Abstract
The enumeration of rare circulating epithelial cells (CEpCs) in the peripheral blood of metastatic cancer patients has shown promise for improved cancer prognosis. Moving beyond enumeration, molecular analysis of CEpCs may provide candidate surrogate endpoints to diagnose, treat, and monitor malignancy directly from the blood samples. Thorough molecular analysis of CEpCs requires the development of new sample preparation methods that yield easily accessible and purified CEpCs for downstream biochemical assays. Here, we describe a new immunomagnetic cell separator, the MagSweeper, which gently enriches target cells and eliminates cells that are not bound to magnetic particles. The isolated cells are easily accessible and can be extracted individually based on their physical characteristics to deplete any cells nonspecifically bound to beads. We have shown that our device can process 9 mL of blood per hour and captures >50% of CEpCs as measured in spiking experiments. We have shown that the separation process does not perturb the gene expression of rare cells. To determine the efficiency of our platform in isolating CEpCs from patients, we have isolated CEpCs from all 47 tubes of 9-mL blood samples collected from 17 women with metastatic breast cancer. In contrast, we could not find any circulating epithelial cells in samples from 5 healthy donors. The isolated CEpCs are all stored individually for further molecular analysis.
Keywords: enrichment of rare cells, magnetic separation, circulating tumor cells
Molecular profiling of rare cells is important in biological and clinical studies. Applications range from characterization of circulating epithelial cells (CEpCs) in the peripheral blood of cancer patients for disease prognosis and personalized treatment (1–7); circulating fetal cells in maternal blood for prenatal diagnosis (8–10); and antigen-specific lymphocytes for immune monitoring (11). The deconvolution of profiling data to extract the relevant biology of rare cells from the mixture of blood leukocytes is challenging, and in most cases impractical (12, 13). Efficient enrichment of these cells of interest is critical before characterization; otherwise, leukocyte contamination would overwhelm any subsequent molecular analyses of rare cells.
Purification of rare cells, defined as cells that comprise <0.01% of a heterogeneous population, presents a technical challenge. Commercially available platforms for isolating rare cells, such as CEpCs, including density-gradient separation of mononucleated cells (14, 15), size filtration (16), and immunomagnetic-based isolation (17–19), can analyze large volumes of blood samples (10–100 mL/hr). Immunomagnetic platforms are currently the lead technology in clinical settings. The major drawback of commercial platforms for CEpC isolation is the lack of purification efficiency (<0.01–0.1%) (18–20). Other alternative methodologies include microfluidic platforms (7, 21) that can efficiently capture CEpCs but suffer from low throughput (<1–2 mL of blood per hour of analysis time) and lack individual cell accessibility for downstream molecular analysis.
Thorough genomic analysis and profiling are therefore dependent on strategies and technologies that improve rare cell sample preparation with high purity. To provide this purity, we have developed a new sample preparation technology, the MagSweeper. The MagSweeper is an automated immunomagnetic separation technology that gently enriches CEpCs by 108-fold from blood. Purified cells can then be individually selected for biochemical analysis. We show that the MagSweeper process keeps cell function intact and does not perturb rare cell gene expression. We also describe the predictive modeling of the capturing efficiency of our device as a function of sweeping parameters, which suggests that further optimization is possible.
Results
Development of the MagSweeper for Enrichment of Rare Cells with High Purity.
MagSweeper technology is based on immunomagnetic separation that gently purifies rare cells present in a mixed population. The final purity of isolated target cells in immunomagnetic-based separation devices depends first on the specificity of the antibodies used to select the desired cells and second on the amount of nonspecific cell capture (i.e., capture of cells not bound via the specific antibody–antigen interactions to magnetic beads). Nonspecific contamination can be from adsorption of background cells to the capturing device or their entrapment within the large excess of magnetic particles needed for labeling rare cells in large volumes.
