To investigate the prevalence of dominant STAT-C resistance mutations in the population we analyzed HCV genome sequences from 507 treatment-naïve HCV genotype 1 infected patients from the US, Germany and Switzerland. Phylogenetic sequence analysis and viral load data were used to identify the possible spread of replication competent, drug resistant viral strains in the population and to infer the consequences of these mutations upon viral replication in vivo. Mutations described to confer resistance to the protease inhibitors Telaprevir, BILN2061, ITMN-191, SCH6 and Boceprevir, the NS5B polymerase inhibitor AG-021541, and to the NS4A antagonist ACH-806 were observed mostly as sporadic, unrelated cases, at frequencies between 0.3% and 2.8% in the population, including two patients with possible multi-drug resistance. Collectively, however, 8.6% of the genotype 1a and 1.4% of the genotype 1b infected patients carried at least one dominant resistance mutation. Viral loads were high in the majority of these patients, suggesting that drug resistant viral strains might achieve replication levels comparable to non-resistant viruses in vivo.
Conclusion: Naturally occurring dominant STAT-C resistance mutations are common in HCV genotype 1 infected treatment-naïve patients. Their influence on treatment outcome should further be characterized to evaluate possible benefits of drug resistance testing for individual tailoring of drug combinations when treatment options are limited due to previous non-response to peginterferon and ribavirin.
Keywords: NS3 protease inhibitor, NS5B polymerase inhibitor, Telaprevir, Boceprevir
Introduction
Hepatitis C virus (HCV) is a global health problem infecting more than 170 million persons worldwide (1). The current standard treatment with pegylated-interferon and ribavirin is complicated by frequent adverse reactions, and a sustained virological response can only be achieved in 50% of patients infected with the most prevalent genotype 1 (2). Mutation F415Y in NS5B has been detected in patients on ribavirin mono-therapy (3, 4) and similar to mutations G404S and E442G in NS5A (5) it has been found to confer ribavirin resistance in vitro (4). However the clinical significance is unclear since patients exhibiting these mutations still achieved sustained virological responses on combination therapy (3, 4). No single site in the HCV genome has consistently been found to be associated with resistance to interferon, most likely because this drug acts more indirectly through several antiviral pathways (6).
With the development of the HCV replicon system a new tool has become available for in vitro testing of drugs that specifically inhibit HCV proteins essential for viral replication. For this new class of specifically targeted antiviral therapies for Hepatitis C (STAT-C) several new compounds have been investigated in recent years, among these the NS3/4A protease inhibitors (PIs) Telaprevir (VX-950), Boceprevir (SCH503034), SCH6, BILN2061, and ITMN-191, the NS4A antagonist ACH-806 (7), along with two types of NS5B RNA-dependent RNA polymerase (RdRp) inhibitors, the nucleoside analogs Valopicitabine (NM283), JTK-109 and R1479, and the non-nucleosides HCV-796, A-837093, A-782759 and AG-021541, some of which have recently attracted attention with promising results in primary clinical trials (reviewed in (8)). However, the rapid selection of viral variants displaying drug resistant phenotypes has been observed in patients experiencing viral rebound during treatment as well as in replicon experiments. STAT-C resistant variants reported to date are summarized in Table 1.
Table 1. STAT-C resistance mutations described in the literature, and prevalence of these in 507 treatment-naïve HCV genotype 1a and 1b infected patients.
Consensus amino acids are numbered according to their position in NS3, NS4A or NS5B (column 2) or the full HCV genome (column 3) relative to the H77 HCV genotype 1a reference sequence. Amino acid substitutions selected during drug treatment in patients are listed separately from substitutions observed to confer drug resistance in replicon experiments or enzymatic assays. Drugs inducing resistance mutations and relevant references are given. Resistance mutations detected as the dominant viral sequence in treatment naïve patients in this study are listed on the right, and their proportion among genotype 1a or 1b infected patients is shown. (Other observed variants not described to confer resistance included A39T, Q41H and S489A/F [NS3], E442D [NS5A] and M423A [NS5B] in genotype 1a, and S489V [NS3], G404C/D/N [NS5A], C316N/S and M414I [NS5B] in genotype 1b.)
| HCV Protein | Drug resistance mutations described in the literature | Detected resistance mutations | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Residue and position | In vivo | In vitro | Drugs | References | Genotype 1a | Genotype 1b | ||||
| Protein | Genome | (N = 362) | % | (N = 145) | % | |||||
| NS3 | C 16 | C 1042 | S | ACH-806 | (7) | |||||
| V 36 | V 1062 | A, M, L, G | A, M, L, G | Telaprevir, Boceprevir | (11, 12, 19, 28) | 2 × M, 6 × L | 2.2 | |||
| A 39 | A 1065 | V | ACH-806 | (7) | 1 × V | 0.3 | ||||
| Q 41 | Q 1067 | R | ITMN-191, Boceprevir | (28, 29) | ||||||
| F 43 | F 1069 | S, C | ITMN-191, Boceprevir, Telaprevir | (28) | ||||||
| T 54 | T 1080 | A, S | A | Telaprevir, Boceprevir | (11–13, 19, 28) | 7 × S | 1.9 | 2 × S | 1.4 | |
| R 109 | R 1135 | K | SCH6 | (18) | ||||||
| S 138 | S 1164 | T | ITMN-191 | (29) | ||||||
| R 155 | R 1181 | K, T, I, M, G, L, S | K, T, Q | Telaprevir, BILN2061, ITMN-191, Boceprevir | (11–13, 28) | 3 × K | 0.8 | |||
| A 156 | A 1182 | S, T, V, I | S, T, V | Telaprevir, BILN2061, SCH6, Boceprevir, ITMN-191 | (11–13, 18, 28) | |||||
| D 168 | D 1194 | A, V, E | BILN2061, ITMN-191 | (13, 29) | 1 × E | 0.3 | ||||
| V 170 | V 1196 | A | Telaprevir, Boceprevir | (13, 28) | ||||||
| S 489 | S 1515 | L | ITMN-191 | (29) | ||||||
| NS4A | V 23 | V 1680 | A | ITMN-191 | (29) | 2 × A | 0.6 | |||
| NS5B | H 95 | H 2515 | Q | A-782759 | (16) | |||||
| S 96 | S 2516 | T | R1479 | (33) | ||||||
| S 282 | S 2702 | T | 2′C-methyl-ribonucleosides (NM283 and others) | (15, 33, 34) | ||||||
| C 316 | C 2736 | Y (in Chimpanzee) | Y | HCV-796, A-837093 | (35, 36) | |||||
| S 365 | S 2785 | T, A | HCV-796 | (36) | ||||||
| N 411 | N 2831 | S | A-782759 | (16) | ||||||
| M 414 | M 2834 | L, T | A-782759 | (16) | ||||||
| M 423 | M 2843 | T, V, I | AG-021541 | (27) | 2 × V, 8 × I | 2.8 | ||||
| Y 448 | Y 2868 | H, C (in Chimpanzee) | H | A-782759, A-837093 | (16, 35) | |||||
| P 495 | P 2915 | L, A | JTK-109 | (37) | ||||||
| G 554 | G 2974 | D, S (in Chimpanzee) | A-837093 | (35) | ||||||
| D 559 | D 2979 | G, S, N (in Chimpanzee) | A-837093 | (35) | ||||||
| Patients with mutations described in vivo, total | 18 | 5.0 | 2 | 1.4 | ||||||
| Patients with mutations described in vivo or in vitro, total | 31* | 8.6 | 2 | 1.4 | ||||||
Includes one patient who carries two mutations (V36M and V23A).
