Abstract
Conformational dynamics are thought to play an important role in ligand binding and catalysis by cytochrome P450 enzymes but few techniques exist to examine them in molecular detail. Using a unique isotopic labeling strategy, we have site specifically inserted a 13C-labeled unnatural amino acid residue, 13C-p-methoxyphenylalanine (MeOF), into two different locations in the substrate binding region of the thermophilic cytochrome P450 enzyme CYP119. Surprisingly, in both cases the resonance signal from the ligand-free protein is represented by a doublet in the 1H,13C-HSQC spectrum. Upon binding of 4-phenylimidazole, the signals from the initial resonances are reduced in favor a single new resonance, in the case of the F162MeOF mutant, or two new resonances, in the case of the F153MeOF mutant. This represents the first direct physical evidence for the ligand-dependent existence of multiple P450 conformers simultaneously in solution. This general approach may be used to further illuminate the role that conformational dynamics plays in the complex enzymatic phenomena exhibited by P450 enzymes.
It has become increasingly clear that protein dynamics are required for enzymatic function.1-3 In fact, the native state of an enzyme can be described as a statistical ensemble of different conformational substates with similar free energies.4 Modern NMR techniques have proved successful at connecting conformational substates and their respective free energies with their individual contributions to catalysis.5 The cytochrome P450 (CYP) enzyme superfamily is a ubiquitous set of heme-containing monooxygenases that typically catalyze the incorporation of one atom of molecular oxygen into a hydrocarbon substrate.6 P450 enzymes are known to exhibit a wide range of dynamic enzymatic behavior, including homotropic and heterotropic cooperativity, sequential oxidation, and substrate inhibition.7 Due to their importance in mammalian drug metabolism and their potential utility as model systems for the study of complex protein dynamics, we report here the application of a unique high-resolution 2D NMR technique for the examination of ligand binding and its contribution to conformational dynamics in this major enzyme family. CYP119 from Sulfolobus solfataricus is a thermophilic P450 that is monomeric and stable at temperatures up to 90°C.8 Ligand-free and ligand-bound crystal structures of this protein suggest that dramatic conformational changes occur upon ligand binding, particularly in the F/G loop region, that result in an overall “closing off” of the active site.8 In order to better understand the structural and thermodynamic contributions of specific residues in this dynamic region of the protein, we have introduced a 13C-labeled p-methoxyphenylalanine (13C-MeOF) unnatural amino acid site-specifically into two unique positions in the F/G loop, F153 and F162, to monitor the effects of ligand binding using 2D 1H,13C-HSQC NMR.9 The advantage of this technique over global labeling of the protein backbone with 15N is that it allows for site-selective incorporation of a single, specific amino acid in vivo, which results in the production of milligram quantities of protein and circumvents the need to make spectral residue assignments. SDS-PAGE analysis of the CYP119-F162-13C-MeOF mutant indicated the presence of a single band of the correct molecular weight (supplemental materials, Figure S1). Whole-protein LC-MS yielded the predicted mass shift of +31 Daltons, demonstrating the incorporation of a single 13C-MeOF residue (Figures S2 & S3). In order to assess the integrity of the labeled protein, a variety of ligands were examined for their ability to bind and induce a spin-state shift in the heme Soret bands (Figure S4). All ligands examined exhibited Kd(app) values similar to those of the wild-type enzyme (Table S1). Surprisingly, initial examination of the 1H,13C-HSQC spectra for both mutants revealed the presence of two symmetrical resonances of equal intensity, separated by ∼0.12 ppm in the 1H dimension (Figure 1, Figure S5 & S6). Since the three protons connected to the 13C-labeled methoxy moiety are chemically equivalent, these two resonances can only be explained by the existence of two distinct conformers in slow chemical exchange, either conformational substates of the protein itself, or specific ring-flip rotomers of the 13C-labeled p-methoxyphenylalanine residue.10 This is further illustrated by coalescence of the two resonances with increasing temperature (Figure S7). Ring-flip rotomers of tyrosine and phenylalanine have previously been shown to be distinguishable on the NMR time scale used in these experiments (i.e., nsec to msec).10,11 Regardless of the origin of the doublet, the signal occurred in all CYP11913CMeOF mutants examined. In order to monitor the dynamic structural changes that occur upon ligand binding, HSQC experiments were performed with a model ligand at a variety of concentrations. The ligand 4-phenylimidazole (4-PI) was chosen due to the existence of a crystal structure and the potential utility of imidazole derivatives as scaffolds for isoform-selective P450 inhibitors.12 Upon addition of the 4-PI ligand to the F162-13C-MeOF labeled mutant, a single new resonance appeared in the spectrum with a chemical shift of 3.67 ppm in the 1H dimension and 57.3 ppm in the 13C dimension (Figure 2b-d, Figure S5). The intensity of the resonance increased with increasing concentration of ligand until reaching saturation at a 1:1 molar stoichiometry. 4-PI is known to form a 1:1 complex with CYP119 with a Kd(app) of ∼120 nM (Table S1).8 In the absence of labeled protein, no resonances are detected in this region of the spectrum, further indicating that this resonance is due to ligand-bound protein (data not shown). It is interesting to note that, even at saturation, the resonances representing the ligand-free species are still present. This phenomenon has been seen previously in a similar study monitoring the binding of a model compound to the human fatty acid synthase.9 This may indicate that the “ligand free” ground state conformation of the enzyme, represented by the corresponding crystal structure, may be accessible even in the presence of saturating amounts of ligand. In comparison, upon titration with 4-phenylimidazole the F153-13C-MeOF mutant initially showed a similar pattern with a single resonance appearing at 1H 3.65 ppm, 13C 58.2 ppm, increasing in intensity with increasing ligand concentration (Figure 3, Figure S6). However, at higher concentrations of ligand, this resonance begins to decrease in intensity as a new resonance appears at 1H 3.65 ppm and 13C 62.3 ppm in the spectrum (Figures S6d,e), suggesting that the 1H 3.65 ppm, 13C 58.2 ppm resonance represents a conformational intermediate on the path to the ground state (crystal structure) conformer. The data suggest that this large scale conformational change proceeds through a stable intermediate that is readily detectable on the timescale of the 1H,13C HSQC NMR experiments. In comparison, the F162-13C-MeOF residue appears to be insensitive to this conformational intermediate. It has recently been demonstrated that mammalian P450s may form an initial, non-productive, “encounter” complex with ligands that leads to secondary conformational changes that transition the ligand into a productive binding orientation within the active site.13 This phenomenon may be represented in the CYP119 F153-13C-MeOF mutant by the appearance of a transient complex at low ligand occupancy.
Figure 1.
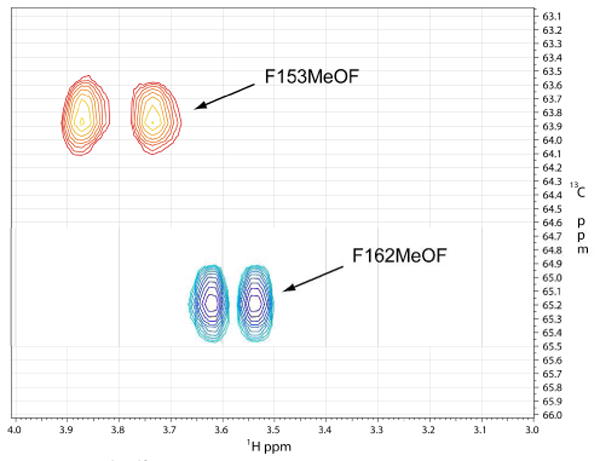
1H,13C-HSQC spectra of CYP119-F153MeOF (red) superimposed upon the 1H,13C-HSQC spectra of CYP119-F162MeOF (blue).
Figure 2.
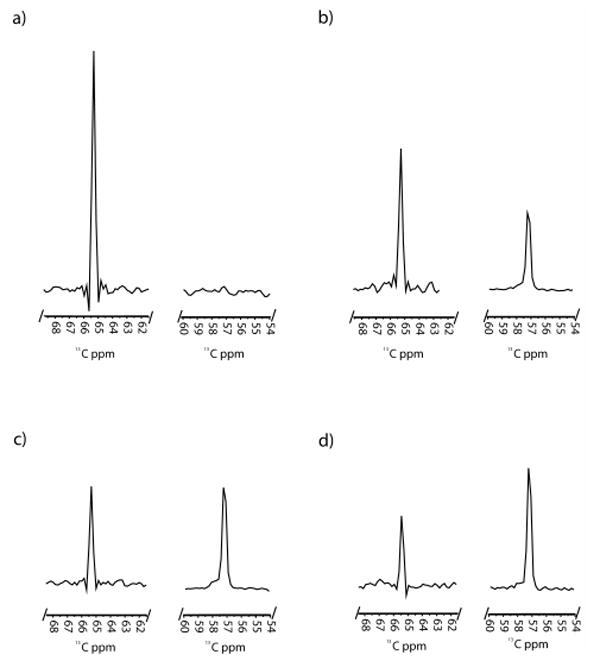
1D slices from the 13C axis of the 1H,13C-HSQC spectra of CYP119-F162-13CMeOF with a) 0 M equivalents of 4-PI, b) 0.3 M equivalents of 4-PI, c) 0.9 M equivalents of 4-PI, and d) 1.2 M equivalents of 4-PI. Slices were taken at the mid-point of the resonance in the 1H dimension. All figures are scaled relative to the same y-axis.
Figure 3.