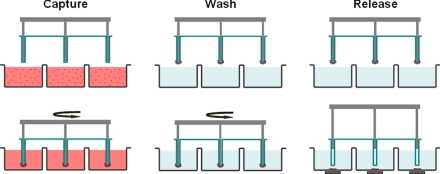
The functional portion of the MagSweeper is a round-bottom, neodymium, magnetic rod covered with an ultrathin (25-μm) nonadherent plastic sheath. This assembly is robotically swept through a well containing the labeled sample (Fig. 1). The plastic material has been optimized to reduce the nonspecific binding of contaminant cells. The rod is 6 mm in diameter with a magnetic flux density of 0.7 T at the rod end. The small rod minimizes the capturing area, and consequently the nonspecific adsorption of undesired cells; however, the magnetic energy gradient in the well also is constrained. To compensate for the reduced magnetic force, the sheathed magnet sweeps the entire well to maximize the capture efficiency of magnetically labeled cells. The sheathed rod is robotically driven to sweep through the well containing the sample in a pattern of overlapping concentric circular loops that cover the entire well area. We experimentally optimized the sweep velocity for (i) cell capture efficiency; (ii) application of sufficient shear force to detach adsorbed nonmagnetically labeled cells; and (iii) prevention of damage to the rare target cells.
Fig. 1.
Schematic of the MagSweeper process. The diluted blood samples, which are prelabeled with magnetic particles, are loaded into the capture wells. The magnetic rods covered with plastic sheaths are swept through the well in concentric circular loops at a level 1.5 mm above the bottom of the wells. After sweeping through the whole area of the capture wells, the sheathed magnets are washed in a circular loop to remove loosely bound contaminating cells. The rods are then immersed into a new buffer solution and disengage from the plastic covers. The external magnets located under the wells facilitate release of labeled cells and excess magnetic particles. Another round of capture-wash-release is performed to eliminate the majority of remaining contaminant cells entrapped within excess magnetic particles.
The rod captures labeled cells and then moves to a wash station containing fresh buffer solution to remove contaminating, unlabeled, and adherent cells. After washing, the cells held by the rod–sheath assembly are moved to a release well. There, the magnetic rod disengages from the plastic sleeve, and an external magnetic field is applied under the well to facilitate release of labeled cells and excess magnetic particles; any unlabeled contaminating cells entrapped among the aggregated magnetic particles also are released. After reengaging the rod into the plastic sheath, the just-released labeled cells are then recaptured with orders of magnitude fewer entrapped contaminating cells. Sequential rounds of capture-wash-release-recapture eliminate background cells that are not specifically labeled with magnetic particles, although at some cost to the capture efficiency.
Simulation of the MagSweeper.
Fundamentally, cell capture occurs when the magnetic force exerted on a labeled cell, Fm, is sufficient to draw the cell to the surface of the magnet. The magnetic force is opposed by a viscous drag force that results from the cell's motion relative to that of the surrounding fluid. As the magnet sweeping speed is increased, the region of cell capture around the magnet (i.e., the cross-sectional capture area) will shrink. The details of cell capture in the MagSweeper system are complex, featuring orbital magnet motion, circular fluid flow, nonuniform magnetic field gradients, and variable particle susceptibility. Thus, to lend insight toward the optimization of the system, we modeled the MagSweeper system to calculate cell trajectories and yield the capture zones.
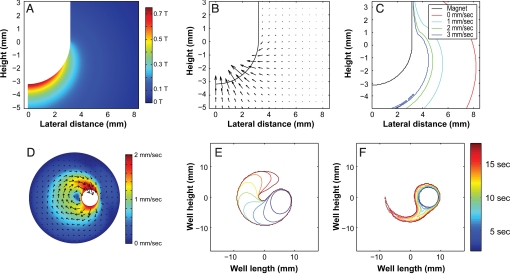
Fig. 2 A and B show the magnetic flux density and the magnetic force-driven cell velocity field, respectively, at the immersed end of the magnet. The velocity field was calculated assuming a stationary magnet and a 10-μm-diameter cell labeled with a single 4.5-μm-diameter magnetic bead. Cell velocities are only appreciable near the rounded tip of the magnet, and they drop off rapidly away from the magnet. This emphasizes the importance of employing thin sleeves to maximize the magnetic force applied to the magnetic particles and, consequently, the overall capture rate. To create a simple model of the MagSweeper, we add a constant and linear sweeping velocity to the magnetic field-driven cell velocity (Fig. 2B) and calculate cell trajectories. Fig. 2C plots the cell capture boundary orthogonal to the direction of motion as a function of magnet velocity. The figures illustrate the key tradeoff between magnet velocity and cross-sectional capture area. Another important observation is the significant decrease in capture area in the region of low magnetic field near the fluid surface.