Due to the high replication rate and the error-prone nature of the viral RdRp, within a given host HCV circulates as a mixture of viral strains each differing slightly from one another, referred to as the viral quasispecies. Two recent reports suggested that resistant variants may already be present at frequencies <1% in the quasispecies population in treatment naïve patients (9, 10), consistent with their dominant emergence only days after treatment initiation (11, 12). However, drug treatment in the setting of resistance mutations may still be beneficial, since a decreased in vitro replicative capacity has been demonstrated for many viral strains resistant to protease or RdRp inhibitors (13–19). Patients may therefore benefit from elimination of the dominant, drug-susceptible viral quasispecies with more than 1000-fold reductions in their viral load, until only drug resistant but replication deficient quasispecies constitute the residual viral population (11, 12). Although in some cases replication levels may later be restored by compensatory mutations, it seems possible that this effect could suffice to achieve treatment success if several drugs were combined to suppress viral replication before compensatory mutations or additional resistance mutations can evolve. Despite the presumed impaired viral replication, however, the PI resistant mutation R155K was recently detected as the dominant quasispecies in a treatment naïve patient (20), raising concerns that drug resistance may not necessarily be associated with reduced viral fitness in the individual in vivo situation. Consequently, preliminary data indicate reduced early treatment responses in patients with ~100% R155K PI resistant quasispecies at baseline, indicating possible primary drug resistance (21).
Since this finding may have important implications for the response rates to new drugs in the population with a possible need for drug resistance screening, here we have investigated the prevalence of dominant mutations described to confer protease or polymerase inhibitor resistance in an international multi-center cohort of treatment-naïve HCV-infected patients. Phylogenetic sequence analysis and viral load data were used to identify the possible spread of replication competent, drug resistant viral strains in the population and to infer the possible consequence of these mutations upon replicative capacity in vivo.
Methods
Patients
Plasma samples were obtained from 507 patients who had been infected with HCV genotype 1a or 1b for at least one year and were naïve to anti-HCV treatment with interferon, ribavirin or any novel or investigational anti-HCV drug. Patients were recruited between 1989 and 2007 from the Massachusetts General Hospital, the Lemuel Shattuck Hospital, and the Fenway Community Health Center (Boston, MA, USA), the University of Tennessee Health Science Center (Memphis, Tennessee, USA), the Center for the Study of Hepatitis C, Weill Cornell Medical College (New York, NY, USA), the Hemophilia Growth and Development Study Cohort (USA) (22), the University of Colorado (Denver, CO, USA), the Swiss HCV Cohort Study (Switzerland) (23), and the Bonn University Hospital (Bonn, Germany). The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the Institutional Review Board at Massachusetts General Hospital. Written informed consent was obtained from each study participant.
Amplification and sequencing of full-length HCV genomes
Viral loads were determined using the Cobas HCV Amplicor Monitor test (Roche), the Cobas Taqman test (Roche), or Versant 2.0 or 3.0 bDNA assays (Bayer). Viral RNA was extracted from plasma samples using the vRNA extraction kit (Qiagen, Hilden, Germany). cDNA was amplified and sequenced using a nested RT-PCR approach as published elsewhere (24), with two additional first round primer pairs 13F_5′-CCTYGCCTACTATTCCATGG-3′, 13R_5′-TGAGCRCGYACRAAGTACGGC-3′ and 14F_5′-TCTAYGGCAARGCCATCCCC-3′, 14R_5′-AGGAGCTTGGACTGGAGCC-3′ to achieve better coverage in genotype 1b samples. At Massachusetts General Hospital 89 samples were sequenced to a final average coverage of 3-fold. For the remaining 418 samples products of the first round RT-PCR were transferred to the Broad Institute NIAID Microbial Sequencing Center. Here, 96 amplicons 500–800 nucleotides in length were generated for each sample and sequenced bidirectionally using an automated high-throughput viral sequencing platform comprising of ABI3730xl sequencers, resulting in an average sequence coverage of 8-fold for each genome.
Sequence analysis and phylogeny
At Massachusetts General Hospital, Sequencher (Gene Codes Corp., Ann Arbor, MI) was used to trim, align and edit sequences. At the Broad Institute, sequences were trimmed for quality and primer sequence, and contigs were assembled using the Broad Institute’s ARACHNE assembler (25) (https://www.broad.mit.edu/wga/). If mixed bases were detected as two different chromatogram peaks at the same residue, after evaluation of all overlapping fragments only the dominant base was called. Sequences with insertions or deletions compared to a reference were validated manually. Genbank accession numbers for all sequences are provided in Supplementary Table 1. Full-length nucleotide sequences were translated into amino acids and aligned using Se-Al v2.0a11 software (http://evolve.zoo.ox.ac.uk). Phylogenetic trees were constructed in ClustalX (26) using the neighbor joining method, and were visualized in FigTree v1.1.2 (http://tree.bio.ed.ac.uk) and Det. (http://research.microsoft.com/vibevis/det.).
