Abstract
St. John’s Wort (SJW; Hypericum perforatum L.) is commonly known for its antidepressant properties and was traditionally used to promote wound healing, but its molecular mechanism of action is not known. Here, chemical-genetic profiling in yeast was used to predict the human intracellular targets of an aqueous extract of SJW. SJW source material was authenticated by TLC, digital microscopy, and HPLC, and further characterized by colorimetric methods for antioxidant activity, protein content, and total soluble phenolic content. SJW extract contained 1.76µg/mL hyperforin, 10.14µg/mL hypericin, and 46.05µg/mL pseudohypericin. The effect of SJW extract on ~5900 barcoded heterozygous diploid deletion strains of Saccharomyces cerevisiae was investigated using high-density oligonucleotide microarrays. Seventy-eight (78) yeast genes were identified as sensitive to SJW and were primarily associated with vesicle-mediated transport and signal transduction pathways. Potential human intracellular targets were identified using sequence-based comparisons and included proteins associated with neurological disease and angiogenesis-related pathways. Selected human targets were confirmed by cell-based immunocytochemical assays. The comprehensive and systematic nature of chemical-genetic profiling in yeast makes this technique attractive for elucidating the potential molecular mechanisms of action of botanical medicines and other bioactive dietary plants.
Keywords: St. John’s Wort, Hypericum perforatum, botanical, dietary supplement, yeast, microarray, wound healing, depression, alternative medicine
1. INTRODUCTION
Hypericum perforatum L., commonly known as St. John’s wort (SJW), is a yellow-flowering perennial herb grown in temperate and subtropical climates that has a long history of use as a medicinal plant for treating wounds and skin ailments, nerve problems, muscle pain, and mood disorders such as depression and anxiety (1). Meta-analysis of several studies found SJW to be effective in the treatment of mild to moderate depression, with fewer side effects than many conventional anti-depressants, but of limited efficacy for cases of moderate to severe depression (2). Various compounds from Hypericum species have also been found to possess inhibitory activity against cancer cell growth (3–5).
Despite a long history of use, the molecular mechanism of action of SJW as a medicinal herb is neither well characterized nor well understood. Although many of the compounds that comprise SJW have been purified and studied, such as the major compounds hypericin, pseudohypericin and hyperforin, it remains unclear whether a single compound or a synergy of compounds is responsible for the bioactive properties of SJW (6, 7). Mechanistic studies of botanical complementary and alternative medicines (CAM) such as SJW are complicated by the fact that the desirable biological functions often seem to result from the synergistic action of multiple constituents. Identification of molecular mechanisms of action is critical for the evaluation and optimization of botanical CAM as therapeutic agents. Currently, however, no standard approach exists for comprehensive and systematic target identification.
A recent technological advance in yeast genomics shows promise as a tool to elucidate the molecular mechanisms affected by botanical-based CAM on a genome-wide scale. To facilitate the analysis of gene function in S. cerevisiae, an international consortium constructed individual gene-deletion mutants for ~95% of the known yeast open reading frames (ORFs), replacing one ORF in each deletion strain with a deletion “cassette” containing the KanMX gene (the expression of which confers geneticin resistance to yeast) flanked by two unique 20-nt sequences (e.g. molecular barcodes). One molecular barcode sequence is located upstream of the KanMX gene in the cassette and is called the ‘UPTAG’, whereas the other molecular barcode sequence is located downstream of the KanMX gene in the cassette and is called the ‘DOWNTAG.’ Using this set of pooled yeast mutants, genes affected by specific growth conditions can be identified without prior knowledge of gene function simply by the monitoring the fitness of each particular gene deletion strain in said growth condition (8, 9). Evaluation of individual strain fitness in a particular growth condition is facilitated by monitoring the hybridization of molecular barcode sequences isolated from the pool of deletion strains to oligonucleotide microarrays. This strategy has been used successfully to identify the molecular mechanisms of individual bioactive agents including anticancer and antifungal compounds (10), radiation (11), and toxic chemicals (12). Here, we demonstrate the further utility of the technique for probing the molecular mechanism of action of a complex, multi-component botanical CAM by identifying the genes essential to the adaptive response of yeast to an aqueous extract of SJW.
By using a chemical-genetic profiling screen, we show that SJW affects yeast genes associated with intra- and intercellular transport and signal transduction. Furthermore, through sequence-based comparison of SJW-sensitive yeast targets, we identify orthologous human proteins implicated in molecular activities associated with neurological function and the formation of new blood vessels, identify potential therapeutic gene targets, and suggest potential mechanisms to explain the wound-healing and neuroprotective activities previously associated with SJW. Our results provide the first comprehensive analysis of a genome-wide functional response to a botanical traditional medicine.
2. MATERIALS AND METHODS
2.1. Chemicals and reagents
Unless otherwise specified all chemicals were purchased from Sigma-Aldrich.
2.2. Plant Material and Authentication
Dried, cut St. John’s wort was obtained from Herbal Advantage (Rogersville, MO). Authentication of the plant material as Hypericum perforatum L. [Clusiaceae] was confirmed by Alkemist Pharmaceuticals, Inc. (Costa Mesa, CA) against reference samples of Hypericum perforatum L. and hypericin using high-performance thin-layer chromatography (HP-TLC) and by the visual identification of hypericum oil glands using digital microscopy.
2.3. Preparation of St. John’s Wort Infusion
Care was taken to prepare an infusion most similar to that typically administered as herbal therapy. Ten grams of dry, cut SJW (Herbal Advantage, Rogersville, MO) was steeped in distilled water for 30 min. The temperature of the water was maintained above 80 °C. Next, the infusion was cooled to room temperature and centrifuged at 2000 rpm for 5 min to remove plant material. The supernatant was filtered through a coffee filter and poured into 50 mL disposable conical tubes, covered with aluminum foil, and stored at 4 °C.
