Abstract
Glucocorticoids (GCs) generally stimulate gene transcription via consensus glucocorticoid response elements (GREs) located in the promoter region. To identify the GRE in the rat tyrosine hydroxylase (TH) gene promoter, we transiently transfected PC12 cells with a 9-kilobase (kb) TH promoter-luciferase (Luc) construct. Dexamethasone (Dex) stimulated Luc activity, which was abolished by mifepristone (RU486). Serial deletion mutations revealed a Dex-responsive 7-base pair (bp) sequence, TGACTAA, located at -5734 to -5728. Deletion of just these seven nucleotides from the 9-kb promoter completely abolished the Dex response and partially reduced the response to phorbol ester but not to forskolin. The Dex response was fully retained in a construct in which most of the 9-kb promoter was deleted, except for 100 bp around the -5.7-kb region, clearly identifying this 7-bp sequence as solely responsible for GC responsiveness. Conversely, deletion of the proximal cAMP-response element (-45/-38) or activator protein-1 (AP-1) (-207/-201) sites in the 9-kb promoter did not affect Dex and phorbol ester responses. A radiolabeled 25-bp promoter fragment bearing the 7-bp TH-GRE/AP-1 showed specific binding to PC12 nuclear proteins. Using antibodies against the glucocorticoid receptors and AP-1 family of proteins and primers for the TH-GRE/AP-1 region, we detected a specific DNA amplicon in a chromatin immunoprecipitation assay. This 7-bp TH-GRE/AP-1 sequence (TGACTAA) does not bear similarity to any known GRE but closely resembles the consensus AP-1 binding site, TGACTCA. Our studies describe for the first time a novel GRE/AP-1 site present in the TH gene promoter that is critical for glucocorticoid regulation of the TH gene.
Glucocorticoids use several mechanisms to bring about transactivation or transrepression of genes (De Bosscher et al., 2003; Rhen and Cidlowski, 2005; Newton and Holden, 2007). One of the main mechanisms includes direct effects on gene expression by the binding of glucocorticoid receptors (GRs) to glucocorticoid responsive elements (GREs) in the promoter region of genes (Scheidereit et al., 1983). The consensus GRE consists of two half sites each consisting of 6 bp separated by three nucleotides (5′-AGAACANNNTGTTCT) (Truss and Beato, 1993). However, there is considerable degeneracy in the GREs, and often one half-site with even a few base changes has been found to be sufficient for activity (Chan et al., 1991; Del Monaco et al., 1997). Glucocorticoids can also exert indirect effects on gene expression through interactions with other transcription factors such as AP-1, Sp1, nuclear factor-κB, Nurr1, or CCAAT/enhancer-binding protein-α (Drouin et al., 1989; Mordacq and Linzer, 1989; Jonat et al., 1990; Yang-Yen et al., 1990; Uht et al., 1997; Rudiger et al., 2002). Thus, GR can act without directly binding to a classic GRE and influence the transcription of genes that do not have a classic DNA binding site for GR (Teurich and Angel, 1995; Rudiger et al., 2002). Similar mechanisms are also known for other steroid receptors, including the progesterone and estrogen receptors (Uht et al., 1997; Owen et al., 1998).
Stimulation of GR is known to increase tyrosine hydroxylase gene transcription. Tyrosine hydroxylase (TH) is an important enzyme that catalyzes the first and rate-limiting step of oxidative hydroxylation of tyrosine in the biosynthesis of catecholamines, norepinephrine, epinephrine, and dopamine. Catecholamine levels are affected by a variety of stimuli, including neurotransmitters, growth factors, estrogen, glucocorticoids, and membrane depolarization, many of which exert regulation at the transcriptional level of the TH gene. TH is expressed in specific neurons of the central nervous system and in the periphery in sympathetic ganglia and adrenal medullary chromaffin cells. TH is also expressed in PC12 cells, a cell line derived from a pheochromocytoma tumor of the rat adrenal medulla that has served as a widely used model to study the molecular mechanisms of TH regulation. Thus, stimulation of GR has been shown to increase TH mRNA in various systems (Fossom et al., 1992; Hagerty et al., 2001a). The promoter of the TH gene has been extensively studied (Kumer and Vrana, 1996), and a number of cis-acting motifs including CRE, AP-1, AP-2, Sp1, and POU/Oct, in the proximal promoter region have been identified (Fung et al., 1992). Lewis et al. (1987) also identified a GRE-like sequence with 10 of 11 bases resembling the pal-indromic GRE (5′-AGTACACTTTGTTCT-3′) at approximately -454 bp in the rat TH 5′-flanking region. However, this sequence turned out to be nonfunctional, having no response to dexamethasone (Dex) either in the context of a TH gene containing -773 to +27 bp fused to the CAT reporter gene or in a heterologous system. In the present study, we used a longer 9-kb DNA of rat TH gene promoter cloned in the pGL3-Basic vector to identify the putative GRE responsible for the stimulatory effect of dexamethasone using the PC12 cell line. Our studies have identified a 7-bp sequence located approximately 5.7 kb from the transcription start site that resembles an AP-1 binding site and is necessary for glucocorticoid responsiveness.