Generation of clonal sequences
Procedures are described in the supplementary methods.
Results
Within a larger cohort study we sequenced the entire HCV proteome from 507 patients infected with HCV genotype 1 recruited from multiple clinical centers in the United States, Germany and Switzerland. Demographic characteristics are given in Table 2. To investigate the prevalence of described drug resistance mutations to established or investigational anti-HCV drugs in the population, we here analyzed dominant amino acid substitutions separately for the 362 genotype 1a and 145 genotype 1b infected patients.
Table 2. Demographic characteristics.
are listed summarized for all 507 patients (leftmost column), and separately for each of the contributing cohorts. Numbers and percentages in the first row summarize patients included from each cohort.
| Total | Bonn (GER) | Boston (MA,USA) | Denver (CO,USA) | HGDS (USA) | Memphis (TN,USA) | New York (NY,USA) | SCCS (CH) | ||
|---|---|---|---|---|---|---|---|---|---|
| N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | ||
| 507 (100) | 12 (2) | 150 (30) | 14 (3) | 62 (12) | 124 (24) | 61 (12) | 84 (17) | ||
|
| |||||||||
| HCV | 1a | 362 (71) | 9 (75) | 120 (80) | 12 (86) | 42 (68) | 95 (77) | 39 (64) | 45 (54) |
| Genotype | 1b | 145 (29) | 3 (25) | 30 (20) | 2 (14) | 20 (32) | 29 (23) | 22 (36) | 39 (46) |
|
| |||||||||
| HIV co-infection | 142 (28) | 3 (25) | 64 (43) | 0 (0) | 50 (81) | 0 (0) | 25 (41) | 0 (0) | |
|
| |||||||||
| Male sex | 310 (61) | 9 (75) | 98 (65) | 12 (86) | 62 (100) | 46 (37) | 41 (67) | 41 (49) | |
|
| |||||||||
| Ethnicity | African | 129 (25) | 0 (0) | 28 (19) | 2 (14) | 9 (15) | 75 (60) | 14 (23) | 2 (2) |
| Caucasian | 321 (63) | 12 (100) | 91 (61) | 12 (86) | 42 (16) | 49 (40) | 38 (62) | 79 (94) | |
| Hispanic | 36 (7) | 0 (0) | 16 (11) | 0 (0) | 10 (68) | 0 (0) | 8 (13) | 2 (2) | |
| Other1 | 21 (4) | 0 (0) | 15 (10) | 0 (0) | 1 (0) | 0 (0) | 1 (2) | 1 (1) | |
|
| |||||||||
| Transmission risk factor | IVDU | 195 (38) | 4 (33) | 76 (51) | 7 (50) | 0 (0) | 57 (46) | 20 (33) | 31 (37) |
| Blood products | 149 (29) | 2 (17) | 13(9) | 0 (0) | 62 (100) | 31 (25) | 16 (26) | 25 (30) | |
| Sexual | 29 (6) | 2 (17) | 11 (7) | 1 (7) | 0 (0) | 9 (7) | 4 (7) | 2 (2) | |
| Other2 | 39 (8) | 0 (0) | 8 (5) | 2 (14) | 0 (0) | 6 (5) | 4 (7) | 19 (23) | |
| Unknown | 95 (19) | 4 (33) | 42 (28) | 4 (29) | 0 (0) | 21 (17) | 17 (28) | 7 (8) | |
Other ethnicities include Asian, Native American and Capeverdian.
Other risk factors include: Tattoo, piercing, occupational infection or needlestick injury, nosocomial infection and intranasal cocaine use. HGDS = Hemophilia growth and development study, SCCS = Swiss HCV Cohort Study.
Prevalence of dominant mutations described to confer resistance to ribavirin, NS5B polymerase and NS3 protease inhibitors in a treatment naïve patient population
In searching for dominant drug resistant variants we first screened sequences for substitutions in positions G404 and E442 in NS5A and position F415 in NS5B, which have been associated with exposure to ribavirin (4, 5). Here, in the genotype 1a infected study population only one (0.3%) patient displayed the dominant F415Y substitution and one (0.3%) patient expressed substitution E442G. Similarly, only two (1.4%) patients with G404S substitutions and two (1.4%) patients exhibiting the E442G mutation were observed among the genotype 1b infected patients. Thus, dominant ribavirin-associated mutations appeared uncommon and therefore unlikely to explain frequent non-response to combination therapy.
Further examination of viral sequences in NS5B revealed that variation in positions previously associated with resistance to the RdRp inhibitors A-837093, A-782759, R1479, 2′-C-Methyl-Ribonucleosides and JTK-109 was similarly rare. With the exception of substitutions C316N/S and M414I in genotype 1b that have not been described as resistance variants, the residues H95, S96, S282, S365, N411, Y448, P495, G554 and D559 were 100% conserved in our patient population. In contrast, however, mutations M423V and M423I conferring a more than 44-fold decrease in susceptibility to the non-nucleoside polymerase inhibitor AG-021541 (27) were present in 10 (2.8%) of the patients with genotype 1a infection (Table 1), including one individual additionally carrying an F415Y mutation associated with ribavirin resistance.
In NS3, among the 362 genotype 1a infected patients we observed three (0.8%) individuals carrying the R155K mutation that exhibits moderate to high levels of drug resistance to the protease inhibitors Telaprevir, BILN2061, ITMN-191 and Boceprevir (11–13, 19, 28). Mutations V36L and V36M conferring low-level resistance were found in six (1.7%) and two (0.6%) patients, respectively, with mutation T54S observed in an additional 7 (1.9%) subjects (12, 13). Of note, none of our study subjects showed substitutions at position A156 that is known to confer the highest level of resistance to Telaprevir, BILN2061, Boceprevir or SCH6 (12, 17, 18), consistent with the reported high impact of these mutations on the replicative capacity in vitro (11–13, 18, 19, 28). Mutations D168E and V23A that confer reduced susceptibility to ITMN-191 (29) were found in one (0.3%) and two (0.6%) patients, respectively. Remarkably, in one of these individuals the V23A substitution was combined with mutation V36M that confers resistance to Telaprevir and Boceprevir (11, 12, 19, 28). Last, mutation A39V associated with resistance to the NS4A antagonist ACH-806 was observed in one (0.3%) patient. Beyond that, no variants associated with drug resistance in vitro were observed in positions Q41, F43, R109, S138, V170 and S489. In the 145 patients infected with genotype 1b, only the T54S substitution was detected in two (1.4%) individuals (Table 1).