2.4. Characterization of St. John’s Wort Extract
Total soluble phenolic content of the SJW infusion was determined as gallic acid equivalents by the Folin-Ciocalteau assay as described previously (13). Antioxidant activity was determined as 2, 2-Diphenyl-1-picrylhydrazyl (DPPH) radical scavenging activity (14). Hypericin, hyperforin, and pseudohypericin concentrations of the infusion were determined by IBC Labs (Tucson, AZ) using HPLC according to the method of Ang et al. (2002) (15), using external standards obtained from ChromaDex (Irvine, CA) and Axxora (San Diego, CA).
2.5. Yeast Strains
The heterozygous diploid gene deletion pool (Yeast Knock-Out version 1) containing 5,936 individual deletion strains (representing deletions of all essential and nonessential genes) and the wild-type parental strain (S. cerevisiae Hansen strain BY4743) were obtained from Invitrogen (Carlsbad, CA)
2.6. Exposure of Yeast Deletion Strains to St. John’s Wort Extract
Clonogenic survival assays with the parental strain, BY4743, were performed to determine the appropriate dose of St John’s wort infusion to result in approximately 30% cell death after 22 h. For experiments with the heterozygous deletion pool, duplicate aliquots of the deletion pool representing approximately 10,000 copies of each of the individual deletion strains, were grown in YPD medium in the presence of St John’s wort infusion (dH2O for controls) on an orbital shaker at 30 °C and 250 rpm for 22 h. The SJW-treated or control cells were then used to inoculate 50 mL of fresh YPD medium and incubated at 30 °C and 250 rpm for an overnight outgrowth period. Cultures were harvested after 18 h. Genomic DNA was extracted from harvested cells using the MasterPure Yeast DNA Purification Kit (Epicentre Biotechnologies, Madison, WI).
2.7. DNA amplification and Microarray Hybridization
DNA amplification was as described (9, 16) with some modifications. Briefly, genomic DNA from the SJW-treated and control pools was used as template in two separate PCR reactions to amplify the UPTAG or DOWNTAG sequences from each strain in the pool using 5’-Cy3-labeled primers complementary to common regions of the gene-deletion cassette. After PCR, blocking oligonucleotides complementary to regions of the PCR product external to the molecular barcode sequences were added to both the UPTAG and DOWNTAG reaction mixes, and then the UPTAG and DOWNTAG reaction mixes were combined. . The combined mixes were briefly heat denatured and then hybridized for 16 h at 42 °C to custom oligonucleotide microarrays (Agilent) of the molecular barcode sequences (16).
2.8. Data Acquisition and Analysis
Arrays were scanned at an emission wavelength of 532 nm using an Axon 4000B scanner (Molecular Devices, Santa Clara, CA). The hybridization intensities of each feature on the array were determined using GenePix 5.0 software (Molecular Devices). GenePix local background measurements were not used for correction of raw signal intensities because this technique was previously found to significantly increase noise (17). For analysis of strain prevalence in the pool, hybridization signal intensities of replicate features for each strain were averaged into a single value. Control/ SJW-treated ratios of signal intensities for each strain were log2 transformed. The fitness of each strain was evaluated using the empirical rule for standard distributions. Logged ratios of the signal intensities were converted to a standard score for each strain by subtracting the mean (xbar) from each log ratio (x) and dividing by the sample standard deviation (s). Standard scores greater than or equal to 2.0 were designated as sensitive strains (representing approximately 2.5% of the pool). Sensitivities for strains represented by both UPTAGS and DOWNTAGS on this final list were averaged. Human orthologs of genes represented by sensitive yeast strains were determined using the NCBI BLASTP weblink at the Saccharomyces Genome Database site (http://db.yeastgenome.org/cgi-bin/bestHits) against Homo sapiens predicated protein sequences and using the NCBI HomoloGene database (http://www.ncbi.nlm.nih.gov/sites/entrez/query.fcgi?db=homologene). Biochemical and physiological pathways linked to human orthologs were determined using Ingenuity Pathways Analysis (Ingenuity Systems, Redwood City, CA).
2.9. Confirmation of Mutant Phenotype by Survival Assay
Our microarray analysis of barcode DNA sequences isolated from the SJW-treated pool identified individual deletion strains whose signal intensity was reduced in comparison the control pool. To determine whether the signal reduction corresponded to SJW-sensitivity leading to cell death or reduced cell growth (and not to a transient initial growth delay), we performed survival experiments on several of the strains that showed reduced signal intensity after SJW treatment. Briefly, selected cultures were grown to OD600nm = 1.0, diluted with fresh YPD media, added to 96-well microplates, and grown in the presence of SJW extract for 19 h. After SJW treatment, the OD600nm was measured and compared with survival of untreated controls.