Materials and Methods
Plasmid DNA and Site-Directed Mutagenesis. We used the 5′-flanking region of the rat TH gene containing -9637 bp of nucleotides upstream of the transcription start site to nucleotide +15, cloned between the XhoI and HindIII sites of the luciferase reporter plasmid, pGL3-Basic (Promega, Madison, WI), bearing the firefly luciferase gene (Min et al., 1994). This construct was used in an earlier study by Kobayashi and coworkers to identify the Nurr1 response element in the TH gene promoter (Iwawaki et al., 2000). We also used several deletion constructs that were derived from this -9-kb construct by restriction enzyme digestion. First, we confirmed the authenticity of the entire sequence of this 9-kb long promoter construct (referred to as the full-length construct) by sequence analysis and compared it with the 5′-flanking sequence of the rat TH gene in the ENSEMBL genome database (Gene ID 25085). We then performed mutations of the 9-kb promoter that deleted specific sequences or entire segments of the 5′-region up to a specified bp using the QuikChange II XL Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). For this, we designed forward and reverse primers upstream and downstream, respectively, of the region to be deleted and used primers purified by polyacrylamide gel electrophoresis or reverse phase chromatography (Integrated DNA Technologies Inc., Coralville, IA) for PCR along with the -9-kb TH-Luc DNA as template, according to the method described by the manufacturer. The entire reaction was subjected to DpnI digestion, which cleaves only the methylated parental DNA, leaving the mutated amplified DNA that was used for transforming XL10-Gold Ultracompetent cells. Plasmid DNA was isolated from several colonies, cut with restriction enzymes, and appropriately sized DNA was verified by sequencing. Selected DNA was then used to transform Escherichia coli DH5α, from which plasmid DNA was isolated after a Maxiprep protocol (Promega) and further purified by polyethylene glycol treatment to remove any contaminating RNA. Additional purification by CsCl gradient centrifugation followed by ethanol precipitation did not yield any greater activity in transfection assays and hence was not routinely used. The purified plasmid DNA was used for transient transfection of cells.
Cell Culture and Transfection. Wild-type (WT) PC12 cells, a cell line established from rat adrenal pheochromocytoma tumors (Van Buskirk et al., 1985), were grown in media consisting of Dulbecco's modified Eagle's medium/Ham's F-12 media (1:1) supplemented with 10% bovine calf serum, 5% horse serum, 2 mM l-glutamine, and antibiotics (20 U/ml penicillin and 20 μg/ml streptomycin) in a humidified 5% CO2 incubator at 37°C. In some experiments, we also used a protein kinase A-deficient PC12 cell line (A126-1B2) (Van Buskirk et al., 1985) grown under identical conditions.
For transient transfection, 24-well cell culture plates coated with poly(l-lysine) were plated with ∼1 × 105 cells/well, and after 18 to 24 h at approximately 80% confluence, cells were moved to OptiMEM (Invitrogen, Carlsbad, CA), a reduced serum medium. Plasmid DNA constructs were delivered into cells using Lipofectamine 2000 reagent (Invitrogen) or TurboFectin 8.0 (Origene Technologies, Rockville, MD) according to the manufacturer's recommendations. Cotransfections were performed with 0.5 μg of DNA per well of the above promoter constructs in the pGL3-Basic vector bearing the firefly luciferase gene along with 0.05 μg/well of a synthetic Renilla reniformis luciferase reporter vector (phRG-Basic; Promega), with the latter used for monitoring transfection efficiency. The total molar amount of DNA per well was kept constant by adjusting the molarity with pBluescript DNA when using shorter constructs. When Lipofectamine 2000 was used for transfection, media containing 2× concentration of sera were added after approximately 5 h, and incubation continued for 12 to 16 h, whereas the cells transfected using TurboFectin were allowed to incubate until the next day without further changes. In later experiments, we switched to using TurboFectin, because these transfected cells seemed healthier. The transfection agent used is mentioned in the figure legends. For treatment, cells were transferred to a serum-free Neurobasal A medium (NBM) supplemented with 2 mM Glutamax and N2 supplement (Invitrogen). Cells were incubated with treatment agents (all from Sigma-Aldrich, St. Louis, MO) including dexamethasone, 12-O-tetradecanoylphorbol 13-acetate (TPA/phorbol ester), or forskolin (EMD Biosciences, San Diego, CA) for up to 24 h. All treatments consisted of replicates of three to four wells per group, and experiments were repeated at least three times. Control cells received appropriate concentrations of vehicles used (ethanol or DMSO at ≤0.01%).