Taken together, various described drug resistance mutations were detectable by bulk sequencing at individual frequencies between 0.3% and 2.8% in treatment-naïve HCV-infected patients. Two subjects each carried two amino acid substitutions conferring resistance to different drugs. Collectively, NS3 protease or NS5B polymerase inhibitor resistance mutations were prevalent at 8.6% (31/362) in genotype 1a and 1.4% (2/145) in genotype 1b infected patients. When the analysis was more conservatively restricted to resistance variants described to be selected during drug treatment in vivo, 5.0% of the genotype 1a isolates and 1.4% of the genotype 1b isolates harbored a resistance variant (Table 1). However, mutations in NS3 position A156 conferring >100-fold resistance to most protease inhibitors were undetectable by bulk population sequencing, consistent with their reported high impact on the replicative capacity in vitro.
Unimpaired in-vivo fitness of viral strains carrying resistance mutations
Since most protease and polymerase inhibitor resistance mutations described to date have been associated with reduced replicative capacity of varying degrees (11–19) it seemed remarkable to find these as dominant viral sequences in treatment naïve patients. Moreover, the emergence of double mutants in positions V36 and R155 has been reported in patients experiencing viral rebound during Telaprevir mono-therapy (11, 12), with double mutant viruses displaying enhanced replication (12) while also exhibiting increased drug resistance compared to single mutant viral strains (12, 28).
To confirm that amino acid substitutions detected in our bulk sequences indeed represented the dominant quasispecies in the patients, and to evaluate whether these were present in combinations with other drug resistance mutations at subdominant levels, we analyzed 153 clones from 11 patients generated from new, independent RT-PCR products spanning the NS3 protease catalytic domain. Here, for all sites of drug resistance the bulk sequence data was correctly representative of 100% of the viral quasispecies in 9/11 patients and of 13/14 and 14/15 clones in the remaining two patients, thus confirming the detected mutations as the predominant viral strain in each patient. Within the limited clonal depth applied here we did not, however, detect any additional combinations of drug resistance mutations on a subdominant level.
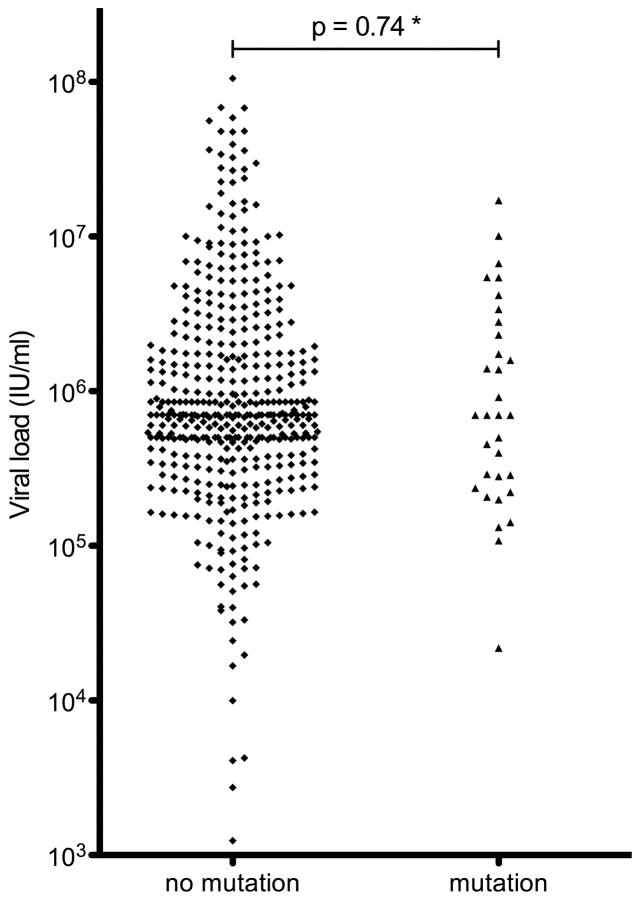
In examining viral replication levels in patients carrying dominant drug resistance mutations 19/33 (58%) of these displayed viral loads in the range of >500,000 − 17×106 IU/ml (Figure 1), including two patients with R155K substitutions and one patient with the V36M/V23A double mutant, indicating that drug resistant strains were not necessarily impaired in their ability to replicate in vivo.
Figure 1. Viral loads compared between patients with and without NS3 protease and NS5B polymerase inhibitor resistance mutations.
The estimated* p-value (Mann-Whitney-U test) shows no significant difference between the two groups, thus arguing against a strong negative impact of these mutations on viral replication in the individual viral strains in vivo, possibly influenced by yet undefined compensatory mutations. (* No mutation group: N = 474, 32 missing values, 65 semi-quantitative values >500,000–850,000 IU/ml above the detection limit of the test; Mutation group: N = 33, 1 missing value, 4 values >500,000 IU/ml).