2.10. Western Immunoblotting
Human targets identified by data analysis as putatively sensitive to SJW were further qualified via Western immunoblot experiments on nuclear and cytoplasmic extracts and culture media of MDA-MB-231 human mammary carcinoma cells treated with SJW. This cell line was chosen because it is known to express HIF1α when grown in low-glucose media. This cell line was also used for preliminary tests of SIRT2 sensitivity, as this protein is known to be expressed in multiple tissues including breast cells. The MDA-MB-231 cell line was provided by the NCI cell line repository. The cells were maintained in high-glucose (4.5 g/L) Dulbecco’s modified Eagle’s Medium (DMEM; Quality Biological, Gaithersburg, MD) supplemented with 10% fetal bovine serum (HyClone Laboratories, Logan, UT), penicillin, streptomycin, 2 mM glutamine at 37 °C, and 5% CO2. To test the effect of SJW extract, 4×105 cells were inoculated in 100×20mm Petri dishes and grown to ~60% confluence in high-glucose DMEM. SJW was added to low-glucose (1g/L) DMEM at a final concentration of 6% (3 mg total phenolic content; 1 µg hyperforin, 6 µg hypericin, 26 µg pseudohypericin) and added to the cells. After 19 hr, the medium was removed and concentrated (for VEGF experiments), while the cells were harvested for isolation of nuclear and cytoplasmic extracts using an NE-PER Nuclear and Cytoplasmic Extraction Kit (Pierce Biotechnologies, Rockford, IL). Equal amounts of nuclear extracts were electrophoresed on SDS-polyacrylamide gels and transferred to nitrocellulose membranes using a semidry blotter (Bio-Rad). Membranes were blocked using Tris-buffered saline with 3% nonfat milk (pH 8.0; Sigma). Blots were then probed with primary anti-HIF1α, anti-VEGF, anti-actin, anti-SIRT2, anti-acetylated α-tubulin, or anti-tubulin (Santa Cruz Biotechnology) in blocking buffer and subsequently by a secondary antibody conjugated to horseradish peroxidase (1:2000). All blots were washed in Tris-buffered saline with Tween 20 (pH 8.0; Sigma) and developed using the ECL procedure (Amersham Biosciences). Anti-rabbit or anti-mouse antibody (Bio-Rad) was used as secondary antibody.
3. RESULTS
3.1. Characterization of St. John’s Wort Extract
Characteristics of the aqueous SJW extract are shown in Table 1. The extract possesses a significant amount of phenolic compounds with a total soluble content just over 5.2 mg/mL, but relatively little protein content (less than 1 µg/ mL). Antioxidant activity of the extract was approximately one-half that of a comparable amount of gallic acid. Hyperforin was present at 0.03% and hypericin at 0.2%. Pseudohypericin was present at 0.9%.
Table 1.
Characteristics of St John's Wort Extract
| Antioxidant Activitya | |
| IC50, Gallic acid | 16.4 µg/ mL |
| IC50, St. John's wort extract | 37.0 µg/ mL |
| Protein Contentb | 0.48 (±0.02) µg/ mL |
| Total Soluble Phenolic Contentc,d | 5249 (±36) µg/ mL |
| Hyperforine | 1.76 µg/ mL |
| Hypericine | 10.14 µg/ mL |
| Pseudohypericine | 46.05 µg/ mL |
Free-radical scavenging
BSA equivalents
Gallic acid equivalents
Determined by spectrophotometry
Determined by HPLC
3.2. Determination of Strain Sensitivity
Signal intensities were obtained for PCR products that hybridized to the sense and antisense UPTAG and DOWNTAG features for each deletion strain represented on the oligonucleotide microarrays. The mean signal intensity was calculated from replicate features for each strain. The mean signal intensity ranged from 106 to 22,581 for the 5,936 strains in the pool. A ratio (untreated/ treated) of the mean signal intensities was calculated for each strain. A ratio of 1 suggests that the SJW had no effect on the strain, whereas a high ratio suggests a growth defect. The ratio was converted into a logarithm, base 2. The logged ratios were filtered using the empirical rule for standard distributions. The logged ratios were converted into standard scores and the cut-off set at +2 standard deviations (positive values represented strains for which SJW-treated signal intensities were lower than control values). At +2 standard deviations the isolated values represent approximately 2.5% of the original pool. At 95%, the confidence interval is 0.020247, or 2.02%. Sensitivities for individual strains identified by both UPTAG and DOWNTAG features were averaged.
3.3. Identification and Confirmation of SJW-Sensitive Strains
In our screen, 78 deletion strains were identified as significantly sensitive to the SJW extract (Table 2). From a search of the primary literature, we have determined that none these strains has been previously described as sensitive to SJW. Due to budget constraints, strains selected for manual confirmation were taken from across the list of sensitive strains, and the results strongly suggest that the remaining strains listed are likely also not false positives. We obtained individual strains for 11 of the 78 putative SJW-sensitive deletion strains, representing the rankings 1–5, 11, 19, 20, 43, 72, and 73 in Table 2. All 11 deletion strains showed significantly reduced growth compared to untreated controls, indicating marked sensitivity to SJW and confirming that their reduced representation on the microarray was not due to an initial growth delay, which would be expected to recover by the end of the 19h incubation period. Table 3 lists the OD600nm readings for these strains after SJW treatment in comparison to untreated controls.
Table 2.
Heterozygous Deletion Strains Sensitive to St. John's Wort Extract.