Dual Luciferase Assay. The Dual-Luciferase Reporter assay system (Promega) was used according to the manufacturer's instructions. Cells were washed with Dulbecco's phosphate-buffered saline and lysed in 100 μl of 1× passive lysis buffer and 20 μl of the lysate used to measure firefly luciferase (Luc), and R. reniformis luciferase activities consecutively using the Berthold Lumat LB 9507 luminometer (Berthold Technologies, Bad Wildbad, Germany). The ratios of firefly to R. reniformis Luc activities were calculated to correct for differences in transfection efficiency. The average ratio of the untreated control group was used to calculate the percentage of change in the reporter activity of treated groups, and results are expressed as the mean ± S.E.M. percentage of control for each individual construct.
Statistics. The luciferase activity ratio data obtained as a percentage of control were subjected to statistical analysis by analysis of variance followed by Newman-Keuls multiple comparison test using the GraphPad Prism 5 software package (GraphPad Software, Inc., San Diego, CA). A p value <0.05 is considered significant.
Electrophoretic Mobility Shift Assay. Nuclear extracts were prepared essentially according to Schreiber et al. (1989). In brief, PC12 cells (∼5 × 106) were plated in 100-mm plates, serum deprived for a day in NBM medium, and treated with 100 nM Dex for 1 h. Thereafter, all operations were carried out at 4°C. Cells were scraped, washed with phosphate-buffered saline, and resuspended in 1 ml of hypotonic buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 200 mM sucrose, and 0.5% Nonidet P-40). After allowing cells to swell on ice for 15 min, they were vortexed vigorously for 10 s and centrifuged at 12,000g for 5 min. The nuclear pellet was resuspended in the equivalent of one packed cell volume of buffer C (20 mM HEPES, pH 7.9, 0.42 M NaCl, 1.5 mM MgCl2, and 0.2 mM EDTA), and the nuclear proteins were extracted by incubation on ice for 30 min, with vortexing every 5 min. The supernatant obtained by centrifugation at 12,000g was diluted by adding an equal volume of buffer D (20 mM HEPES, pH 7.9, 100 mM KCl, 0.2 mM EDTA, and 20% glycerol) and then divided into aliquots and used for electrophoretic mobility shift assay (EMSA). To buffers A, C, and D just before use were added 0.5 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, and 1% protease inhibitor cocktail (P-8340; Sigma). Protein concentration in the nuclear extracts was determined using the Bio-Rad protein assay reagent (Bio-Rad, Hercules, CA).
As a probe for the EMSA, the following 25-bp nucleotide sequence (-5743/-5719) of TH promoter including the seven bases (underlined) identified as TH GRE/AP1, was used: 5′-CCTTGTGAGTGACTAATGGGAACTG (TH-GRE/AP1). The following are the sense sequences of other oligonucleotides used, with regions of interest underlined: mutated TH-GRE/AP-1, 5′-CCTTGTGAGCGTATTATGGGAACTG, where only the underlined 7 bp were scrambled; TH-AP1 (-207/-201), 5′-CTGAGGGTGATTCAGAGGCAG; TH-CRE (-46/-39), 5′-GGGCTTTGACGTCAGCCTGG; Sp1, 5′-TTCGATCGGGGCGGGGCGAG; TATGRE, 5′-ATTACTAGAACATCCTGTACAGTCGAC; and consensus GRE, 5′-CTAGCGGTACATTTTGTTCTAGAACAAAATGTACCGGTACATTTT. Complementary strands of these DNA oligonucleotides were annealed by heating in 1× buffer containing 50 mM NaCl, 1 mM EDTA, and 10 mM Tris at 95°C for 5 min and cooling at ambient temperature. Probes were then labeled with [γ-32P]ATP (3000 Ci/mmol; PerkinElmer Life and Analytical Sciences, Waltham, MA) and T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) and purified on G-25 Sephadex Quick Spin columns (Roche, Indianapolis, IN). For EMSA assays, nuclear extract (10 μg of protein) was incubated for 20 min at room temperature with ∼5 × 104 cpm of the 32P-labeled DNA probe in 20 μl of binding buffer consisting of 20 mM HEPES, pH 7.6, 100 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 12% glycerol, and 1 μg of poly(d)(I-C) (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK). For competition, nuclear extracts were incubated with 100× molar excess of unlabeled double-stranded nucleotides for 10 min at 4°C before the addition of the radiolabeled probe. After the reaction, the samples were resolved on a 4% nondenaturing polyacrylamide gel in 0.5× Tris borate-EDTA buffer. Gels were dried and exposed to Kodak BioMax film to visualize radioactivity captured by the Fotodyne imaging system (Eastman Kodak, Rochester, NY).