Fig. 2.
Numerical simulations for assessing cell transport and capture. (A) The magnetic flux density magnitude along a radial slice of the magnet immersed in the solution (−5 mm < z < 3 mm). Note the peak magnetic flux density is 0.75 T, which exceeds the specified surface field for the blunt-tip magnet by 0.25 T. Thus, the rounded tip both increases the surface field and the surface area for capture relative to the blunt-tip magnet. (B) The resulting magnetic velocity field for 10-μm-diameter cells labeled with a single 4.5-μm-diameter superparamagnetic particle. For scale, the maximum plotted velocity vector is 7.8 mm/sec (at r = 2 mm, z = −2.5 mm). Note, the true maximum occurs adjacent to the magnet surface at the tip (≈12 mm/sec). (C) Lateral trapping boundaries for an idealized fluid-porous magnet as a function of magnet velocity. We see that particles at the bottom of the well are captured at a distance of 4 mm at 1 mm/sec, but only 2 mm at 3 mm/sec (note that the 0 mm/sec case assumes a capture time of 20 sec). (D) Fluid velocity within the well calculated for a magnet velocity of 2 mm/sec and orbit radius of 6 mm. The vectors indicate the instantaneous flow velocity in the xy-plane near the top of the curved section of the magnet (z = −0.2 mm), and the color gives the flow magnitude. The fluid velocity field was used to calculate particle trajectories and capture times. (E) Cell-trapping profiles in the xy-plane located beneath the magnet near the bottom of the well (z = −4.8 mm). (F) Cell-trapping profiles at the fluid surface (z = 3.5 mm).
In actual practice, the magnet follows a circular path and induces fluid flow within the well (Fig. 2D). Fluid in the path of the magnet experiences a pressure force and is pushed away from the magnet. Fluid adjacent to the magnet encounters a shear force, which imparts momentum to the fluid, thus creating a wake where the fluid follows the magnet. The resulting 3D fluid motion has a significant impact on the particle capture. For the plane beneath the magnet, the capture zone resembles the zone from the linear model, except with a curved path (Fig. 2E). Note that there is a gap in the annular capture zone, even at the completion of a full orbit. This is a consequence of the fluid motion, which pushes particles ahead of the magnet. At the higher z-planes, where magnetic fields are weak, the capture area is reduced >4-fold (Fig. 2F). Here, particles mostly travel around the magnet through the wake without being captured.
Enrichment of Rare Immunological Model Cells from a Mixed Sample.
To study the performance of MagSweeper in isolating rare cells from a mixed population, we spiked 50 HLA-A2-positive human peripheral blood mononuclear cells (PBMCs) as target cells into a solution containing differing amounts of HLA-A2-negative PBMCs. The target cells (HLA-A2) were labeled with 4.5-μm magnetic beads functionalized with an anti-HLA-A2 antibody. The capture rate and purity of the targeted cells isolated by MagSweeper after 2 rounds of capture-wash-release are shown in Fig. 3. The capture efficiency of target cells by the MagSweeper was ≈60%, regardless of the number of background cells. The purity of isolated HLA-A2 cells was 100% until the background cells were in excess of 2 × 105 and was 89% ± 2% (mean ± SD) when the number of background cells was 2 × 106 (Fig. 3). In addition, these data indicate the enrichment of target cells by 2.5 × 105-fold when the background cells are as high as 2 × 107.
Fig. 3.
The capturing efficiency and purity of HLA-A2 target cells from a solution containing different amounts of background cells. A total of 50 target cells were mixed with 20,000, 200,000, 2 million, and 20 million background cells. Error bars show 1 SD (n = 3). For ease of detection, the target HLA-A2 cells and background cells are stained with green and red fluorescent dyes, respectively.
After visual inspection of the captured cells under fluorescent microscopy, we found that the majority of contaminant cells were attached to magnetic beads. This indicates that the purity is limited by the antibody specificity or reagent quality (nonspecific sticking of background cells to the beads) per our experimental observation.