| Sensitivity Rank | ORF | Gene Name | Biological process |
|---|---|---|---|
| 1 | YCL061C | MRC1 | cell cycle (DNA replication) |
| 2 | YOR198C | BFR1 | meiosis |
| 3 | YLR180W | SAM1 | methionine metabolism |
| 4 | YKR059W | TIF1 | telomere maintenance |
| 5 | YLR120C | YPS1 | protein processing |
| 6 | YGL049C | TIF4632 | translational initiation |
| 7 | YBR120C | CBP6 | translation |
| 8 | YDR397C | NCB2 | regulation of transcription from polymerase II promoter |
| 9 | YOR036W | PEP12 | Golgi to vacuole transport |
| 10 | YGL028C | SCW11 | cytokinesis, completion of separation |
| 11 | YNL098C | RAS2 | sporulation |
| 12 | YGR162W | TIF4631 | ribosome biogenesis and assembly |
| 13 | YNR034W | SOL1 | tRNA export from nucleus |
| 14 | YER044C | ERG28 | ergosterol biosynthesis |
| 15 | YGR078C | PAC10 | tubulin folding |
| 16 | YGL249W | ZIP2 | synapsis |
| 17 | YIR033W | MGA2 | regulation of transcription from polymerase II promoter |
| 18 | YIL002C | INP51 | cell wall organization |
| 19 | YDR191W | HST4 | chromatin silencing at telomere |
| 20 | YOR339C | UBC11 | protein monoubiquitination |
| 21 | YDR192C | NUP42 | mRNA export from nucleus |
| 22 | YCR011C | ADP1 | transport |
| 23 | YBL019W | APN2 | base-excision repair |
| 24 | YLL054C | unknown | |
| 25 | YDL232W | OST4 | protein amino acid N-linked glycosylation |
| 26 | YOL088C | MPD2 | protein folding |
| 27 | YIL050W | PCL7 | regulation of glycogen biosynthesis |
| 28 | YLR372W | SUR4 | telomere maintenance |
| 29 | YLL021W | SPA2 | pseudohyphal growth |
| 30 | YMR129W | POM152 | mRNA export from nucleus |
| 31 | YOR023C | AHC1 | histone acetylation |
| 32 | YBR274W | CHK1 | protein amino acid phosphorylation |
| 33 | YBR111C | YSA1 | unknown |
| 34 | YPL018W | CTF19 | chromosome segregation |
| 35 | YDR507C | GIN4 | protein amino acid phosphorylation |
| 36 | YLR213C | CRR1 | spore wall assembly |
| 37 | YGL212W | VAM7 | telomere maintenance |
| 38 | YBR229C | ROT2 | cell wall organization |
| 39 | YER167W | BCK2 | G1/S transition of mitotic cell cycle |
| 40 | YOR038C | HIR2 | regulation of transcription from polymerase II promoter |
| 41 | YAL054C | ACS1 | histone acetylation |
| 42 | YDR293C | SSD1 | response to drug |
| 43 | YJL128C | PBS2 | protein amino acid phosphorylation |
| 44 | YPR072W | NOT5 | regulation of transcription from polymerase II promoter |
| 45 | YOR298C-A | MBF1 | regulation of transcription from polymerase II promoter |
| 46 | YGR006W | PRP18 | nuclear mRNA splicing, via spliceosome |
| 47 | YGR042W | telomere maintenance | |
| 48 | YDR500C | RPL37B | translation |
| 49 | YBR236C | ABD1 | mRNA capping |
| 50 | YGL195W | GCN1 | regulation of translational elongation |
| 51 | YIL160C | POT1 | fatty acid beta-oxidation |
| 52 | YPR093C | ASR1 | response to ethanol |
| 53 | YDL081C | RPP1A | translational elongation |
| 54 | YDR137W | RGP1 | retrograde transport, endosome to Golgi |
| 55 | YBL028C | ribosome biogenesis and assembly | |
| 56 | YPR138C | MEP3 | nitrogen utilization |
| 57 | YOR140W | SFL1 | regulation of transcription from polymerase II promoter |
| 58 | YAL059W | ECM1 | cell wall organization |
| 59 | YML061C | PIF1 | telomere maintenance |
| 60 | YDL181W | INH1 | ATP synthesis coupled proton transport |
| 61 | YBR115C | LYS2 | lysine biosynthesis |
| 62 | YBL042C | FUI1 | uridine transport |
| 63 | YOR009W | TIR4 | unknown |
| 64 | YBR152W | SPP381 | nuclear mRNA splicing, via spliceosome |
| 65 | YOR292C | unknown | |
| 66 | YJL109C | UTP10 | ribosome biogenesis and assembly |
| 67 | YEL061C | CIN8 | mitotic sister chromatid segregation |
| 68 | YDL056W | MBP1 | DNA replication |
| 69 | YMR035W | IMP2 | mitochondrial protein processing |
| 70 | YBR176W | ECM31 | pantothenate biosynthesis |
| 71 | YOR109W | INP53 | cell wall organization |
| 72 | YOR260W | GCD1 | regulation of translational initiation |
| 73 | YEL030W | ECM10 | protein refolding |
| 74 | YCR036W | RBK1 | ribose metabolism |
| 75 | YDR346C | SVF1 | response to oxidative stress |
| 76 | YPL254W | HFI1 | telomere maintenance |
| 77 | YPR189W | SKI3 | mRNA catabolism |
| 78 | YDR298C | ATP5 | ATP synthesis coupled proton transport |
Table 3.
Manual Confirmation of Selected SJW-sensitive a Deletion Strains
| Sensitivity Rank | Gene/ Strain | Control b | SJW-treated b |
|---|---|---|---|
| 1 | MRC1 | 1.39 | 1.17 |
| 2 | BFR1 | 1.37 | 0.93 |
| 3 | SAM1 | 1.37 | 1.12 |
| 4 | TIF1 | 1.38 | 1.15 |
| 5 | YPS1 | 1.39 | 1.14 |
| 11 | RAS2 | 1.55 | 1.36 |
| 20 | HST4 | 1.53 | 1.31 |
| 21 | UBC11 | 1.52 | 1.18 |
| 45 | PBS2 | 1.50 | 1.21 |
| 76 | GCD1 | 1.53 | 1.20 |
| 77 | ECM10 | 1.49 | 0.72 |
Strains were cultured in SJW extract in YPD broth.
OD600nm reading (median of 8) taken after 19hrs growth.
3.4 Identification of Human Orthologs
Computational analyses using the NCBI BLASTP and HomoloGene databases revealed that 52 of the 78 SJW-sensitive yeast genes have human orthologs. Table 4 shows the list of 52 human orthologs ranked according to the SJW-sensitivity of their yeast counterpart.
