Abstract
Determining how an organism responds to its environment by altering gene expression is key to understanding its ecology. Here, we used RNA-seq to comprehensively and quantitatively assess the transcriptional response of the bacterial opportunistic cystic fibrosis (CF) pathogen and endemic soil dweller, Burkholderia cenocepacia, in conditions mimicking these 2 environments. By sequencing 762 million bases of cDNA from 2 closely related B. cenocepacia strains (one isolated from a CF patient and one from soil), we identified a number of potential virulence factors expressed under CF-like conditions, whereas genes whose protein products are involved in nitrogen scavenging and 2-component sensing were among those induced under soil-like conditions. Interestingly, 13 new putative noncoding RNAs were discovered using this technique, 12 of which are preferentially induced in the soil environment, suggesting that ncRNAs play an important role in survival in the soil. In addition, we detected a surprisingly large number of regulatory differences between the 2 strains, which may represent specific adaptations to the niches from which each strain was isolated, despite their high degree of DNA sequence similarity. Compared with the CF strain, the soil strain shows a stronger global gene expression response to its environment, which is consistent with the need for a more dynamic reaction to the heterogeneous conditions of soil.
Keywords: cystic fibrosis, RNA-seq, soil, transcriptomics
To study differential expression of an organism in 2 different environments, we took advantage of the uniquely disparate natural ecologies of Burkholderia cenocepacia, a bacterium routinely isolated from soils and plant rhizospheres (1). B. cenocepacia can also lead to a necrotizing pneumonia or death in cystic fibrosis (the most common lethal genetic disorder in Caucasian populations) or immunocompromised patients (2). Understanding how B. cenocepacia sense and respond to their environment may aid in influencing their niche behavior, whether this is a pathogenic relationship within a human respiratory tract or a mutually beneficial interaction with plant roots. By measuring transcriptional responses, we can identify putative targets for a broad range of environmental, medical, and agricultural uses.
Using direct high-throughput Illumina sequencing of cDNAs (Illumina RNA-seq), the transcriptomes of several eukaryotes have been recently reported, but no such investigation has yet been reported for a bacterial transcriptome probably due to the lack of mRNA polyA tails in bacteria, which prevents specific targeting of the mRNA versus the much larger rRNA pool. We present here the first use of RNA-seq to investigate bacterial transcriptomes. We used enrichment of mRNA by an rRNA pulldown approach plus the much greater sequencing depth of the Illumina platform to overcome the barrier of lack of polyA tails.
We investigated 2 closely related B. cenocepacia strains, AU1054 and HI2424, as models for niche response and evolutionary divergence studies. The multireplicon genomes of these 2 strains share a 99.8% average nucleotide identity in their conserved genes [ANI (3)] but were isolated from 2 very disparate habitats, a cystic fibrosis patient (AU1054) and soil from an agricultural field (HI2424) (4, 5), both habitats that commonly harbor this species. AU1054 contains 50 genes not found in HI2424, whereas HI2424 carries 470 genes absent in AU1054. Although AU1054 is considered an epidemic strain, there have been few studies that examine its mechanism of virulence and none to investigate the regulatory differences between these 2 strains even though they represent an excellent pair for investigating the genetic and evolutionary basis of niche response and pathogenicity because of their genetic similarity and disparate sources.
In this study, we also identify potential B. cenocepacia virulence factors at a global scale under conditions mimicking a natural pathogenic niche, the human lung. We identified a large number of genes that were induced under CF conditions and found that several of them have been implicated in virulence in other bacterial pathogens. The 2 strains showed a marked difference in their regulated gene response to the 2 conditions, suggesting that adaptations that have occurred since these 2 strains diverged play a role in survival in their natural environments. Interestingly, the differentially regulated genes are unevenly distributed along the 3 chromosomes of this species suggesting that adaptations to niche may be selective to certain replicons.