Enrichment of MCF7 Cancer Cells Spiked in Blood Samples.
To test the performance of the MagSweeper in isolating CEpCs, first we spiked 50 stained cancer cells from the human breast cancer cell line MCF7 into 1 mL of peripheral blood drawn from healthy human volunteers. For ease of detection, the target MCF7 cells and background blood cells were stained with SNARF-1 (Invitrogen) and CFDA SE (Invitrogen) fluorescent dyes, respectively, according to the manufacturer's recommendation. The MCF7 cells were labeled within the blood with 4.5-μm paramagnetic beads functionalized with antibodies against the epithelial cell adhesion molecule (EpCAM), which is expressed on the cell membrane of epithelial cells but not on leukocytes or red blood cells (22–24). The 4.5-μm magnetic beads permit isolation of target epithelial cells, even with only 1 bead attached to the cells, which makes the procedure suitable for isolating CEpCs with moderate to low EpCAM expression.
We found the capturing efficiency of MCF7 cells by MagSweeper to be 62% ± 7%, with a purity of 51% ± 18%, when the separation and labeling process is carried out at 4 °C. Enriched MCF7 cells were individually accessible and could be further manipulated based on their physical and morphological features to increase the purity. To test the efficiency of this protocol, we spiked up to 50 GFP-expressing MCF7 cells into 3 mL of peripheral blood drawn from healthy human volunteers. The MCF7 cells were labeled with anti-EpCAM beads and enriched with MagSweeper. MCF7 cells then were extracted by pipetting all captured cells with a diameter larger than 20 μm (including the magnetic beads). After this step, all of the extracted cells were confirmed for their green fluorescent emission of GFP (Fig. S1). In replicate experiments (n = 9), the combination of MagSweeper enrichment with visual inspection and extraction of cells based on their size purified the MCF7 cells to 100% purity, with a cell capture rate of 59% ± 27%. As a control, we repeated the same protocol on blood samples from 5 healthy volunteers that contained no spiked cancer cells. No candidate cells were found for extraction in any of the healthy controls.
Perturbation of Cellular Gene Expression as a Function of MagSweeper Process.
To assess whether the MagSweeper protocol perturbs the gene expression profile of the CEpCs during the isolation process, we interrogated the genome-wide RNA expression profile of MCF7 cells by using microarray analysis. The expression profiling of 20,000 MCF7 cells grown in culture media was compared with a similar number of MCF7 cells incubated for 30 minutes with anti-EpCAM magnetic beads before and after MagSweeper isolation. We chose 20,000 MCF7 cells as starting materials for our analysis because all steps from RNA isolation, whole-genome amplification of extracted RNA, and microarray hybridization are well standardized and induce minimal technical variation in the sample processing for this number of cells.
The experiment with these 3 conditions was repeated 3 times. The Pearson correlation coefficients between arrays were all >0.98 Table S1), with very similar intragroup correlation (0.9851 ± 0.0035) and cross-correlation of samples (0.9849 ± 0.0047), suggesting high consistency between the conditions. In addition, coefficients of variation of gene expression between cultured cells and after MagSweeper isolation were comparable, suggesting little evidence of increased variation due to isolation. Moreover, gene expression fold changes between cultured cells and MagSweeper were analyzed. A total of 42% of probe sets had changes <10%, another 35% had changes 10–25%, and an additional 17% had between 25% and 50%. Thus, 94% of probe sets had changes <50% (Fig. S2). Only 289 of 54,675 probe sets, corresponding to 174 genes, had a change between 2- and 4-fold. Statistical analysis of gene expression between the culture cells and MagSweeper reveals that none of the changes are significant at a 5% false-discovery rate, indicating that the MagSweeper isolation protocol does not induce any significant perturbation in the gene expression profile of the cells during the isolation process.
Enrichment of CEpCs from Breast Cancer Patients.