Chromatin Immunoprecipitation Assay. Chromatin immunoprecipitation (ChIP) assays were performed using the EZ-Magna ChIP-A kit (Millipore, Billerica, MA) according to the manufacturer's instructions. In brief, PC12 cells (∼1 × 107 cells) were cultured in 150-mm plates, and after 24 h in serum-free NBM medium, they were treated with 100 nM Dex for 1 h. Cells were treated with 1% formaldehyde for 10 min at room temperature to cross-link chromatin and were collected and lysed on ice. The nuclear pellet was recovered and subjected to sonication for 3 pulses of 10 s each to achieve a size of sheared DNA between 200 and 1000 bp. Aliquots of sheared DNA-protein complexes were subjected to immunoprecipitation using the following antibodies: anti-GR (monoclonal antibody SC12763 and rabbit polyclonal antibody GR-1002X; Santa Cruz Biotechnology, Santa Cruz, CA); antibodies to the AP-1 family of proteins, including cFos, FosB, cJun, JunD, and JunB (all from Active Motif, Carlsbad, CA); and anti-Phospho CREB (Millipore). All ChIP assays always included nonimmune rabbit IgG (Millipore) as a control to assess nonspecific interactions. After immunoprecipitation for 2 h at 4°C, the immune complexes were collected using protein A-conjugated magnetic beads, which were extensively washed, and the DNA was eluted and subjected to reverse crosslinking with proteinase K at 62°C for 2 h. The eluted DNA was purified on spin columns and used for analysis by PCR. The PCR primer set was designed to yield a 135-bp product at approximately the -5772/-5638 region of the TH promoter, encompassing the TH-GRE/AP1 region (-5734/-5728). For semiquantitative PCR, serial dilutions of DNA were used as template using the GoTaq green HotStart Taq polymerase master mix (Promega). PCR was conducted for 35 cycles, the reactions were analyzed on 2% agarose gel, and bands were visualized by staining the gel with GelRed DNA dye (Biotium, Hayward, CA). For quantitative real-time PCR, the same DNA templates and primers were used along with a reaction mix containing SyBr Green dye master mix (Applied Biosystems, Foster City, CA), and a standard amplification of 40 cycles was performed in an ABI Prism 7900 machine. The results were normalized using the IgG control, and the fold enrichment with other antibodies was calculated from the Ct values obtained.
Results
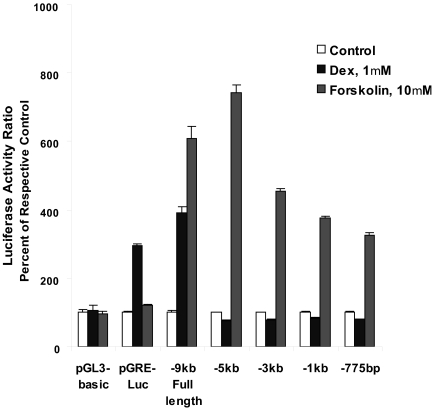
Luciferase Activities of Rat TH Promoter Constructs. Wild-type PC12 cells transfected with the -9-kb full-length rat TH-luciferase construct and treated with Dex (1 μM) or forskolin (10 μM) for 20 h showed a 4- to 6-fold increase in luciferase activity over vehicle-treated control cells (Fig. 1). However, in cells transfected with promoter-Luc constructs measuring 5 kb or shorter, the response to Dex was absent, whereas the forskolin response, which works via the CRE present in the proximal promoter region, was intact. As a positive control, we also used the pGRE-luciferase vector (Clontech, Mountain View, CA), which has multiple copies of the GRE enhancer fused to a TATA-like region of the HSV-TK promoter cloned upstream of the luciferase gene. Dexamethasone, but not forskolin, stimulated luciferase activity in cells transfected with the pGRE construct, whereas the empty pGL3-Basic vector, used as negative control, did not respond to any treatment (Fig. 1). Based on this experiment, we used the -9-kb full-length construct for all mutation studies.
Fig. 1.
Luc activities of TH promoter constructs. WT PC12 cells were transfected using Lipofectamine 2000 with TH-Luc DNA constructs bearing different lengths of the rat TH 5′-flanking sequence. As controls, we used the pGL3-Basic empty vector (negative control) and the pGRE-Luc vector (positive control) that had multiple copies of the GRE enhancer fused to a TATA-like promoter of the thymidine kinase cloned upstream of the firefly Luc gene. Dual Luc assays were performed 20 h after treatment without or with dexamethasone (1 μM) or forskolin (10 μM). Results are presented as the percentage of respective controls are the mean ± S.E.M. of triplicate samples from three separate experiments.