Results and Discussion
To understand the expression of B. cenocepacia in its 2 ecological settings, human sputum and the soil, we grew the strains under conditions that mimic those habitats. We used a synthetic CF sputum medium that nutritionally mimics the components of CF sputum but lacks the patient-to-patient variability found in actual sputum samples (6). The soil medium (SE) was made from a hot water extract of a maize soil where B. cenocepacia has been previously isolated (1). Biological replicates of B. cenocepacia strains were grown at body temperature (37 °C) or typical summer daytime temperature (22 °C) in CF or SE media, respectively (Fig. 1A) before RNA extraction, purification, mRNA enrichment, and conversion to cDNA.
Fig. 1.
Experimental details for this study. (A) Growth of B. cenocepacia AU1054 and HI2424 under CF and SE conditions. Arrows indicate when cells were harvested for RNA purification. (B) Illumina reads mapped onto the GroES and GroEL genes. Numbers indicate position on HI2424 chromosome 1. Black and red arrows above the x axis correspond to single reads from CF or SE conditions, respectively. Asterisks indicate pillars of reads. (C) Relative quantitative real-time PCR results compared with Illumina data for 8 unlinked genes.
Sequencing of the Microbial Transcriptome.
The number of mapped cDNA reads, which varied between 1.7 and 4.5 million reads per sample, totaled 761,954,880 bases of sequenced B. cenocepacia cDNA (Table 1). ≈95% of the transcripts were assignable to the genome, which illustrates the suitability of using RNA-seq for bacterial transcriptomic studies. Although some of the samples yielded high numbers of non-rRNA reads (up to 54.2%), in other samples the mRNA enrichment was less substantial (Table 1), which may be attributable to different fragmentation states of the RNA samples, because the mRNA enrichment protocol we used performs better with unfragmented rRNA. Overall, 83,940,138 bp of cDNA solely from mRNA transcripts (63,000 - 908,423 reads per sample) were sequenced. Therefore, the next generation sequencing approach resulted in massive amount of non-rRNA informative reads, to a level where the vast majority of all genes were covered by at least 1 read. Fig. 1B shows an example of the distribution of reads mapped along a protein-coding gene in the HI2424 strain. The stacks of reads along the gene may correspond to sequencing hotspots (7).
Table 1.
Summary of B. cenocepacia cDNA samples sequenced using the Illumina genome analyzer
| Sequenced sample* | Read size, bp | No. of total mapped reads | No. of total mapped bps (× 106) | No. of mapped mRNA reads | No. of mapped mRNA bp (× 106) | mRNA reads (% of all mapped reads) |
|---|---|---|---|---|---|---|
| AU1054, CF (1) | 41 | 3,154,189 | 129.3 | 88,991 | 3.6 | 2.8% |
| AU1054, CF (2) | 27 | 2,007,792 | 54.2 | 787,800 | 21.3 | 39.2% |
| AU1054, Soil (1) | 41 | 2,204,756 | 90.4 | 118,540 | 4.9 | 5.4% |
| AU1054, Soil (2) | 27 | 1,674,586 | 45.2 | 908,423 | 24.5 | 54.2% |
| HI2424, CF (1) | 39 | 2,029,242 | 79.1 | 335,664 | 13.1 | 16.5% |
| HI2424, CF (2) | 27 | 2,887,442 | 78.0 | 87,687 | 2.4 | 3.8% |
| HI2424, Soil (1) | 39 | 2,875,024 | 112.1 | 76,599 | 3.0 | 2.7% |
| HI2424, Soil (2) | 39 | 4,450,939 | 173.6 | 286,860 | 11.2 | 6.4% |
| Sum | 21,283,970 | 762.0 | 2,690,564 | 83.9 | 12.6% |
*Indicated are the strain (B. cenocepacia AU1054 or HI2424), growth medium (cystic fibrosis or soil extract media), and replicate number (1 or 2).