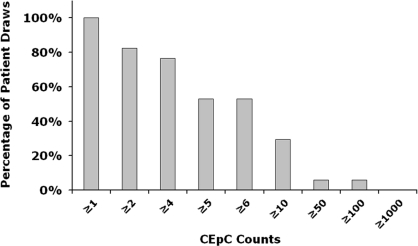
To determine the efficiency of MagSweeper technology in isolating CEpCs from patients with epithelial cancers, we obtained blood samples from 17 women with metastatic breast cancer on one or more occasions and 5 healthy volunteers with informed consent. All patients were under treatment at the time of blood draws. In total, we analyzed 52 tubes of 9-mL blood samples (47 from cancer patients and 5 from healthy donors). We found CEpCs in 47 of 47 samples derived from patients with metastatic disease, which contained 12 ± 23 CEpCs per 9 mL of blood. In contrast, we did not identify any circulating epithelial cells in any samples from healthy donors. The complementary cumulative distribution of isolated CEpC counts is shown in Fig. 4. The number of CEpCs isolated from each patient is detailed in Table S2. These cells were then frozen for subsequent molecular characterization.
Fig. 4.
Complementary cumulative distribution of the number of CEpCs purified by MagSweeper for each patient draw sample. The CEpC counts are reported for a tube of blood sample (9 mL).
Discussion
Since the beginning of 20th century, it has been the dream of generations of cancer researchers to diagnose malignancy from the peripheral blood, especially at an early stage. Despite tremendous efforts, we have not yet achieved that dream. In the age of targeted therapies, treatment selection for individualized therapy has become an important goal for cancer care. Circulating tumor epithelial cells circulate in the peripheral blood and may represent intermediate cells between primary tumors and metastases. Showing promise for improved cancer prognosis, CEpC numbers are reported to better correlate with cancer progression than imaging, lymph node biopsy, and serum bio marker analysis (3, 25). In addition to CEpC enumeration, successful molecular analysis of CEpCs—including detecting DNA aberrations, profiling gene expression, or determining degree of phenotype heterogeneity of the cells—should provide candidate surrogate endpoints for evaluation in clinical trials.
Molecular analysis of rare CEpCs in heterogeneous blood samples requires a sample preparation step to enrich the cells of interest among other nucleated background cells. Commercially available platforms can generally achieve just a 104-fold to 105-fold enrichment, which is not sufficient and leaves the isolated CEpCs at only 0.01–0.1% purity (18–20). Here, we report a device and protocol for magnetic isolation of CEpCs that achieves the necessary purity. First, we have shown the performance of the MagSweeper in isolating rare target cells in a model system (Fig. 3). Then, we have shown that the MagSweeper enriches MCF7 cancer cells spiked into blood by 108-fold by eliminating all cells that are not bound to anti-EpCAM beads. Further, the isolated target cells can be extracted individually based on their physical properties to eliminate cells nonspecifically bound to beads, if indeed there are any. The ability to individually extract CEpCs provides opportunities for the detailed characterization of single cells for subpopulation studies on heterogeneous CEpCs.
Using the presented protocol, we have demonstrated the efficient isolation of breast CEpCs from blood cells of metastatic cancer patients to a very high degree of purity. Beyond labeling with antibody-coated magnetic beads, no sample processing of blood is required before MagSweeper use, which decreases operator hands-on time and risk for perturbing the CEpCs. We have shown that MagSweeper can process 9 mL of blood per hour with 3–5 minutes total hands-on time, and can capture >50% of CEpCs as measured in spiking experiments without any significant changes in the gene expression of the captured cells. Another advantage of MagSweeper is the flexibility in the starting sample volume and process scalability. The throughput of the device can be increased by sweeping an array of sheathed magnetic rods through multiple samples in parallel with a single motion-controlled system. We have also successfully purified 1 million to 1.5 million neutrophils/mL from whole blood, which suggests that the MagSweeper is capable of purifying cells across a wide dynamic range of cell numbers.
Our modeling of the MagSweeper capturing process suggests opportunities for further optimization of the device. In particular, we have shown that in regions of high magnetic field gradient (e.g., near the rounded tip of the magnet), labeled cells are efficiently captured, resulting in a large sweep area. As noted above, the main influence of the fluid motion in this region is the presence of a small gap of uncaptured particles left following a single orbit (Fig. 2E). A simple first optimization is the extension of the sweep to overlap a portion of the circumference (≈10%), thus capturing the remaining particles. We also have shown that the effect of fluid motion is strong in regions of low magnetic field gradient, leading to a significant reduction in capture cross-section (Fig. 2F). In retrospect, we note that our current protocol addresses this reduced cross-section with its small steps (1-mm) in orbit radius. To further optimize CEpC recovery, we intend to engineer the shape of the magnet to yield a more uniform magnetic field gradient with, consequently, a larger and more uniform capture region along the length of the magnet.