Table 4.
Human Orthologs of SJW-sensitive Yeast Genes.
| Yeast Gene | Yeast Rank | Human Ortholog | Human Biological Function | Associated Disease |
|---|---|---|---|---|
| SAM1 | 3 | MAT2A | Alpha methionine adenosyltransferase II | |
| TIF1 | 4 | EIF4A2 | Eukaryotic translation initiation factor 4A isoform 2 | |
| TIF4632 | 6 | EIF4G1 | Eukaryotic translation initiation factor 4 gamma 1 | |
| NCB2 | 8 | DR1 | Down-regulator of transcription 1 | |
| PEP12 | 9 | STX7 | Syntaxin 7; nervous system-specific protein implicated in docking of synaptic vesicles at presynaptic plasma membrane | |
| RAS2 | 11 | KRAS | Kirsten rat sarcoma viral oncogene | |
| TIF4631 | 12 | EIF4G1 | Eukaryotic translation initiation factor 4 gamma, 1 | |
| SOL1 | 13 | PGLS | 6-Phosphogluconolactonase | |
| PAC10 | 15 | VBP1 | von Hippel-Lindau binding protein 1 | |
| HST4 | 19 | SIRT2 | Silent mating type information regulation 2 homolog | |
| UBC11 | 20 | UBE2C | Ubiquitin-conjugating enzyme E2C | |
| ADP1 | 22 | ABCG2 | ATP-binding cassette, sub-family G (Xenobiotic transport) | |
| APN2 | 23 | APEX2 | APEX nuclease 2 (DNA repair) | |
| SUR4 | 28 | ELOVL7 | Unclassified | |
| SPA2 | 29 | GIT1 | G protein-coupled receptor kinase interactor 1, Regulator of membrane trafficking | Huntingtin Disease |
| CHK1 | 32 | CHEK1 | CHK1 checkpoint homolog | |
| YSA1 | 33 | NUDT5 | Nucleoside diphosphate linked moiety X-type motif 5 | |
| GIN4 | 35 | BRSK1 | BR serine/threonine kinase 1 | |
| VAM7 | 37 | SNX12 | Sorting nexin 12 | |
| ROT2 | 38 | GANAB | Alpha glucosidase, neutral AB | |
| BCK2 | 39 | MYO18B | Myosin XVIIIB | |
| HIR2 | 40 | HIRA | HIR histone cell cycle regulation defective homolog A | DiGeorge Syndrome |
| ACS1 | 41 | ACSS1 | Acyl-CoA synthetase short-chain family member 1 | |
| SSD1 | 42 | DIS3 | DIS3 mitotic control homolog | |
| PBS2 | 43 | MEK2 | Mitogen-activated protein kinase kinase 2 | |
| NOT5 | 44 | CNOT3 | CCR4-NOT transcription complex, subunit 3 | |
| MBF1 | 45 | EDF1 | Endothelial differentiation-related factor 1 | |
| PRP18 | 46 | PRPF18 | PRP18 pre-mRNA processing factor 18 homolog | |
| RPL37B | 48 | RPL37 | Ribosomal protein L37 | |
| ABD1 | 49 | RNMT | RNA (guanine-7-) methyltransferase | |
| GCN1 | 50 | GCN1L1 | GCN1 general control of amino-acid synthesis 1-like 1 | |
| POT1 | 51 | ACAA1 | Acetyl-Coenzyme A acyltransferase 1 | Psuedo-Zellweger Syndrome |
| ASR1 | 52 | TRIM2 | Unclassified | |
| RPP1A | 53 | RPLP1 | Large ribosomal protein, P1 | |
| MEP3 | 56 | RHAG | Rh-associated glycoprotein | |
| SFL1 | 57 | HSF4 | Heat shock transcription factor 4 | |
| PIF1 | 59 | PIF1 | PIF1 5'-to-3' DNA helicase homolog | |
| LYS2 | 61 | AASDH | 2-Aminoadipic 6-semialdehyde dehydrogenase | |
| SPP381 | 64 | GABPB2 | GA binding protein transcription factor, beta subunit 2 | |
| YOR292C | 65 | MPV17 | Mitochondrial inner membrane protein | Navajo neurohepatopathy |
| UTP10 | 66 | HEATR1 | Unclassified | |
| CIN8 | 67 | KIF11 | Kinesin family member 11 | |
| MBP1 | 68 | DAPK1 | Death-associated protein kinase 1 | |
| IMP2 | 69 | IMMP2L | Inner mitochondrial membrane peptidase-like | |
| INP53 | 71 | SYNJ2 | Synaptojanin 2 | |
| GCD1 | 72 | EIF2B3 | Eukaryotic translation initiation factor 2B, subunit 3 gamma | |
| ECM10 | 73 | HSPA5 | Heat shock 70kDa protein 5 (glucose-regulated) | Alzheimer's Disease, Bipolar Disorder, Neurodegeneration |
| RBK1 | 74 | RBKS | Ribokinase | |
| SVF1 | 75 | IFNA14 | Interferon, alpha 14 (Immune response) | |
| HFI1 | 76 | PHKG1 | Phosphorylase kinase, gamma 1 | |
| SKI3 | 77 | KIAA0372 | Unclassified | |
| ATP5 | 78 | ATP5O | H+-ATP synthase, mitochondrial F1 complex, O subunit |
3.5. Up-regulation of HIF1α and VEGF Protein Expression by SJW Extract in MDA-MB-231 Cells
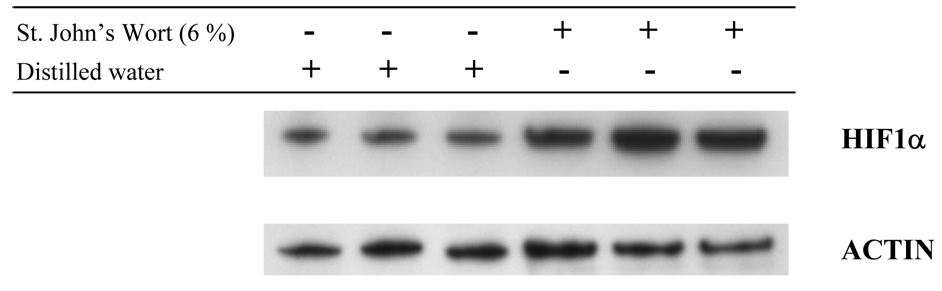
Computational comparative genomic studies of the yeast gene-deletion microarray data implicated the human HIF1α and VEGF signaling pathways as putative targets of SJW. Human UBE2C , an ortholog of the SJW-sensitive yeast gene UBC11, is involved in HIF1α degradation, while activity of human EIF2B3, KRAS, and MEK2, orthologs of the SJW-sensitive yeast genes GCD1, RAS2, and PBS2, respectively, can modulate VEGF signaling (in addition to other activities). To investigate whether SJW extract can affect the protein expression of endogenous HIF1α and VEGF in a human cell line, we treated MDA-MB-231 cells with SJW extract and examined HIF1α and VEGF levels by Western immunoanalysis. Our results revealed that HIF1α protein expression increased 2-3-fold with SJW after 19 h of treatment (Fig 1). VEGF protein was absent from control media samples but was strongly detected after SJW treatment (Fig 2).