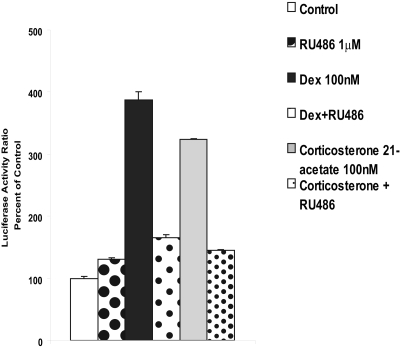
Because the stimulation of promoter activity was similar for both glucocorticoids, corticosterone 21-acetate and dexamethasone (Fig. 2), we chose to use Dex throughout this study. In the dose-response study with Dex using the full-length construct, a significant increase in Luc activity was seen at 10 nM and was maximal at 100 nM (data not shown). Therefore, 100 nM Dex was used in all subsequent experiments. To determine whether the glucocorticoid response was via the glucocorticoid receptor (GR), we used RU486 (1 μM) before the addition of Dex or corticosterone 21-acetate. Measurement of Luc activities 20 h later indicated that inclusion of RU486 abolished the stimulation seen with both Dex and corticosterone (Fig. 2). Although the concentration of RU486 used may inhibit receptors for both glucocorticoids (GRs) and mineralocorticoids, it has been reported that expression of mineralocorticoid receptor is absent in PC12 cells (Lai et al., 2005), suggesting that the response to glucocorticoids shown in Fig. 2 was mediated by a classic GR.
Fig. 2.
Glucocorticoid effects are mediated via the glucocorticoid receptor. WT PC12 cells were transfected with the -9-kb full-length promoter DNA using Lipofectamine 2000 and after 24 h were treated with 100 nM Dex or corticosterone 21-acetate. Some cells were pretreated with 1 μM RU486 for 30 min before the addition of glucocorticoids. Dual Luc assays were performed 20 h after treatment; results represent the mean ± S.E.M. of triplicate samples in two experiments.
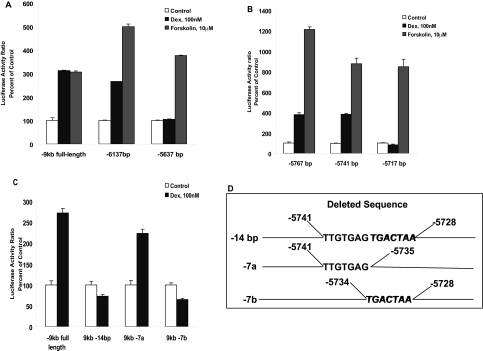
Deletion Analysis of TH Promoter. When deletion was carried out sequentially to remove 1-kb segments from the 5′ end of the full-length promoter, we found that the Dex response was not affected by the deletion of nearly 3.5 kb of the upstream sequence. As shown in Fig. 3A, Luc activity was intact for both Dex and forskolin in the -6137-bp construct, whereas deletion of the next 500 bp (-5637-bp construct) resulted in the loss of only Dex but not the forskolin response. Additional deletions narrowed the Dex response to 24 bp between -5741 and -5717 (Fig. 3B). Of these 24 bp, further analysis of the 14-bp region (-5741 to -5728, TTGTGAGTGACTAA; Fig. 3D) indicated that the GR stimulatory activity was associated with the second 7-bp sequence, TGACTAA (designated 7b), and not with the first 7 bp (designated 7a) (Fig. 3C). These experiments clearly established that the GRE resides at -5734 to -5728 bp of rat TH promoter. Because these 7 bp bear resemblance to the AP-1 element, we refer to them as TH-GRE/AP-1 (Fig. 4A).
Fig. 3.
Deletion analysis of the TH promoter region. A, cells were transfected with the -9-kb full-length, -6.1 or -5.6 kb TH-Luc constructs using Lipofectamine 2000. Dual Luc assays were performed 20 h after treatment with 100 nM Dex or 10 μM forskolin. B, Dex and forskolin response of -5.7-kb deletion constructs for examining the GRE activity in 500 bp between -5767 to -5717. C, comparison of the Dex response of the full-length and constructs with only 14 or 7 bp deletions from the -9-kb promoter as depicted in D.
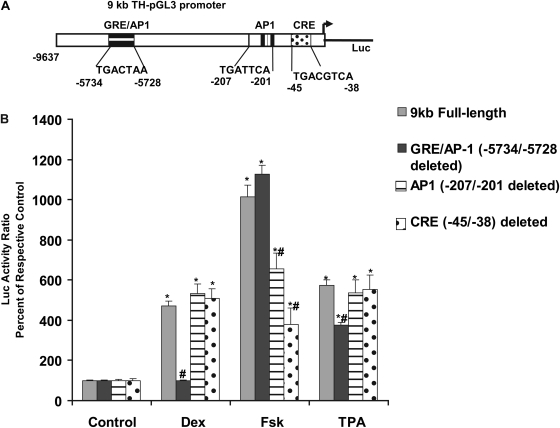
Fig. 4.