Methods
Cell Culture.
MCF7 breast cancer cell lines were purchased from American Type Culture Collection. Cells were cultured in Dulbecco modified Eagle medium high glucose supplemented with 10% FBS and 100 U/mL penicillin–streptomycin (Invitrogen Corp.). The cells were grown at 37 °C and 5% CO2 in a humidified atmosphere.
Labeling and Processing MCF7 Cancer Cell Line.
Cells were trypsinized from their plates and suspended in media. Cells were then centrifuged at 300 × g for 10 minutes at room temperature, and media were removed. Approximately 1,000 cells were resuspended in 1 mL of PBS buffer (pH 7.4 with 0.1% BSA). A known number of cells in the presence of 10 μL of anti-EpCAM Dynabeads (CELLection Epithelial Enrich; Invitrogen, and Dynal) were then spiked into blood samples from healthy donors. Samples were incubated for 45–60 minutes with gentle mixing. After incubation, the blood samples were diluted to 9 mL each with PBS buffer (pH 7.4). Samples were then added to wells of a 6-well plate (Falcon and Becton Dickinson) and processed through the MagSweeper protocol.
Labeling and Processing HLA-A2 Model Cells.
To test the efficiency and purity of MagSweeper capture, human PBMCs expressing HLA-A2 were used as the model system. Both HLA-A2-positive and HLA-A2-negative human blood samples were obtained under an approved institutional review board from Stanford Blood Center. The samples were generally used without prior freezing, but on occasion frozen–thawed samples were used. Human leukocyte cells harvested from leukocyte reduction system filters were further purified by using standard Ficoll gradient techniques (Ficoll-Paque Plus; 17–1440-03; GE). HLA-A2-negative cells and positive cells were stained with SNARF-1 (Invitrogen) and CFDA SE (Invitrogen), respectively, according to the manufacturer's recommendation. Dynabeads goat anti-mouse IgG (Invitrogen) was conjugated to mouse anti-HLA-A2 antibodies (BD Biosciences) according to the manufacturer's protocol. Approximately 1,000 HLA-A2-positive cells were resuspended in 1 mL of PBS buffer (pH 7.4 with 0.1% BSA) and incubated with 10 μL of the functionalized Dynabeads for ≈60 minutes. HLA-A2-negative cells were used as background. Cell quantity was counted by using a hemacytometer. The mixtures were then added into the capture wells with 9 mL of PBS.
Labeling of CEpCs in Patient Samples.
Patients with known metastatic breast cancer or healthy normal controls gave consent before sample collection in accordance with Stanford's Human Subjects Research Compliance Board and HIPAA regulations. Blood was collected in 10 mL of BD Vacutainer plastic EDTA tubes (Becton Dickinson). Blood was collected by venipuncture or from implanted venous access ports. At least the first 9 mL from each blood draw was discarded to prevent contamination by skin epithelial cells from the needle puncture site. All blood samples were processed within 3 hr of collection. The blood was split into 3 samples, diluted to 6 mL each with PBS and labeled with 10 μL of CELLection Epithelial Enrich Dynabeads (Invitrogen), with constant mixing for 45–60 minutes. These samples were added to wells of a 6-well plate, brought up to 10 mL each with PBS, and then processed by the MagSweeper.
RNA Extraction and Microarray Analysis.