The correlation between read proportions for the vast majority of the genes was similar between replicates indicating high reproducibility. The best reproducibility was observed in biological replicates with a similar number of mapped reads (Fig. S1). We validated the Illumina data using quantitative real-time PCR (qRT-PCR) and in 12 of 16 comparisons the Illumina gene expression ratios were not statistically different from the qRT-PCR ratios (Fig. 1C).
Global Interpretations.
We observed that 99.8% of the genes were represented by at least a single read in each replicate sample. In fact, a continuum in gene expression levels is observed when examining ordered expression levels for all genes within a sample rather than a breakpoint between “true” versus “background” expression levels. The large amount of low level transcriptional signal in our data suggests that gene expression is stochastic in natural settings of clonal bacterial populations.
To quantitatively compare the number of reads per gene in each condition, the replicate data for each gene was pooled and normalized for gene size and sample size. The resulting number, the Gene Expression Index (GEI) (expressed as reads per kilobase), allows for comparisons between genes and between growth conditions.
Two methods were used to globally assess whether strains AU1054 and HI2424 respond differently to each condition. The Kolmogorov–Smirnov 2-sample test was performed to examine log2-transformed GEIs for all genes. Under both conditions, the distribution of reads was significantly different between the 2 strains (Fig. 2). Compared with AU1054, HI2424 has a greater number of very highly expressed genes, which may indicate evolutionary adaptations have occurred in HI2424 which allow this strain to react strongly and quickly to out-compete its nearest neighbors in the nutrient-fluctuating soil environment. Additionally, a 2-factor ANOVA on the data showed that both strain and condition have a significant effect (P values <0.05) on gene expression and further suggests that these samples are significantly different in their gene expression responses.
Fig. 2.
Kolmogorov–Smirnov plots show the sample distribution of log2-transformed Gene Expression Indices for all genes in genome under each condition. Between strains under CF and SE conditions, the D values are 0.19 and 0.58, respectively.
Differential gene expression was established by performing χ2 testing on GEI values between strains under the 2 conditions or between strains under the same condition. Under CF conditions compared with SE conditions, 139 and 263 induced genes were identified in AU1054 and HI2424, respectively (Fig. 3 and Table S1); conversely, under SE conditions compared with CF conditions, 185 and 326 genes were induced, respectively (Fig. 3 and Table S2). The large difference in induced gene number between strains again illustrates that HI2424 has a stronger response to changes in environmental conditions than does AU1054.
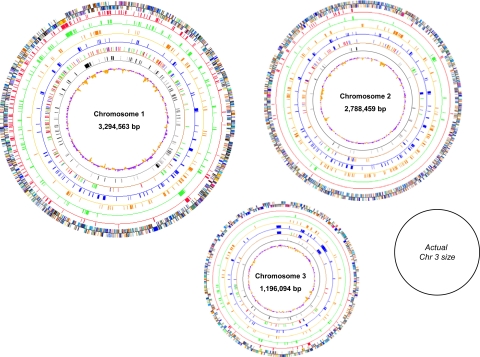
Fig. 3.
Distribution of differentially expressed genes. Outermost rings indicate all coding regions in the genomes colored according to COG designation; COG color and functional designations are described in Table S4. Red and green rings correspond to AU1054 and HI2424 genes induced under CF conditions, respectively. Orange and blue rings correspond to AU1054 and HI2424 genes induced under SE conditions, respectively. The brown ring indicates genes differentially regulated between the strains colored by condition of induction. The gray ring indicates genes whose expression is conserved under CF (black) or SE conditions (gray) in both strains. The innermost ring shows either high (purple) or low (orange) GC content.
The distribution of differentially regulated genes was not proportional to the number of genes present on each of the 3 chromosomes. For both strains under CF conditions, the number of induced genes was proportionally greater for chromosome 1 (Fig. 3 and Table S3). This disparate distribution suggests that under CF conditions, B. cenocepacia responds primarily by expressing core housekeeping genes that are found mainly on chromosome 1 (8). A different pattern occurs under SE conditions, where the genes differentially regulated between the 2 strains and those induced in HI2424 were more abundant on chromosomes 2 and 3, suggesting that they may play a role in fitness for the soil environment.