Figure 1. HIF1α protein expression in response to St. John’s Wort extract in MDA-MB-231 cells.
Cells were treated with St. John’s Wort as indicated for 19 h. Cell nuclear extracts were analyzed for HIF1α expression by Western blot analysis using antibody to HIF1α. Blots for actin confirm equal loading of the samples.
Figure 2. VEGF protein expression in response to St. John’s Wort extract in MDA-MB-231 cell culture medium.
Cells were treated with St. John’s Wort as indicated for 19 h. Cell culture medium was concentrated and analyzed for VEGF expression by Western blot analysis using antibody to VEGF. The blot was stripped and stained with Coomassie Brilliant Blue to verify equal loading of the samples.
3.6. Modulation of SIRT2 Protein Expression and α-Tubulin Acetylation by SJW Extract in MBA-MB-231 Cells
Computational comparative genomic studies of the yeast gene-deletion microarray data implicated human SIRT2, an ortholog of the SJW-sensitive yeast gene HST4, as a putative target of SJW. We further investigated whether SJW extract can modulate SIRT2, a deacetylase, by examining the actual protein expression of endogenous SIRT2 and acetylated α tubulin in MDA-MB-231 cells, where both proteins are expressed. Our results revealed that SIRT2 protein expression decreased to background level after SJW exposure (Fig 3). Acetylated α-tubulin levels increased from background level to a strong signal after 19 h of treatment with SJW (Fig 3), further suggesting reduced SIRT2 deacetylase activity.
Figure 3. SIRT2, acetylated α-tubulin, and α-tubulin protein expression in response to St. John’s Wort extract in MDA-MB-231 cells.
Cells were treated with St. John’s Wort as indicated for 19 h. Cell cytoplasmic extracts were analyzed for expression of each protein by Western blot analysis using antibody to SIRT2, acetylated α-tubulin, or α-tubulin.
4. DISCUSSION
The present study demonstrates the power of chemical-genetic profiling in yeast as a model system to predict potential human molecular targets of bioactive botanical products. First, using SJW, we show that the use of a high-density yeast barcode microarray has considerable power to identify SJW-sensitive yeast deletion strains not previously described as sensitive to SJW. Second, using a microarray containing multiple replicate barcode features, we show that the strains can be ranked according to their sensitivity to SJW. Finally, using computational comparative genomics techniques and human cell-based immunocytochemical assays, we demonstrate that the identification of SJW-sensitive genes in yeast with the barcode microarray facilitated the identification of SJW-sensitive orthologous human genes linked to angiogenesis, a process involved in wound recovery and tumor growth, and genes linked to the predisposition of neurodegenerative and psychiatric genetic diseases.
Yeast has long been used as a human genetic model system. It is easy to culture, genetically tractable, and has a genome with approximately 44% homology to the human genome. Comparative genomics techniques that can integrate and interrogate yeast and human molecular information can be used to elucidate the answers to functional genomics questions. Therefore, identification of botanical molecular targets in yeast may help identify potential orthologous targets in humans, based on the conservation of homologous genes and proteins throughout phylogeny. Interestingly, although sequence and structural aspects may be conserved throughout evolution, that is not always the case for molecular function, such that orthologs of molecular targets which mediate a protective function in yeast may have a dramatically different function in human physiology.
We identified 78 genes in yeast represented by SJW-sensitive strains for which the signal intensity in the untreated sample was higher than that for the treated sample (Table 2). Biological processes represented by genes in this list include transport, vesicle-mediated transport, signal transduction, protein modification, lipid metabolism, transcription, and translation, among others. In yeast, these genes are identified as serving an important protective role or essential function in the adaptive response(s) to the cytotoxic challenge incurred by SJW exposure.
Using sequence-based comparison techniques, we determined that 52 of the 78 SJW-sensitive yeast genes have human orthologs (Table 4). The top molecular functions were associated with cellular growth, development, assembly, and organization and cell death of brain and central nervous system cells. It should be noted that the yeast genome does not contain genes for cytochrome P450 enzymes, which may explain the absence of such genes from the list of human orthologs.