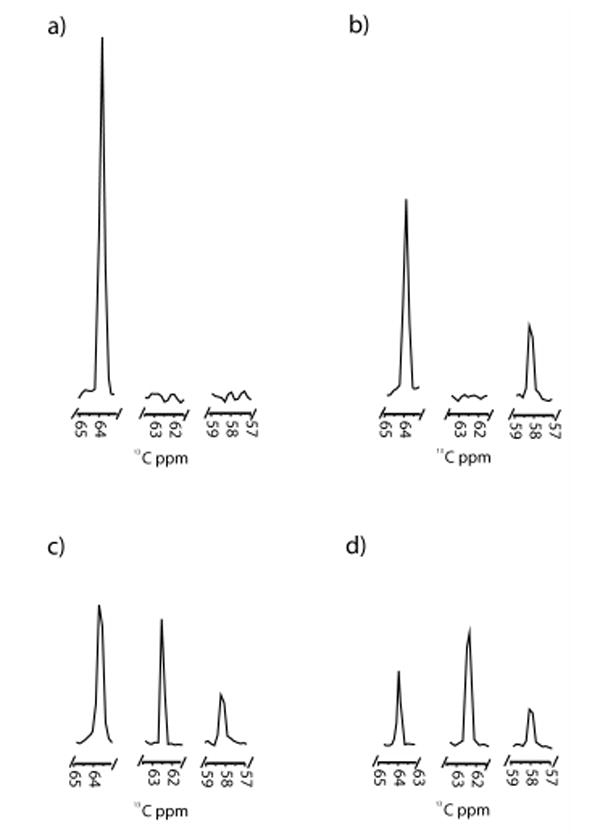
1D slices from the 13C axis of the 1H,13C-HSQC spectra of CYP119-F153-13CMeOF with a) 0 M equivalents of 4-PI, b) 0.3 M equivalents of 4-PI, c) 0.6 M equivalents of 4-PI, and d) 1.2 M equivalents of 4-PI. Slices were taken at the mid-point of the resonance in the 1H dimension. All figures are scaled relative to the same y-axis.
In summary, here we have demonstrated the efficacy of a unique NMR approach utilizing isotopically-labeled artificial amino acids to examine ligand-dependent conformational substates of a model cytochrome P450. In the future, this approach may also shed additional light on the role conformational dynamics plays in the complex enzymatic phenomena exhibited by mammalian cytochrome P450 enzymes, such as heterotropic cooperativity and substrate promiscuity. Moreover, the approach may be universally applicable in studying complex enzymatic behavior in a variety of systems.
Supplementary Material
Experimental procedures, UV-vis and NMR data and full reference 9. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
We thank Peter Schultz for the unnatural amino acid expression system. This work as supported by National Institute of Health grant GM25515.
References
- 1.Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M, Kern D. Nature. 2007;450:913–6. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 2.Pochapsky SS, Pochapsky TC, Wei JW. Biochemistry. 2003;42:5649–5656. doi: 10.1021/bi034263s. [DOI] [PubMed] [Google Scholar]
- 3.Kijac AZ, Li Y, Sligar SG, Rienstra CM. Biochemistry. 2007;46:13696–703. doi: 10.1021/bi701411g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.James LC, Tawfik DS. Trends Biochem Sci. 2003;28:361–8. doi: 10.1016/S0968-0004(03)00135-X. [DOI] [PubMed] [Google Scholar]
- 5.Mittermaier A, Kay LE. Science. 2006;312:224–8. doi: 10.1126/science.1124964. [DOI] [PubMed] [Google Scholar]
- 6.Ortiz de Montellano PR. Cytochrome P450: Structure, Mechanism, and Biochemistry. 3rd. Kluwer Academic/Plenum Publishers; New York: 2005. [Google Scholar]
- 7.Atkins WM. Annu Rev Pharmacol Toxicol. 2005;45:291–310. doi: 10.1146/annurev.pharmtox.45.120403.100004. [DOI] [PubMed] [Google Scholar]
- 8.Yano JK, Koo LS, Schuller DJ, Li H, Ortiz de Montellano PR, Poulos TL. J Biol Chem. 2000;275:31086–92. doi: 10.1074/jbc.M004281200. [DOI] [PubMed] [Google Scholar]
- 9.Cellitti SE, et al. J Am Chem Soc. 2008;130:9268–81. doi: 10.1021/ja801602q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Battiste JL, Li R, Woodward C. Biochemistry. 2002;41:2237–45. doi: 10.1021/bi011693e. [DOI] [PubMed] [Google Scholar]
- 11.Wagner G, Wuthrich K. Nature. 1978;275:247–8. doi: 10.1038/275247a0. [DOI] [PubMed] [Google Scholar]
- 12.Verras A, Kuntz ID, Ortiz de Montellano PR. J Med Chem. 2004;47:3572–9. doi: 10.1021/jm030608t. [DOI] [PubMed] [Google Scholar]
- 13.Isin EM, Guengerich FP. J Biol Chem. 2006;281:9127–36. doi: 10.1074/jbc.M511375200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental procedures, UV-vis and NMR data and full reference 9. This material is available free of charge via the Internet at http://pubs.acs.org.