Gene-Based Interpretations.
Differentially expressed genes were classified into functional categories based on clusters of orthologous genes (COG) designations. The functional profiles of these gene sets differed greatly between the 2 conditions (Fig. 4) indicating that the environmental cues and subsequent gene expression patterns are quite unique for each condition.
Fig. 4.
Functional classification of differentially regulated genes. Profiles of the functional classes are shown as a percentage of all genes in each condition or gene set. “Both” indicates genes induced in either or both strains between conditions; “Different” indicates genes significantly differentially regulated between the 2 strains under the same condition. COG designations are described in Table S4. Asterisks indicate statistically significant differences exist between conditions.
Under CF compared with SE conditions, translation and energy production and conversion genes were induced, which correlate well with growth rates and medium composition. We also observed many genes whose protein products display similarity to virulence factors found in other bacterial pathogens (Table S1) including molecular chaperones, iron acquisition proteins, proteins expressed in macrophages, and immunogenic proteins. Another pathway highly induced under CF conditions is involved in phenylacetate (Paa, a byproduct of aromatic catabolism) metabolism although Paa was not detectable in our medium before growth so the reason for this induction is not obvious. However, it was recently shown that mutations in this pathway disrupted Burkholderia virulence in a C. elegans model system (9). Members of the general secretion (Sec) pathway were also induced under CF conditions. Of the genes induced in CF conditions, 56 have predicted Sec leader sequences including genes encoding putative porins, extracellular binding proteins, siderophore receptors, translocases, and an adhesin; some of which could be targets/models for therapeutics.
Different dominant functional classes were expressed under soil conditions, such as transcription, signal transduction, and uncharacterized proteins (Fig. 4). Genes annotated as hypothetical proteins were expressed in excess under SE conditions, which demonstrates how little we understand regarding life in the soil. Several genes involved in nitrogen acquisition were also found to be induced under SE conditions including RpoN, the sigma factor (σ54) involved in regulation of nitrogen uptake in many bacteria (10). This is concordant with results obtained by chemical extraction, where nitrate and ammonium concentrations were 124- or 525-fold lower in the SE medium compared with CF medium, respectively. The structural components of the twin-arginine signal (Tat) secretion pathway were also found to be induced (Fig. S2 and Table S2). Of the genes induced under SE conditions, 48 contain predicted Tat leader sequences. These data suggest B. cenocepacia utilizes 2 different protein export systems with the Sec system used under CF conditions and the Tat system used in SE conditions.
A published microarray-based study compared the gene expression response of a related B. cenocepacia epidemic strain, J2315, in a dilute CF sputum medium versus a minimal medium. This study found 760 differentially expressed J2315 genes (11), of which 554 have orthologs in the strains used in our study. Only 9 genes were found to be induced under CF-like conditions in both studies (Table S1). This small number of similarly acting genes could be the result of several key differences between the 2 studies (i.e., strains used, media composition and temperature, statistical analysis of the data).
Because RNA-seq is not limited to specific probes as are microarrays, an added advantage of the RNA-seq approach is the ability to efficiently discover putative noncoding RNA genes (ncRNAs). We searched intergenic regions showing differential expression between CF and SE conditions and identified 13 putative ncRNAs (e.g., Fig. S3). Of these, 12 were induced under SE conditions (Table S5). These potential ncRNAs were scanned for presence of putative promoter −10 and −35 regions, and were predicted to have a high degree of secondary structure (e.g., Fig. S3). The expression of 4 unique ncRNAs was verified by Northern Blot (e.g., Fig. S3) in 4 related strains of B. cenocepacia (AU1054, HI2424, MC0–3, J2315) indicates that these intergenic regions indeed are expressed and are conserved within the species. The high tendency of these ncRNAs to be activated in SE conditions supports previous observations that ncRNAs are frequently expressed under suboptimal growth conditions (13). A previous attempt to find ncRNAs in the related B. cenocepacia strain J2315 using computationally-derived predictions yielded 213 putative ncRNAs (14). None of these were identified in our study as differentially regulated. This discrepancy may exist because 12 of the 13 ncRNAs we detected were induced under the soil condition, which was not tested in the previously published study. Although the exact biological role of the B. cenocepacia ncRNAs we identified remains to be determined, our discovery illustrates yet another application of high throughput sequencing of bacterial transcriptomes.