Four of the 52 human orthologs are associated with angiogenesis, the formation of new blood vessels. These orthologs include UBE2C, KRAS, MEK2, and EIF2B3. Computational analysis associated UBE2C with angiogenesis at the level of HIF1α degradation. The potential for HIF1α modulation by SJW was investigated by Western blot analysis of SJW-treated MDA-MB-231 cells. Protein expression of HIF1α increased after 19 h of exposure to SJW (Fig. 1). Computational analysis associated KRAS, MEK2, and EIF2B3 with angiogenesis at the level of VEGF signaling. The potential for VEGF modulation by SJW was investigated by Western blot analysis of the culture medium of MDA-MB-231 cells treated with SJW. VEGF protein levels in the culture medium of MDA-MB-231 cells increased after 19 h of treatment with SJW (Fig. 2). The ability of SJW to activate HIF1α and VEGF protein expression suggests a potential for SJW to promote wound healing, recovery, or cell growth processes through the activation of angiogenesis to increase blood flow through new blood vessel formation.
Six of the 52 identified human orthologs were associated with neurological diseases or psychiatric disorders. These genes include GIT1, HIRA, ACAA1, MPV17, HSPA5, and SIRT2. Our focus was attracted to SIRT2, which encodes an NAD (+)-dependent protein deacetylase involved in alpha-synuclein-mediated toxicity in cellular models of Parkinson's disease (25). The potential for SIRT2 modulation by SJW was investigated by Western blot analysis of SJW-treated MDA-MB-231 cells. SIRT2 protein levels decreased after 19 h of treatment with SJW (Fig. 3). Provided results similar to ours can be repeated in neuronal cells, the ability of SJW to deactivate SIRT2 may have implications for potential therapeutic use of SJW in treatment of Parkinson Disease, as SIRT2 inactivation has been found to restrict Lewy body formation and lead to the rescue of neuronal cells (25).
It should be noted that the bioavailability of SJW compounds within the body and at physiological target sites will play an important role in the evaluation of potential mode of action models generated by genetic studies. Unfortunately, while many reports exist which describe the effect of SJW on the bioavailability of drugs taken concomitantly, few studies have been conducted to investigate the pharmacokinetic profile of SJW compounds in humans after oral ingestion. However, in the case of our suggestion herein that SJW may promote wound healing through the activation of angiogenic activities, our hypothesis is bolstered by the understanding that traditionally SJW salves were applied topically to wounds, which may allow SJW compounds direct access to target cells at the wound site.
In summary, we have shown that chemical-genetic profiling in yeast represents a powerful tool for the identification of human intracellular targets of dietary, bioactive botanical products, such as the medicinal plant SJW. We identified 78 genes previously undescribed as essential to the adaptive response to SJW. Fifty-two of these yeast genes have human orthologs, some of which have been implicated in neurological diseases, psychiatric disorders, angiogenesis, and human cancer. Three putative human intracellular targets predicted by chemical-genetic profiling in yeast (HIF1α, VEGF, and SIRT2) were confirmed by cell-based assays of SJW-treated human cells. Future research will seek to better understand the role of individual constituents of SJW in the adaptive responses observed using the whole SJW extract. We envision that the approach described here may be useful to the functional food and agricultural research community in the determination of molecular mechanisms of action for bioactive botanical products as whole extracts, the form in which they are most commonly used in traditional medicine, prior to the identification of singular bioactive constituents. Subsequently, individual compounds and/ or synergies among constituents that are responsible for various aspects of the overall molecular mechanism can be elucidated by comparison of individual chemogenomic profiles to that of the whole extract.
Acknowledgments
P.M. acknowledges funding support through a Director’s Fellowship from the National Center for Complementary and Alternative Medicine (NCCAM).
LITERATURE CITED
- 1.Charrois TL, Sadler C, Vohra S. Complementary, holistic, and integrative medicine: St. John's wort. Pediatr Rev. 2007;28:69–72. doi: 10.1542/pir.28-2-69. [DOI] [PubMed] [Google Scholar]
- 2.Linde K, Berner M, Egger M, Mulrow C. St John's wort for depression: meta-analysis of randomised controlled trials. Br. J. Psychiatry. 2005;186:99–107. doi: 10.1192/bjp.186.2.99. [DOI] [PubMed] [Google Scholar]
- 3.Schempp CM, Kirkin V, Simon-Haarhaus B, Kersten A, Kiss J, Termeer CC, Gilb B, Kaufmann T, Borner C, Sleeman JP, Simon J. Inhibition of tumour cell growth by hyperforin, a novel anticancer drug from St. John's wort that acts by induction of apoptosis. Oncogene. 2002;21:1242–1250. doi: 10.1038/sj.onc.1205190. [DOI] [PubMed] [Google Scholar]
- 4.Jayasuriya H, McChesney JD, Swanson SM, Pezzuto JM. Antimicrobial and cytotoxic activity of rottlerin-type compounds from Hypericum drummondii. J. Nat. Prod. 1989;52:325–331. doi: 10.1021/np50062a018. [DOI] [PubMed] [Google Scholar]