Total RNA was isolated and DNA removed by using a Qiagen RNeasey mini kit and protocol from samples of 20,000 MCF7 cells. The amount of RNA in the sample was quantified by using Nanodrop analysis (ThermoFisher Scientific). The quality of the RNA isolated was tested by using Agilent pico RNA chips and an Agilent 2100 machine and software (Agilent Technologies). The Agilent chip was prepared as required by the manufacturer, and 1 μL of the 20 μL of undiluted sample was loaded on the chip for analysis. Because of the limited amount of total RNA isolated from the 20,000 cells, total RNA was amplified to cDNA by using the Ribo-SPIA amplification system developed by NuGEN Technologies Inc.. Biotin-labeled amplified cDNA targets were generated starting from 5 ng of total RNA using the WT-OvationPicoRNA Amplification system (NuGEN Technologies Inc.). Five micrograms of the amplified materials was hybridized onto GeneChip Human Genome U133A v2 arrays (Affymetrix). The arrays were washed and scanned as recommended by the WT-Ovation PicoRNA Amplification system v1 System User Guide. Robust multichip average was used to calculate expression values of the arrays (26). Expression values were transformed to log2 for analysis. Differentially expressed genes between cultured cells and MagSweeper isolations were identified by t test, with control of multiple tests by false-discovery rate using q value.
Simulations and Computational Resources.
Numerical simulations of particle capturing were developed by using a finite element-based simulation package (COMSOL Multiphysics) and Matlab (Mathworks). The magnetic flux (B) and magnetic energy density (Um = H·B/2, where H is the magnetic field) were calculated in COMSOL by using the magnetostatic application mode and an axisymmetric model of the full magnet within a large domain with relative magnetic permeability of unity. The magnetization (0.81e6 A/m) of the magnet was selected to yield a surface magnetic flux density equal to the manufacturer's specification (0.5 T) at the blunt end. This produced a peak flux of 0.75 T at the rounded tip. The fluid flow in response to the magnet motion was modeled in COMSOL by using the incompressible Navier–Stokes application mode and a 3D model of the fluid-filled portion of a single well, including the wetted magnet tip. The fluid properties were for water at 20 °C (density of 1,000 kg/m3 and viscosity of 1.0e-3 Pa.s). The velocity field was solved in the frame of motion of the magnet, where the magnet was rotating in place and the well rotated about the magnet. Hence, the boundary conditions for the magnet surface and well walls were chosen as inlet boundary conditions with a tangential fluid velocity equal to the appropriate wall velocity within the rotating frame. The free fluid surface was assigned a slip boundary condition.
The total particle velocity field (VT) is equal to the sum of the liquid velocity (VL) and the magnetic field-driven velocity of the superparamagnetic particle (Fm), which is determined using the magnetic force and Stokes drag formula, as follows:
Here, rcell is the cell radius, rp is the particle radius, ν is the viscosity, and χeff is the effective susceptibility, which is a function of the local magnetic flux. This formulation for the magnetic force (depending on the gradient of the magnetic energy density) assumes a curl-free magnetic field, and the velocity formulation assumes the case of hardest particle to capture, where a cell is labeled with a single superparamagnetic particle. A custom Matlab code was developed which imports the scalar |B| and Um fields simulated in Comsol Multiphysics and calculates the total particle velocity field. |B| was used exclusively to calculate the magnetic susceptibility based on a spline fit to the bulk magnetization curve provided by the particle manufacturer (Invitrogen). Given the total velocity field, a second Matlab code calculated particle trajectories and capture times by using a finite difference-based time stepping scheme.
Supplementary Material
Acknowledgments.
Dr. Mostafa Ronaghi, Dr. Haiyu Zhang, and Dr. Kurosh Ameri provided us with useful discussions for which we are grateful. We also thank Julie Wilhelmy for extracting and processing the RNA samples and performing chip hybridization. This work was supported in part by National Institutes of Health Grant P01 HG000205 (to R.W.D.) and the California Breast Cancer Research Program of the University of California, Grant 11IB-0175 (to S.S.J.). The original MagSweeper prototype was generously funded by Vladimir and Natalie Ermakoff.
Footnotes
Conflict of interest statement: A.H.T., A.A.P., R.F.P., M.N.M., S.S.J., and R.W.D. have applied for a patent relating to the method described in this study. The other authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813188106/DCSupplemental.