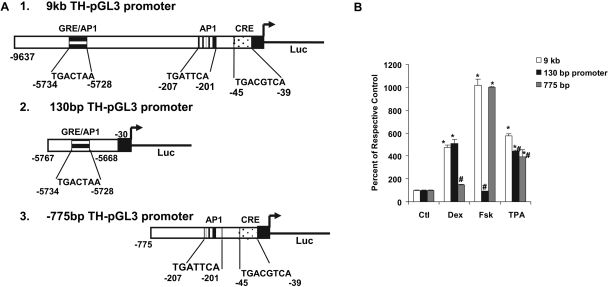
Luciferase responses of the 9-kb full-length TH-Luc promoter and constructs bearing deletions of the 7-bp TH-GRE/AP-1 sequence (-5734/-5728), or the proximal AP-1 site (-207/-201) or mutation of the CRE (-45/-38) from TGACGTCA to CGAGATTA in WT PC12 cells. A, depicts the 9-kb full-length rat TH-pGL3 construct identifying various regions of interest. B, after overnight transfection with promoter constructs using TurboFectin 8.0, WT PC12 cells were treated in serum-free Neurobasal A medium for 20 h until dual Luc assay without (control) or with Dex (100 nM), forskolin (Fsk, 10 μM), or TPA/phorbol ester (100 nM). Results are the mean ± S.E.M. of the percentage of respective controls from three or more experiments, each with three to four wells. *, p < 0.05 compared with respective control group for each construct; #, p < 0.05 compared with the corresponding treatment for the 9-kb promoter.
We also tested the response of the 9-kb full-length and the 7-bp TH-GRE/AP-1 deletion constructs in a mutant PC12 cell line, A126-1B2, deficient in protein kinase A (PKA-PC12). These cells responded to Dex by a 3-fold increase in Luc activity when transfected with the 9-kb promoter. Like in the WT cells, deletion of the 7-bp TH-GRE/AP1 completely eliminated the response to Dex (data not shown). These cells did not respond to forskolin as expected, because the latter response is known to require an intact PKA/CRE activity.
As depicted in Fig. 4A, earlier studies have identified CRE and AP-1 sites in the rat TH promoter proximal to the transcription start site. In the present study, using the 9-kb promoter construct as the template, we also made deletion of either the proximal AP-1 site, TGATTAA at -207/-201 or mutated the CRE site at -45/-38, from TGACGTCA to CGCGATTA and tested their responses to Dex and other appropriate agents. In WT PC12 cells transfected with the 9-kb construct, all of the treatments produced significant increases in Luc activity (Fig. 4B). Dex response was abolished only by the deletion of 7-bp TGACTAA at -5734/-5728 but not by the deletion/mutation of the proximal AP-1 TGATTCA (-207/-201) or CRE sites. With the 7-bp TH-GRE/AP-1 deletion construct, in addition to Dex, the response to TPA was also significantly reduced compared with the 9-kb parent construct, although it was not completely abolished, as seen for Dex (Fig. 4B).
Forskolin response was unaltered by the deletion of 7 bp at -5734/-5728 but was significantly reduced in the CRE or proximal AP-1 site mutant constructs. Mutating the CRE (-45 to -38 bp) caused ∼60% reduction in forskolin response but had little effect on Dex or TPA response. Likewise, deletion of the proximal AP-1 site also caused a ∼30% reduction in response to forskolin but not to Dex or phorbol ester (TPA), with the latter responses being comparable with those seen with the 9-kb construct (Fig. 4B).
In an attempt at heterologous expression, we cloned the 7-bp TH-GRE/AP-1, TGACTAA, upstream of a partial SV40 TK (thymidine kinase) promoter flanking the luciferase gene in the pGL3 vector (pTk-Luc), and tested responses to the agonists in WT PC12 cells. The addition of just the 7 bp did not alter any responses, because no differences were seen between this construct and the control pTk-Luc plasmid in their responses to Dex, forskolin, or TPA (data not shown).
We then made another mutation construct by sequential deletion of the 9-kb parent promoter deleting most of the sequences, including the proximal AP-1 and CRE sites, but keeping only 100 bp from -5767 to -5668 at the 5′ end joining the sequences up to -30 bp that included the TATA box (TTTAAA, -29/-24), upstream of the Luc gene (Fig. 5A). This 130-bp promoter construct retained full activity to Dex, but not to forskolin, perhaps because of the absence of the proximal CRE and AP-1 sites (Fig. 5B). TPA stimulated the activity of this construct, although the response was slightly (∼20%), but significantly lower compared with the 9-kb construct. We also tested another shorter TH-Luc construct that included only 775 bp of the proximal promoter region upstream of the transcription start site flanking the Luc gene (Fig. 5A). Dex response was completely absent with this construct, whereas it retained full response to forskolin (also shown in Fig. 1), and it also responded to TPA (Fig. 5B). Here again, this TPA response was significantly blunted compared with the 9-kb construct.