- 5.Ferraz A, Faria DH, Benneti MN, da Rocha AB, Schwartsmann G, Henriques A, von Poser GL. Screening for antiproliferative activity of six southern Brazilian species of Hypericum. Phytomedicine. 2005;12:112–115. doi: 10.1016/j.phymed.2003.07.015. [DOI] [PubMed] [Google Scholar]
- 6.Patocka J. The chemistry, pharmacology, and toxicology of the biologically active constituents of the herb Hypericum perforatum L. J. Appl. Biomed. 2003;1:61–70. [Google Scholar]
- 7.Ioannides C. Pharmacokinetic interactions between herbal remedies and medicinal drugs. Xenobiotica. 2002;32:451–478. doi: 10.1080/00498250210124147. [DOI] [PubMed] [Google Scholar]
- 8.Shoemaker DD, Lashkari DA, Morris D, Mittmann M, Davis RW. Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nat. Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- 9.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 10.Giaever G, Flaherty P, Kumm J, Proctor M, Nislow C, Jaramillo DF, Chu AM, Jordan MI, Arkin AP, Davis RW. Chemogenomic profiling: identifying the functional interactions of small molecules in yeast. Proc. Natl. Acad. Sci. U. S. A. 2004;101:793–798. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birrell GW, Giaever G, Chu AM, Davis RW, Brown JM. A genome-wide screen in Saccharomyces cerevisiae for genes affecting UV radiation sensitivity. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12608–12613. doi: 10.1073/pnas.231366398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doostzadeh J, Davis RW, Giaever GN, Nislow C, Langston JW. Chemical genomic profiling for identifying intracellular targets of toxicants producing Parkinson's disease. Toxicol. Sci. 2007;95:182–187. doi: 10.1093/toxsci/kfl131. [DOI] [PubMed] [Google Scholar]
- 13.McCue P, Zheng Z, Pinkham JL, Shetty K. A model for enhanced pea seedling vigour following low pH and salicylic acid treatments. Process Biochem. 2000;35:603–613. [Google Scholar]
- 14.McCue P, Kwon YI, Shetty K. Anti-diabetic and anti-hypertensive potential of sprouted and solid-state bioprocessed soybean. Asia Pac. J. Clin. Nutr. 2005;14:145–152. [PubMed] [Google Scholar]
- 15.Ang CY, Cui Y, Chang HC, Luo W, Heinze TM, Lin LJ, Mattia A. Determination of St. John's wort components in dietary supplements and functional foods by liquid chromatography. J. AOAC Int. 2002;85:1360–1369. [PubMed] [Google Scholar]
- 16.Yuan DS, Pan X, Ooi SL, Peyser BD, Spencer FA, Irizarry RA, Boeke JD. Improved microarray methods for profiling the Yeast Knockout strain collection. Nucleic Acids Res. 2005;33:e103. doi: 10.1093/nar/gni105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peyser BD, Irizarry RA, Tiffany CW, Chen O, Yuan DS, Boeke JD, Spencer FA. Improved statistical analysis of budding yeast TAG microarrays revealed by defined spike-in pools. Nucleic Acids Res. 2005;33:e140. doi: 10.1093/nar/gni138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarimón J, Bertranpetit J, Boada M, Tàrraga L, Comas D. HSP70-2 (HSPA1B) is associated with noncognitive symptoms in late-onset Alzheimer"s disease. J. Geriatr. Psychiatry Neurol. 2003;16:146–150. doi: 10.1177/0891988703256051. [DOI] [PubMed] [Google Scholar]
- 19.Wu YR, Wang CK, Chen CM, Hsu Y, Lin SJ, Lin YY, Fung HC, Chang KH, Lee-Chen GJ. Analysis of heat-shock protein 70 gene polymorphisms and the risk of Parkinson's disease. Hum. Genet. 2004;114:236–241. doi: 10.1007/s00439-003-1050-1. [DOI] [PubMed] [Google Scholar]
- 20.Fung HC, Chen CM, Wu YR, Hsu WC, Ro LS, Lin JC, Chang KH, Wang HK, Lin SJ, Chan H, Lin YY, Wei SL, Hsu Y, Hwang JC, Tung LC, Lee-Chen GJ. Heat shock protein 70 and tumor necrosis factor alpha in Taiwanese patients with dementia. Dement. Geriatr. Cogn. Disord. 2005;20:1–7. doi: 10.1159/000085067. [DOI] [PubMed] [Google Scholar]
- 21.Pae CU, Kim TS, Kwon OJ, Artioli P, Serretti A, Lee CU, Lee SJ, Lee C, Paik IH, Kim JJ. Polymorphisms of heat shock protein 70 gene (HSPA1A, HSPA1B and HSPA1L) and schizophrenia. Neurosci. Res. 2005;53:8–13. doi: 10.1016/j.neures.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Pae CU, Mandelli L, Serretti A, Patkar AA, Kim JJ, Lee CU, Lee SJ, Lee C, De Ronchi D, Paik IH. Heat-shock protein-70 genes and response to antidepressants in major depression. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2007;31:1006–1011. doi: 10.1016/j.pnpbp.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 23.Goehler H, Lalowski M, Stelzl U, Waelter S, Stroedicke M, Worm U, Droege A, Lindenberg KS, Knoblich M, Haenig C, Herbst M, Suopanki J, Scherzinger E, Abraham C, Bauer B, Hasenbank R, Fritzsche A, Ludewig AH, Büssow K, Coleman SH, Gutekunst CA, Landwehrmeyer BG, Lehrach H, Wanker EE. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol. Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 24.Karadimas CL, Vu TH, Holve SA, Chronopoulou P, Quinzii C, Johnsen SD, Kurth J, Eggers E, Palenzuela L, Tanji K, Bonilla E, De Vivo DC, DiMauro S, Hirano M. Navajo neurohepatopathy is caused by a mutation in the MPV17 gene. Am. J. Hum. Genet. 2006;79:544–548. doi: 10.1086/506913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, Volk CB, Maxwell MM, Rochet JC, McLean PJ, Young AB, Abagyan R, Feany MB, Hyman BT, Kazantsev AG. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science. 2007;317:516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]