In addition to medium-specific differential regulation, we also identified genes differentially regulated between the 2 strains in a single condition. Based on the high amount of nucleotide identity conservation in homologous genes (i.e., 99.8% ANI), we anticipated very few genes to be differentially regulated. However, 179 and 120 homologous genes had significantly different expression between the strains in CF or SE conditions, respectively (Table S6). These large numbers suggest that the number of transcriptional differences is not necessarily reflected in coding region identity and thus our current measures of relatedness between strains are insensitive to the volume of regulatory differences between these strains. These differentially regulated genes likely contribute to adaptations to the natural niches of these strains.
Differentially regulated genes between AU1054 and HI2424 under CF conditions function primarily in translation and energy production (Fig. 4 and Table S6) and those induced uniquely in AU1054 under CF conditions (38 genes) may yield a further defined list of potential virulence factors for examination. Those genes differentially regulated in SE conditions include primarily uncharacterized proteins (Fig. 4 and Table S6). Within each gene found differentially regulated between the 2 genomes, we examined the nucleotide sequence identity of the area surrounding each coding region and identified 57 regulatory units (encompassing 92 genes) that contain single nucleotide polymorphisms (SNPs) or insertion/deletions possibly responsible for the observed difference in expression (Fig. S2 and Table S7). The small number of potentially affected genes compared with the large number of expression differences from the RNA-seq data (432) indicates that the majority of gene expression changes are at a global regulatory scale. This is supported by the large number of transcription-related genes differentially expressed between the strains, including 2 sigma factors (Table S7).
The ability to mimic 2 distinct B. cenocepacia ecological niches is a strong advantage in shedding light into what genes might be important for each strain's survival within each niche. We have uncovered a surprising number of strain-specific responses to each environment, which may aid in the identification of therapeutic targets for new vaccines or antibiotics, which is important medically because this organism is resistant to a broad spectrum of antibiotics. Although further investigation is required to tease apart the precise roles of the identified differentially regulated B. cenocepacia genes and ncRNAs in either niche, this study establishes RNA-seq as a powerful method for the quantitative and qualitative examination of bacterial transcriptomes.
Materials and Methods
Strains and Growth Conditions.
The 2 Burkholderia cenocepacia strains used in this study, AU1054 and HI2424, were isolated from a CF patient (4) and from bulk soil from a histosol (muck) agricultural field in New York state (5), respectively. All cells were grown on a rotary shaker in 125-mL flasks containing 30 mL of medium. The cultures grown on synthetic CF sputum medium (6) at 37 °C were inoculated from cells grown on LB agar plates. The inoculum for the soil extract medium (SE) was grown overnight in 1/10th tryptic soy broth at 22 °C, centrifuged at 4500 × g at room temperature, and the pellets were resuspended in 2 mL of spent supernatant. One ml was diluted into 30 mL of moderate soil medium (15), which contains 10% soil extract (prepared by autoclaving 400 g of sieved maize rhizosphere soil per liter of water for 15 min, centrifuging, and filtering to remove particulates) and supplemented with 3 mM glucose (equal to the CF medium). A significant portion of degradable carbon entering agricultural soils from plants flows through glucose, making glucose a common energy resource for soil populations (16, 17). The soil cultures were incubated at 22 °C.
RNA Harvesting.