Sporadic nature of naturally occurring drug resistance
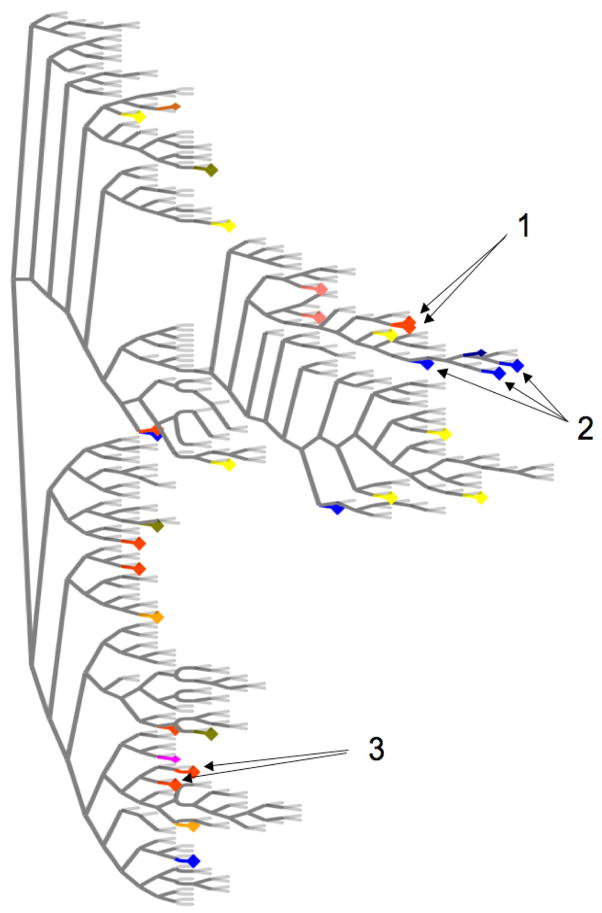
We next asked whether the presence of naturally occurring protease or polymerase inhibitor resistance mutations was influenced by transmission of resistant viral strains, high-risk behaviour, HIV co-infection, race, sex or geographic location. In examining relationships between amino acid sequences from patients with identical drug resistance mutations, only two viral strains carrying the M423I mutation resistant to the polymerase inhibitor AG-021541 clustered closely together in a phylogenetic analysis (Figure 2). Upon closer examination these differed in 50/3012 (1.7%) amino acid residues, arguing against a recent transmission event, but compatible with transmission or infection from a common source further in the past (24). Both samples came from intravenous drug users from the same small town in Switzerland and were drawn three years apart from each other, suggesting possible transmission and long-term stability of drug resistance mutations. More distant relationships were observed in two other clusters in the tree between sequences from two patients from Boston sharing the M423I mutation, and between sequences from three patients from different cities in Switzerland carrying the V36L mutation (Figure 2). These differed by 105–130 amino acids and were thus not suggestive of direct transmission. Beyond that no other viral strains sharing resistant variants displayed similar close relationships, with resistant mutants spread across the entire tree (Figure 2). Among patients carrying drug resistance mutations the proportions of transmission risk factors, HIV/HCV co-infection, male sex and ethnic groups were similar to those observed in the entire cohort, and drug resistance mutations were present in patients from all involved international HCV cohorts from the United States, Germany and Switzerland. Thus, while transmission of a resistance mutation seemed possible between two patients in our cohort, the majority of resistant variants represented sporadic cases rather than locally spread, closely related viral strains or mutants specifically selected within a certain subgroup of patients.
Figure 2. Phylogenetic tree comparing full length HCV amino acid sequences from 362 genotype 1a infected patients.
Viral strains carrying identical drug resistance mutations are marked with identical colors. Only two strains carrying the M423I mutation in NS5B are directly related (Arrows 1), whereas other mutations clustering together are more distant (Arrows 2 and 3), with the majority of drug resistance mutations spread across the entire tree, thus excluding a bias for resistance mutations due to closely related variants in a certain subpopulation of the cohort.
Discussion
In this study we analyzed the prevalence and distribution patterns of dominant drug resistance mutations in 507 treatment naïve HCV genotype 1 infected patients. Dominant mutations were mostly observed as sporadic, unrelated cases at frequencies between 0.3% and 2.8% in the population. Taken together, however, 8.6% of genotype 1a and 1.4% of genotype 1b infected patients exhibited at least one drug resistance mutation, including two cases with possible multi-drug resistance. Viral loads were high in the majority of patients carrying these mutations, suggesting that resistant viruses might achieve replicative capacities comparable to non-resistant strains in vivo.
Specifically targeted antiviral therapies for HCV (STAT-C) offer new opportunities to overcome limited response rates to interferon and ribavirin. Treatment success as measured by decreasing viral loads represents a balance between the achievable drug concentration in the plasma, the frequency and degree of drug resistance among the viral quasispecies (21), and the replication efficiency of drug resistant viral strains. Numerous studies have investigated the impact of amino acid substitutions on viral replication and drug susceptibility to investigational NS5B polymerase or NS3/4A protease inhibitors in the replicon model. Here, a pronounced reduction in the replicative capacity was described for the highly resistant A156 variants in NS3 and M423 variants in NS5B, and also to a lesser extent for the low-level PI resistant R155, T54 and V36 mutations (12, 13, 19, 27). It has been speculated that these lower replication levels could facilitate eradication through combination treatment (11), as STAT-C resistant viral strains appear to remain sensitive to interferon and ribavirin (19). However, the high viral loads observed in patients with dominant drug resistance mutations in our study demonstrate that viral fitness may be unimpaired in the individual in vivo situation, suggesting possible limitations of in vitro studies for the assessment of viral fitness. Possibly, as yetunknown secondary mutations may also be present which might compensate any replication defects. Unfortunately, the low number of individual resistance mutations in our study prevented the identification of candidate residues for compensatory mutations.
As far as comparative data on drug susceptibility are available, mutations observed in our study at residues M423 in NS5B (27), and in some cases R155 in NS3 (13, 17–19, 28) were described to confer high levels of drug resistance compared to wild-type virus in the replicon model, whereas amino acid substitutions at residues V36, T54 and D168 resulted in relatively moderate resistance (13, 19, 28). In a recent clinical trial the in vivo relevance of these findings was supported by the observation that weakly Telaprevir resistant variants arose predominantly in patients who only achieved lower plasma drug levels, whereas increasing drug concentrations selected for mutants that were also highly resistant in vitro (12). Although this suggests that viruses carrying low-level resistance mutations can be eradicated at high drug concentrations, the clinical impact of dominant amino acid substitutions observed in our study is unclear, since in vivo data published so far have originated from patients with resistance only in subdominant quasispecies. However, preliminary results indicate that dominant resistance mutations can potentially reduce the early treatment response to STAT-C drugs (21), and it must be assumed that low-level resistant strains will sustain viral replication in patients who do not achieve optimal drug levels with standard dosing, in cases with poor adherence, or when dose reductions are inevitable due to adverse events.