References
- 1.Cristofanilli M, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 2.Hayes DF, et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res. 2006;12:4218–4224. doi: 10.1158/1078-0432.CCR-05-2821. [DOI] [PubMed] [Google Scholar]
- 3.Budd GT, et al. Circulating tumor cells versus imaging–predicting overall survival in metastatic breast cancer. Clin Cancer Res. 2006;12:6403–6409. doi: 10.1158/1078-0432.CCR-05-1769. [DOI] [PubMed] [Google Scholar]
- 4.Moreno JG, et al. Circulating tumor cells predict survival in patients with metastatic prostate cancer. Urology. 2005;65:713–718. doi: 10.1016/j.urology.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 6.Cohen SJ, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 7.Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bianchi DW, Flint AF, Pizzimenti MF, Knoll JH, Latt SA. Isolation of fetal DNA from nucleated erythrocytes in maternal blood. Proc Natl Acad Sci USA. 1990;87:3279–3283. doi: 10.1073/pnas.87.9.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herzenberg LA, Bianchi DW, Schroder J, Cann HM, Iverson GM. Fetal cells in the blood of pregnant women: Detection and enrichment by fluorescence-activated cell sorting. Proc Natl Acad Sci USA. 1979;76:1453–1455. doi: 10.1073/pnas.76.3.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung MC, Goldberg JD, Kan YW. Prenatal diagnosis of sickle cell anaemia and thalassaemia by analysis of fetal cells in maternal blood. Nat Genet. 1996;14:264–268. doi: 10.1038/ng1196-264. [DOI] [PubMed] [Google Scholar]
- 11.Moon JJ, et al. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smirnov DA, et al. Global gene expression profiling of circulating tumor cells. Cancer Res. 2005;65:4993–4997. doi: 10.1158/0008-5472.CAN-04-4330. [DOI] [PubMed] [Google Scholar]
- 13.Calvano SE, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 14.Berthold F. Isolation of human monocytes by Ficoll density gradient centrifugation. Blut. 1981;43:367–371. doi: 10.1007/BF00320315. [DOI] [PubMed] [Google Scholar]
- 15.Kahn HJ, et al. Enumeration of circulating tumor cells in the blood of breast cancer patients after filtration enrichment: Correlation with disease stage. Breast Cancer Res Treat. 2004;86:237–247. doi: 10.1023/B:BREA.0000036897.92513.72. [DOI] [PubMed] [Google Scholar]
- 16.Vona G, et al. Isolation by size of epithelial tumor cells: A new method for the immunomorphological and molecular characterization of circulatingtumor cells. Am J Pathol. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allard WJ, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 18.Racila E, et al. Detection and characterization of carcinoma cells in the blood. Proc Natl Acad Sci USA. 1998;95:4589–4594. doi: 10.1073/pnas.95.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin VM, et al. Immunomagnetic enrichment of disseminated epithelial tumor cells from peripheral blood by MACS. Exp Hematol. 1998;26:252–264. [PubMed] [Google Scholar]
- 20.Zieglschmid V, Hollmann C, Bocher O. Detection of disseminated tumor cells in peripheral blood. Crit Rev Clin Lab Sci. 2005;42:155–196. doi: 10.1080/10408360590913696. [DOI] [PubMed] [Google Scholar]
- 21.Cheng X, et al. A microfluidic device for practical label-free CD4(+) T cell counting of HIV-infected subjects. Lab Chip. 2007;7:170–178. doi: 10.1039/b612966h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Momburg F, Moldenhauer G, Hammerling GJ, Moller P. Immunohistochemical study of the expression of a Mr 34,000 human epithelium-specific surface glycoprotein in normal and malignant tissues. Cancer Res. 1987;47:2883–2891. [PubMed] [Google Scholar]
- 23.Balzar M, Winter MJ, de Boer CJ, Litvinov SV. The biology of the 17–1A antigen (Ep-CAM) J Mol Med. 1999;77:699–712. doi: 10.1007/s001099900038. [DOI] [PubMed] [Google Scholar]
- 24.Rao CG, et al. Expression of epithelial cell adhesion molecule in carcinoma cells present in blood and primary and metastatic tumors. Int J Oncol. 2005;27:49–57. [PubMed] [Google Scholar]
- 25.Cristofanilli M, et al. Circulating tumor cells in metastatic breast cancer: Biologic staging beyond tumor burden. Clin Breast Cancer. 2007;7:471–479. [PubMed] [Google Scholar]
- 26.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.