Cell growth was monitored in triplicate 30-mL cultures of each strain under each condition. When the O.D.600 reached ≈1.0 or 0.8 for CF and SE conditions, respectively, cells were centrifuged at 4500 × g at 4 °C and cell pellets were resuspended in 3 mL of PBS. One milliliter of culture was subjected to RNA purification by RNeasy Midi Kit (Qiagen) and eluted in 500 μL of RNase-free water. Samples were treated with DNaseI to remove any residual DNA and purified by phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation.
mRNA Purification and cDNA Synthesis.
Ten micrograms from each total RNA sample was subjected to further purification by enriching for mRNA via the MICROBExpress kit (Ambion) according to the manufacturer's instructions. Samples were resuspended in 15 μL of RNase-free water. cDNA was generated according to instructions given in Invitrogen's SuperScript Double-Stranded cDNA Synthesis Kit. Briefly, each mRNA sample was mixed with 100 pmol of random hexamers (Integrated DNA Technologies), incubated at 70 °C for 10 min, chilled on ice, mixed with 5 μL of First-Strand Reaction Buffer (Invitrogen), 2.5 μL of 0.1 M DTT, 1.25 μL of 10 mM RNase-free dNTP mix, 3 μL of SuperScript III reverse transcriptase, and incubated at 50 °C for 2 h. To generate the second strand, the following Invitrogen reagents were added: 86 μL of RNase-free water, 30 μL of second-strand reaction buffer, 3 μL of 10 mM RNase-free dNTP mix, 10 units Escherichia coli DNA Ligase, 40 U E. coli DNA Polymerase, 2 U E. coli RNase H, and incubated at 16 °C for 2 h. Ten units of T4 DNA Polymerase (Invitrogen) were added and the reactions were incubated for an additional 5 min at 16 °C. Reactions were stopped by adding 10 μL of 0.5 M RNase-free EDTA and purified by phenol-chloroform extraction and ethanol precipitation. Residual RNA was removed by treating with RNaseH and RNaseA followed by phenol-chloroform extraction and ethanol precipitation.
Illumina Sequencing and Analysis.
cDNA samples were submitted to the Joint Genome Institute for sequencing using Illumina Genome Analyzer. Libraries were prepared for sequencing according to the manufacturer's instructions. Sequencing reads were mapped to the genome using BlastN with a threshold e value of 0.0001 and using the “−F F” parameter. These parameters allow mapping of reads to the genome with up to 5 mismatches. Reads mapped to rRNA and reads not mapped under the mentioned parameters were removed from further analysis. The number of reads overlapping each gene based on GenBank annotation was recorded. Reads from replicate samples were pooled, and the number of reads per gene was normalized according to the total number of reads in each library and the gene size. The resulting number is the gene expression index (GEI) (expressed as normalized reads per kilobase). Reproducibility was assessed by plotting replicate GEI values against each other. Raw data files can be accessed at www.weizmann.ac.il/molgen/Sorek/files/Burkholderia/data.html.
Further Characterization of Differentially Expressed Genes.
The χ2 test was performed on GEIs to determine genes with differential expression between conditions or between strains. The Bonferroni multiple testing correction was applied to χ2 values and those genes with adjusted P values <0.05 and with GEI ratios greater than or equal to 2 were considered significant. Descriptive statistics and the Kolmogorov–Smirnov test were performed on all log2 GEI ratios for each sample using the Microsoft Excel add-in XLStat (Addinsoft) with default settings. The distribution of overexpressed genes were analyzed using either GenomeViz software (18) or 2-factor ANOVA with replicates in Microsoft Excel.
Overexpressed genes were assigned into cluster of orthologous genes (COG) categories based on National Center for Biotechnology Information classifications and plotted as percentages of total induced genes in a given gene list. For genes induced under CF or SE conditions, the amino acid sequences of the putative ORFs were entered into the SignalP and TatP prediction servers (19–20), respectively, and SignalP-NN or TatP-NN (Neural Network) D scores (the average of the S-mean and Y-max scores) were used as indicators of signal sequence presence or absence.
Identification of Mutations Near Differentially Regulated Genes.