Importantly, continued viral replication in the presence of the selecting drug would put the patient at risk to develop additional resistance mutations. Here, observations from HIV infection suggest that baseline resistance even against only one drug in a multidrug antiviral regimen may jeopardize treatment success (12), and further indicate that apart from single mutations conferring high level resistance, stepwise accumulation of subtle but synergistically acting resistance mutations may also eventually lead to treatment failure (30). Data from a recent study using Telaprevir has indicated a similar pathway in HCV infections, where combination of the low-level resistance mutations V36M and R155K resulted in a highly drug resistant phenotype, the appearance of which coincided with viral breakthrough (11, 12). In our treatment naïve patients we did not detect any combinations of resistance mutations against the same drug, neither in the bulk sequences nor by clonal analysis. However, while the odds for simultaneous occurrence of two specific resistance mutations in the same viral strain has been calculated at 10−8 to 10−10, the chance to add a single mutation to a pre-existing, dominant quasispecies is at least 1,000-fold higher (19). In this context, our finding that up to 8.6% genotype 1a infected patients carry dominant low-level resistance mutations (including 3.0% with V36M/L or R155K) may be relevant, since these patients would be expected to have a significantly lower genetic barrier to developing full resistance during drug treatment (30). Therefore, a combination of drugs might be necessary to rapidly suppress viral replication before additional resistance mutations can evolve. Unfortunately, no combination therapy is currently available for peginterferon non-responders, the patient group that needs new treatment options most urgently. Novel STAT-C drugs as alternative treatment combinations appear to be prone to viral resistance mutations, likely due to their direct interaction with HCV proteins. Since many mutations in NS3 confer cross-resistance to several protease inhibitors, the options to combine drugs from this currently more advanced STAT-C class are limited. This problem might be overcome by the addition of polymerase inhibitors or NS4A antagonists. However, with M423I/V and A39V we have already identified dominant resistance mutations also to these classes of drugs in treatment naïve patients. In addition, combinations of mutations V36M/V23A or M423I/F415Y that confer resistance to the protease inhibitors Telaprevir and ITMN-191 or to the polymerase inhibitor AG-021541 and ribavirin indicate possible multi-drug resistance.
In HIV infection resistance testing was calculated to be cost-effective when the prevalence of drug resistance becomes >1% at baseline (31) and is currently recommended in areas with more than 5% prevalence of resistant strains, and in all cases of treatment failure (32). For novel STAT-C drugs important factors such as their cost and treatment response rates are not yet available to derive similar calculations, as is detailed information about the effectiveness of these therapies in the face of pre-existing resistance mutations. However, with a baseline frequency of drug resistance mutations between 5.0% and 8.6% in genotype 1a infected patients in our study, and possibly increasing rates due to transmission of resistant variants once new drugs are widely used, it seems possible that pre-treatment screening for baseline resistance mutations might be warranted to enable the individual tailoring of specific drug combinations for those patients whose treatment options are limited because of previous non-response to peginterferon and ribavirin.
Different rates of resistance mutations between HCV genotype 1a and 1b have previously been observed in patients on Telaprevir monotherapy (11, 12). Here, mutations V36M or R155K require only one nucleotide change from the genotype 1a consensus sequence, but two substitutions in genotype 1b, resulting in a relative genetic barrier to resistance (11). Different constraints on viral evolution due to different protein structures between viral subtypes may provide alternative explanations. Although clonal sequence analysis in a subset of patients demonstrated a high concordance with the bulk sequencing results it is also likely that our approach underestimates the number of patients with relevant proportions of resistance mutations in their quasispecies, which may have randomly skewed mutational frequencies towards genotype 1a. Finally, mutations T54S, R155K and M423I/V fall within the described CD8+ T-cell epitopes CINGVCWTV, HAVGLFRAA and ARMILMTHF, and it would be intriguing to speculate that viral evolution driven by escape from CD8+ T-cell pressures might differentially influence the prevalence of drug resistance mutations.
In summary, dominant mutations described to confer resistance to STAT-C drugs in vivo or in vitro can be observed as sporadic, unrelated cases in up to 8.6% of treatment-naïve HCV genotype 1 infected patients from North America and Europe. High viral loads in patients carrying drug resistance mutations suggest unimpaired viral replication, thus questioning previous speculations about maintained drug efficiency in the setting of pre-existing drug resistance mutations. These data provide a rationale to further characterize possible correlations between dominant STAT-C resistance mutations at baseline and treatment outcome, to evaluate a possible need for drug resistance testing and individual tailoring of drug combinations. Sequences published with this manuscript may be used as a baseline reference for cost-effectiveness calculations for current and future anti-HCV drugs.
Supplementary Material
Acknowledgments
We thank the Broad Institute’s Genome Sequencing Platform, Special Projects Group, Toby Bloom, Michael Fitzgerald, Keenan Ross, Heidi Spurling, Sampath Settipalli, Adam Brown and Lucia Alvarado for their contributions to building a high-throughput HCV sequencing and annotation pipeline. We also acknowledge Raymond Peterson and Queenie Brown, M.S. for assistance with data collection and sample procurement. We thank Caroline A. Riely for her work with the UTHSC (Memphis) HCV cooperative.