Fig. 5.
Comparison of the reporter activities of shorter promoter constructs with the 9-kb full-length construct. A, the 130-bp construct was made by sequential deletion mutations using the 9-kb TH-pGL3 construct. It contains 100 bp of the distal region (from -5767 to -5668 bp), adjoining -30 bp of the proximal region flanking the transcription start site (denoted by the arrow); deletion was made just after the CRE site but retaining the TATA box at -30 and the rest of the pGL3-Basic vector. The -775-bp construct contained only the sequences from +15 to -775 bp of rat TH promoter cloned into the pGL3 plasmid. B, WT PC12 cells were transfected using TurboFectin 8.0 with pGL3 vector bearing either the full-length or the shorter promoter sequences as shown in A. Cells were treated the next day with agents at concentrations given in the legend for Fig. 4B and assayed for dual Luc activity. *, p < 0.05 compared with respective control group for each construct; #, p < 0.05 compared with the corresponding treatment for the 9-kb promoter.
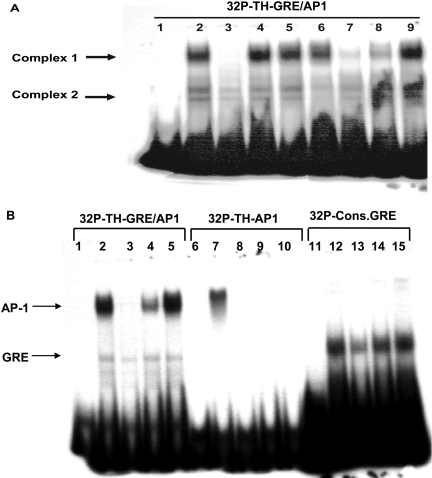
Analysis of Binding by EMSA. When the 25-bp double-stranded oligonucleotide sequence (-5743 to -5719), which included the 7-bp TH-GRE/AP1, TGACTAA, was used as the radiolabeled probe in EMSA, it showed distinct binding to PC12 nuclear extract (Fig. 6). A major DNA protein band (complex 1) was seen at the top of the gel, along with two minor/weaker bands (complex 2) at approximately the middle of the gel (Fig. 6A, lane 2). Inclusion of molar excess of unlabeled TH-GRE/AP-1 completely eliminated binding to complex1 and also competed at complex 2, albeit less effectively (lane 3). A mutant form of this 25-bp sequence, in which only the 7-bp TGACTAA sequence was scrambled to CGTATTA without changing other bases, did not compete for probe binding (lane 4). The binding of the probe at complex 1 was not inhibited by inclusion of excess unlabeled TAT-GRE, the GRE sequence identified in the enzyme tyrosine aminotransferase (TAT) (lane 5) or consensus GRE (lane 6), but the lower complex 2 bands were reduced by the consensus GRE (lane 6). The complex 1 band was effectively reduced by the inclusion of unlabeled AP-1 (lane 7) or CRE (lane 8) sequences, but not by Sp1 (lane 9). Complex 2 bands were also reduced by AP-1 to a similar extent as with TH-GRE/AP-1.
Fig. 6.
EMSA with TH-GRE/AP-1, TH-AP-1, or consensus GRE probes. Nuclear extracts (NE) prepared from WT PC12 cells treated with 100 nM Dex for 1 h were used in all experiments. A, the 25-bp double-stranded oligonucleotide (5′-CCTTGTGAGTGACTAATGGGAACTG, -5743/-5719 of rat TH promoter, with the 7-bp TH-GRE/AP-1 underlined in the middle) was used as the radiolabeled probe. All lanes contained the same amount of radiolabeled probe (5 × 104 cpm), and all lanes except lane 1 received 10 μg of NE protein. In lanes 3 to 9, the following unlabeled double-stranded oligonucleotides at 100-fold molar excess were included (sequence of only the sense strand is shown and relevant parts of the sequence are underlined): lane 3, TH-GRE/AP-1 (same as the radiolabeled probe, 5′-CCTTGTGAGTGACTAATGGGAACTG); lane 4, mutated TH-GRE/AP-1, 5′-CCTTGTGAGCGTATTATGGGAACTG; lane 5, TAT-GRE, 5′-ATTACTAGAACATCCTGTACAGTCGAC; lane 6, consensus GRE, 5′-CTAGCGGTACATTTTGTTCTAGAACAAAATGTACCGGTACATTTT; lane 7, TH-AP1 (-207 to -201, 5′-CTGAGGGTGATTCAGAGGCAG; lane 8, TH-CRE (-45 to -38), 5′-GGGCTTTGACGTCAGCCTGG; and lane 9, Sp1, 5′-ATTCGATCGGGGCGGGGCGAGC. Free probe is seen at the bottom of the gel, and the shifted bands are indicated by arrows. A representative image from three experiments is shown. B, EMSA with 32P-labeled probes: TH-GRE/AP-1 (lanes 1-5) or TH-AP-1 (lanes 6-10) or 32P-consensus GRE probes (lanes 11-15). Lanes 1, 6, and 11 received only the respective probes. All other lanes included 10 μg of Dex-treated NE protein. Lanes 2 (TH-GRE/AP-1), 7 (TH-AP-1), and 12 (consensus GRE) show the specific binding of the probes to the NE in the absence of unlabeled nucleotides. The following unlabeled nucleotides (100× molar excess) were included along with the respective probes: lanes 3, 9, and 14, TH-GRE/AP-1; lanes 4, 8, and 15, TH-AP-1; lanes 5 and 13, consensus GRE; lane 10, CRE. Free probe is at the bottom of the gel, and the shifted bands are indicated by the arrows, identifying AP-1 and GRE binding sites.