Genomic differences potentially responsible for the observed differences in expression between AU1054 and HI2424 were detected using MUMmer genome alignment software (21) and manually examining the genetic environment of the differentially regulated gene. Any single nucleotide polymorphisms (SNPs), insertion/deletions (indels), or IS elements within 1,000 bp of a differentially expressed gene (or operon) promoter were recorded.
ncRNA Identification and Verification.
Read coverage in regions having coverage higher than the least expressed 20% of all genes were further analyzed. To determine the boundaries of ncRNAs within each candidate intergenic region, a sliding window of 20 bp was used to optimize subregion continuity of expression, requiring the lowest expression to be at least 30% of the highest expression in the range (or higher than the expression of the median of the gene coverage). Candidates sized 100 bp or more were further considered. To prevent misclassification of untranslated regions (UTRs) as ncRNAs, candidates having expression similar to one of the flanking genes were discarded. 100-bp regions flanking the predicted ncRNA were examined for promoter −10 and −35 regions and putative Shine-Dalgarno sequences (ribosomal binding sites). RNAfold was used to predict secondary structure for all differentially regulated predicted ncRNAs (12).
Verification of 4 ncRNAs was performed by Northern Blot using RNA samples harvested from B. cenocepacia strains J2315 (epidemic CF isolate), AU1054, HI2424, and MC0–3 (corn rhizosphere isolate) under CF conditions as described above. Either BioReagents RiboLadder RNA Standard (Fisher) or 10 ugs of each total RNA sample was loaded into each lane of a 1% agarose gel. Ambion's NorthernMax Kit was used for electrophoresis, blotting, and hybridization according to manufacturer's recommendations. Single-strand RNA probes were generated using PCR primers designed against predicted small RNA sequences with all 5′ primers containing a T7 minimal promoter tag and all 3′ primers contain SP6 minimal promoter tag. The primers were then used to amplify the predicted small RNA from genomic DNA. Ambion's MAXIscript Kit SP6/T7 and [α-32P]-uridine 5′-triphosphate (Perkin–Elmer) were used to generate 32P -labeled single strand RNA probe either from T7 promoter or SP6 promoter according to manufacturer's recommendations.
Quantitative PCR.
Unique primers were designed for 100 bp and 500 bp segments from Bcen_3737 (Formamidase), Bcen_0378 (GroEL), Bcen_1135 (SpoVR-like), Bcen_1133 (Serine protein kinase, PrkA), Bcen_2037 (Succinyl Co-A synthetase alpha subunit), Bcen_0196 (50S ribosomal protein L13), Bcen_0560 (OmpA/MotB), and Bcen_2866 (Flagellin) using Primer Express software (Applied Biosystems, Foster City, IA). Triplicate qPCR reactions were set up using 1X concentration of SybrGreen PCR Master Mix (Applied Biosystems), optimized concentrations of appropriate primers, and one of the following templates: 105, 104, 103, 102, 101, 100 molecules of 500 bp PCR-generated standard curve template, 20 ng of cDNA (both cDNA samples used for Illumina sequencing replicate 2 and new samples prepared in parallel to replicate 2 sample preparation), 80 ng of genomic DNA, or 100 ng of total RNA. Reactions were loaded into a ABI Prism 7900HT Sequence Detection System and the PCR was performed with the following conditions: 50 °C for 2 min, 95 °C for 10 min, (95 °C for 15 sec, 60 °C for 1 min) × 40 cycles. After optimization, average relative concentrations were calculated using Microsoft Excel.
Supplementary Material
Acknowledgments.
We thank Feng Chen, Dorothy Lang, Stephanie Malfatti, Wei Wang, and Dr. Robert Tempelman for help in sample preparation, figure preparation, and data analysis. R.S. was supported by the Y. Leon Benoziyo Institute for Molecular Medicine; the work at Lawrence Berkeley National Laboratory and Lawrence Livermore National Laboratory was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program, under contract number DE-AC02-05CH11231 and DE-AC52-07NA27344; and the Michigan State University work supported by National Science Foundation Grant 516252 and National Institutes of Health Grant R21HL087833.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813403106/DCSupplemental.