Members of the Swiss HCV Cohort Study (SCCS) are: Philippe Bürgisser (Lausanne), Gieri Cathomas (Liestal), Andreas Cerny (Lugano), Rolf Dubs (Zürich), Jean-François Dufour (Berne), Patrick Francioli (Lausanne), Meri Gorgievski (Berne), Antoine Hadengue (Geneva), Markus Heim (Basel), Beat Helbling (Zürich), Hans Hirsch (Basel), Laurent Kaiser (Geneva), Raffaele Malinverni (Neuchâtel), Gladys Martinetti (Bellinzona), Virginie Masserey Spicher (Berne), Christa Meyenberger (St. Gallen), Darius Moradpour (Lausanne), Beat Müllhaupt (Zürich), Francesco Negro (Geneva), Martin Rickenbach (Lausanne), Laura Rubbia-Brandt (Geneva), Detlev Schulze (St. Gallen)
Financial support
This project has been funded in whole or in part with funds from the Deutsche Forschungsgemeinschaft grant DFG KU2250/1-1 (TK), and with Federal funds from the National Institutes of Health grant RO1-AI067926-01 (TMA), U19-AI666345 (TMA, GML, BDW) and the National Institute of Allergy and Infectious Disease, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN266200400001C (BWB). The Swiss Hepatitis C Cohort Study (SCCS) is supported by grant No. 3347C0-108782/1 from the Swiss National Science Foundation, No. 03.0599 from the Swiss Federal Office for Education and Sciences, and No. LSHM-CT-2004-503359 from the European Commission. The SCCS is a member of the European Vigilance Network for the Management of Antiviral Drug Resistance (VIRGIL Project). The Hemophilia Growth and Development Study was funded by the National Institute of Child Health and Human Development 1, R01 HD41224. The Hepatitis C Cooperative at the University of Tennessee Health Science Center is supported by NIH grants AI048216 and AI066316. The Center for the Study of Hepatitits C at Weill Cornell Medical College NY was supported by the Greenberg Medical Research Foundation (IMJ, GLB, BRE, AHT) and by funds received through a commitment made at the Clinton Global Initiative (AHT).
Abbreviations
- PI
Protease inhibitor
- HCV
Hepatitis C Virus
- RdRp
RNA dependent RNA polymerase inhibitor
- STAT-C
Specifically targeted antiviral therapies for HCV
References
- 1.Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med. 2001;345:41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 2.Hadziyannis SJ, Sette H, Jr, Morgan TR, Balan V, Diago M, Marcellin P, Ramadori G, et al. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann Intern Med. 2004;140:346–355. doi: 10.7326/0003-4819-140-5-200403020-00010. [DOI] [PubMed] [Google Scholar]
- 3.Lutchman G, Danehower S, Song BC, Liang TJ, Hoofnagle JH, Thomson M, Ghany MG. Mutation rate of the hepatitis C virus NS5B in patients undergoing treatment with ribavirin monotherapy. Gastroenterology. 2007;132:1757–1766. doi: 10.1053/j.gastro.2007.03.035. [DOI] [PubMed] [Google Scholar]
- 4.Young KC, Lindsay KL, Lee KJ, Liu WC, He JW, Milstein SL, Lai MM. Identification of a ribavirin-resistant NS5B mutation of hepatitis C virus during ribavirin monotherapy. Hepatology. 2003;38:869–878. doi: 10.1053/jhep.2003.50445. [DOI] [PubMed] [Google Scholar]
- 5.Pfeiffer JK, Kirkegaard K. Ribavirin resistance in hepatitis C virus replicon-containing cell lines conferred by changes in the cell line or mutations in the replicon RNA. J Virol. 2005;79:2346–2355. doi: 10.1128/JVI.79.4.2346-2355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wohnsland A, Hofmann WP, Sarrazin C. Viral determinants of resistance to treatment in patients with hepatitis C. Clin Microbiol Rev. 2007;20:23–38. doi: 10.1128/CMR.00010-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang W, Zhao Y, Fabrycki J, Hou X, Nie X, Sanchez A, Phadke A, et al. Selection of replicon variants resistant to ACH-806, a novel hepatitis C virus inhibitor with no cross-resistance to NS3 protease and NS5B polymerase inhibitors. Antimicrob Agents Chemother. 2008;52:2043–2052. doi: 10.1128/AAC.01548-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cholongitas E, Papatheodoridis GV. Review article: novel therapeutic options for chronic hepatitis c. Aliment Pharmacol Ther. 2008 doi: 10.1111/j.1365-2036.2008.03644.x. [DOI] [PubMed] [Google Scholar]
- 9.Cubero M, Esteban JI, Otero T, Sauleda S, Bes M, Esteban R, Guardia J, et al. Naturally occurring NS3-protease-inhibitor resistant mutant A156T in the liver of an untreated chronic hepatitis C patient. Virology. 2008;370:237–245. doi: 10.1016/j.virol.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Lu L, Mo H, Pilot-Matias TJ, Molla A. Evolution of resistant M414T mutants among hepatitis C virus replicon cells treated with polymerase inhibitor A-782759. Antimicrob Agents Chemother. 2007;51:1889–1896. doi: 10.1128/AAC.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kieffer TL, Sarrazin C, Miller JS, Welker MW, Forestier N, Reesink HW, Kwong AD, et al. Telaprevir and pegylated interferon-alpha-2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology. 2007;46:631–639. doi: 10.1002/hep.21781. [DOI] [PubMed] [Google Scholar]
- 12.Sarrazin C, Kieffer TL, Bartels D, Hanzelka B, Muh U, Welker M, Wincheringer D, et al. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology. 2007;132:1767–1777. doi: 10.1053/j.gastro.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 13.He Y, King MS, Kempf DJ, Lu L, Lim HB, Krishnan P, Kati W, et al. Relative replication capacity and selective advantage profiles of PI-resistant HCV NS3 protease mutants in the HCV genotype 1b replicon system. Antimicrob Agents Chemother. 2007 doi: 10.1128/AAC.01149-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu L, Pilot-Matias TJ, Stewart KD, Randolph JT, Pithawalla R, He W, Huang PP, et al. Mutations conferring resistance to a potent hepatitis C virus serine protease inhibitor in vitro. Antimicrob Agents Chemother. 2004;48:2260–2266. doi: 10.1128/AAC.48.6.2260-2266.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludmerer SW, Graham DJ, Boots E, Murray EM, Simcoe A, Markel EJ, Grobler JA, et al. Replication fitness and NS5B drug sensitivity of diverse hepatitis C virus isolates characterized by using a transient replication assay. Antimicrob Agents Chemother. 2005;49:2059–2069. doi: 10.1128/AAC.49.5.2059-2069.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mo H, Lu L, Pilot-Matias T, Pithawalla R, Mondal R, Masse S, Dekhtyar T, et al. Mutations conferring resistance to a hepatitis C virus (HCV) RNA-dependent RNA polymerase inhibitor alone or in combination with an HCV serine protease inhibitor in vitro. Antimicrob Agents Chemother. 2005;49:4305–4314. doi: 10.1128/AAC.49.10.4305-4314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong X, Chase R, Skelton A, Chen T, Wright-Minogue J, Malcolm BA. Identification and analysis of fitness of resistance mutations against the HCV protease inhibitor SCH 503034. Antiviral Res. 2006;70:28–38. doi: 10.1016/j.antiviral.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Yi M, Tong X, Skelton A, Chase R, Chen T, Prongay A, Bogen SL, et al. Mutations conferring resistance to SCH6, a novel hepatitis C virus NS3/4A protease inhibitor. Reduced RNA replication fitness and partial rescue by second-site mutations. J Biol Chem. 2006;281:8205–8215. doi: 10.1074/jbc.M510246200. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Y, Bartels DJ, Hanzelka BL, Muh U, Wei Y, Chu HM, Tigges AM, et al. Phenotypic Characterization of Resistant Val36 Variants of Hepatitis C Virus NS3-4A Serine Protease. Antimicrob Agents Chemother. 2008;52:110–120. doi: 10.1128/AAC.00863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colson P, Brouk N, Lembo F, Castellani P, Tamalet C, Gerolami R. Natural presence of substitution R155K within hepatitis C virus NS3 protease from a treatment-naive chronically infected patient. Hepatology. 2008;47:766–767. doi: 10.1002/hep.22122. [DOI] [PubMed] [Google Scholar]
- 21.Bartels DJ, Zhou Y, Zhang E, Marcial M, Byrn RA, Adiwijaya B, Lin C, et al. Natural prevalence of HCV variants with decreased susceptibility to NS3-4A protease inhibitors in treatment-naive subjects. J Hepatol. 2008;48(Supp 2):S316. doi: 10.1086/591141. [DOI] [PubMed] [Google Scholar]
- 22.Hilgartner MW, Donfield SM, Willoughby A, Contant CF, Jr, Evatt BL, Gomperts ED, Hoots WK, et al. Hemophilia growth and development study. Design, methods, and entry data. Am J Pediatr Hematol Oncol. 1993;15:208–218. doi: 10.1097/00043426-199305000-00009. [DOI] [PubMed] [Google Scholar]
- 23.Prasad L, Spicher VM, Zwahlen M, Rickenbach M, Helbling B, Negro F. Cohort Profile: the Swiss Hepatitis C Cohort Study (SCCS) Int J Epidemiol. 2007;36:731–737. doi: 10.1093/ije/dym096. [DOI] [PubMed] [Google Scholar]
- 24.Kuntzen T, Timm J, Berical A, Lewis-Ximenez LL, Jones A, Nolan B, Schulze zur Wiesch J, et al. Viral sequence evolution in acute hepatitis C virus infection. J Virol. 2007;81:11658–11668. doi: 10.1128/JVI.00995-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaffe DB, Butler J, Gnerre S, Mauceli E, Lindblad-Toh K, Mesirov JP, Zody MC, et al. Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res. 2003;13:91–96. doi: 10.1101/gr.828403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi ST, Herlihy KJ, Graham JP, Fuhrman SA, Doan C, Parge H, Hickey M, et al. In Vitro Resistance Study of AG-021541, a Novel Nonnucleoside Inhibitor of the Hepatitis C Virus RNA-Dependent RNA Polymerase. Antimicrob Agents Chemother. 2008;52:675–683. doi: 10.1128/AAC.00834-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tong X, Bogen S, Chase R, Girijavallabhan V, Guo Z, Njoroge FG, Prongay A, et al. Characterization of resistance mutations against HCV ketoamide protease inhibitors. Antiviral Res. 2008;77:177–185. doi: 10.1016/j.antiviral.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Seiwert S. Sequence variation of NS3 and NS4A in hepatitis C virus (HCV) replicons following exposure to ITMN-191 concentrations likely to encompass those achieved in human liver following clinical dosing. First international workshop in hepatitis C resistance and new compounds; Boston, MA, USA. 2006; 2006. [Google Scholar]
- 30.Kuritzkes DR. Preventing and managing antiretroviral drug resistance. AIDS Patient Care STDS. 2004;18:259–273. doi: 10.1089/108729104323076007. [DOI] [PubMed] [Google Scholar]
- 31.Sax PE, Islam R, Walensky RP, Losina E, Weinstein MC, Goldie SJ, Sadownik SN, et al. Should resistance testing be performed for treatment-naive HIV-infected patients? A cost-effectiveness analysis. Clin Infect Dis. 2005;41:1316–1323. doi: 10.1086/496984. [DOI] [PubMed] [Google Scholar]
- 32.Hirsch MS, Brun-Vezinet F, Clotet B, Conway B, Kuritzkes DR, D’Aquila RT, Demeter LM, et al. Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society-USA Panel. Clin Infect Dis. 2003;37:113–128. doi: 10.1086/375597. [DOI] [PubMed] [Google Scholar]
- 33.Le Pogam S, Jiang WR, Leveque V, Rajyaguru S, Ma H, Kang H, Jiang S, et al. In vitro selected Con1 subgenomic replicons resistant to 2′-C-methyl-cytidine or to R1479 show lack of cross resistance. Virology. 2006;351:349–359. doi: 10.1016/j.virol.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 34.Migliaccio G, Tomassini JE, Carroll SS, Tomei L, Altamura S, Bhat B, Bartholomew L, et al. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J Biol Chem. 2003;278:49164–49170. doi: 10.1074/jbc.M305041200. [DOI] [PubMed] [Google Scholar]
- 35.Chen CM, He Y, Lu L, Lim HB, Tripathi RL, Middleton T, Hernandez LE, et al. Activity of a potent hepatitis C virus polymerase inhibitor in the chimpanzee model. Antimicrob Agents Chemother. 2007;51:4290–4296. doi: 10.1128/AAC.00723-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCown MF, Rajyaguru S, Le Pogam S, Ali S, Jiang WR, Kang H, Symons J, et al. The HCV replicon presents a higher barrier to resistance to nucleoside analogs than to non-nucleoside polymerase or protease inhibitors. Antimicrob Agents Chemother. 2008 doi: 10.1128/AAC.01317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomei L, Altamura S, Bartholomew L, Biroccio A, Ceccacci A, Pacini L, Narjes F, et al. Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J Virol. 2003;77:13225–13231. doi: 10.1128/JVI.77.24.13225-13231.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.