References
- 1.Ramette A, LiPuma JJ, Tiedje JM. Species abundance and diversity of Burkholderia cepacia complex in the environment. Appl Environ Microbiol. 2005;71:1193–1201. doi: 10.1128/AEM.71.3.1193-1201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LiPuma JJ. Burkholderia and emerging pathogens in cystic fibrosis. Semin Respir Crit Care Med. 2003;24:681–692. doi: 10.1055/s-2004-815664. [DOI] [PubMed] [Google Scholar]
- 3.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen JS, Witzmann KA, Spilker T, Fink RJ, LiPuma JJ. Endemicity and inter-city spread of Burkholderia cepacia genomovar III in cystic fibrosis. J Pediatr. 2001;139:643–649. doi: 10.1067/mpd.2001.118430. [DOI] [PubMed] [Google Scholar]
- 5.LiPuma JJ, Spilker T, Coenye T, Gonzalez CF. An epidemic Burkholderia cepacia complex strain identified in soil. Lancet. 2002;359:2002–2003. doi: 10.1016/S0140-6736(02)08836-0. [DOI] [PubMed] [Google Scholar]
- 6.Palmer KL, Aye LM, Whiteley M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol. 2007;189:8079–8087. doi: 10.1128/JB.01138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chain PS, et al. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc Natl Acad Sci USA. 2006;103:15280–15287. doi: 10.1073/pnas.0606924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Law R.J., et al . A functional phenylacetic acid catabolic pathway is required for full pathogenicity of Burkholderia cenocepacia in the Caenorhabditis elegans host model. J Bacteriol. 2008;109:7209–7218. doi: 10.1128/JB.00481-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thony B, Hennecke H. The −24/-12 promoter comes of age. FEMS Microbiol Rev. 1989;5:341–357. doi: 10.1016/0168-6445(89)90028-4. [DOI] [PubMed] [Google Scholar]
- 11.Drevinek P, et al. Gene expression changes linked to antimicrobial resistance, oxidative stress, iron depletion and retained motility are observed when Burkholderia cenocepacia grows in cystic fibrosis sputum. BMC Infect Dis. 2008;8:121. doi: 10.1186/1471-2334-8-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toledo-Arana A, Repoila F, Cossart P. Small noncoding RNAs controlling pathogenesis. Curr Opin Microbiol. 2007;10:182–188. doi: 10.1016/j.mib.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Coenye T, et al. Identification of putative noncoding RNA genes in the Burkholderia cenocepacia J2315 genome. FEMS Microbiol Lett. 2007;276:83–92. doi: 10.1111/j.1574-6968.2007.00916.x. [DOI] [PubMed] [Google Scholar]
- 15.Kilmer VJ, Hanson AA. Handbook of Soils and Climate in Agriculture. CRC Series in Agriculture. Boca Raton, FL: CRC; 1982. [Google Scholar]
- 16.Kamilova F, et al. Organic acids, sugars, and L-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol Plant Microbe Interact. 2006;19:250–256. doi: 10.1094/MPMI-19-0250. [DOI] [PubMed] [Google Scholar]
- 17.Kuiper I, Kravchenko LV, Bloemberg GV, Lugtenberg BJ. Pseudomonas putida strain PCL1444, selected for efficient root colonization and naphthalene degradation, effectively utilizes root exudate components. Mol Plant Microbe Interact. 2002;15:734–741. doi: 10.1094/MPMI.2002.15.7.734. [DOI] [PubMed] [Google Scholar]
- 18.Ghai R, Hain T, Chakraborty T. GenomeViz: Visualizing microbial genomes. BMC Bioinformatics. 2004;5:198. doi: 10.1186/1471-2105-5-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 20.Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. Prediction of twin-arginine signal peptides. BMC Bioinformatics. 2005;6:167. doi: 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.