To further examine the nature of these binding sites (Fig. 6B), we compared the pattern of binding of labeled AP-1 (lanes 6-10) and consensus GRE sequences (lanes 11-15) and their competition with unlabeled oligonucleotides with that of [32P]TH-GRE/AP-1 (lanes 1-5). When AP-1 was used as the probe, only the top band corresponding to complex 1 was seen (lane 7). This band was competed out by both unlabeled TH-GRE/AP-1 (lane 8) and AP-1 (lane 9) but not by the mutated sequences (data not shown). This result confirmed that the complex 1 in Fig. 6A is the AP-1 binding site. Similar to the TH-GRE/AP-1 probe, excess unlabeled CRE also competed for the labeled AP-1 binding (lane 10). In comparing the sequences of the probes and the competing ligands, we notice that they all share the first three bases TGA, which perhaps are important for binding to the AP-1 site.
When we used consensus GRE (lanes 11-15) as the 32P-labeled probe, it showed binding to the nuclear extract only near the middle of the gel, corresponding to bands identified as complex 2 in Fig. 6A (lane 12). This band was competed effectively by the unlabeled consensus GRE (lane 13), but not by unlabeled AP-1 (lane 15), allowing us to identify complex 2 as the GR binding site. Excess unlabeled TH-GRE/AP-1 also competed for the lower band, albeit weakly (lane 14), whereas it completely eliminated binding to the upper band (AP-1 site, lanes 3 and 9).
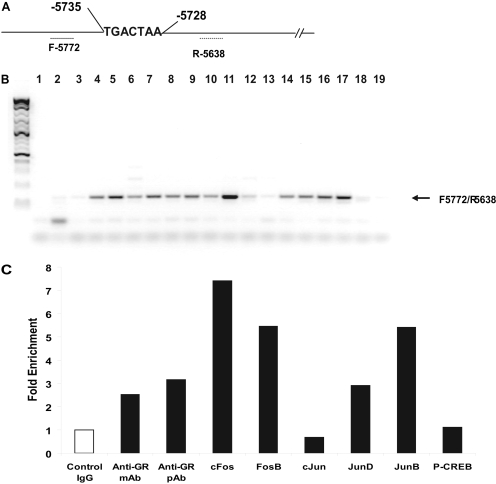
ChIP Analysis. To further elucidate the proteins associated with binding to the GRE/AP-1 region in the TH promoter, we performed ChIP analysis. Using primer sets (F -5772/R -5638) that were designed to include the TH-GRE/AP-1 sequence (-5734 to -5728, Fig. 7A), we obtained a band of the predicted size (135 bp) by semiquantitative PCR in cross-linked DNA fragments immunoprecipitated using antibodies to GR and to the AP-1 family of proteins (Fig. 7B). Compared with the control IgG (lanes 2 and 3), we observed a specific PCR band in ChIP with antibodies against GR (monoclonal anti-GR, lanes 4 and 5, and polyclonal anti-GR, lanes 6 and 7), and AP-1 family proteins, including cFos (lanes 8 and 9), FosB (lanes 10 and 11), JunD (lanes 14 and 15), and JunB (lanes 16 and 17), but not cJun (lanes 12 and 13). Neither did we see this band in the ChIP with anti-phospho-CREB (lanes 18 and 19), with the latter used as a control antibody unrelated to AP-1 proteins; these latter bands were of similar intensity as seen with the control IgG (lanes 2 and 3). Quantitative PCR confirmed these data, showing a several-fold enrichment of these proteins relative to the control IgG with all antibodies tested, except for cJun and P-CREB (Fig. 7C). We observed a similar pattern in semiquantitative PCR using another primer set (F -5902/R -5638) designed around the TH-GRE sequence to give a 283-bp product (data not shown), further confirming the ChIP PCR data.
Fig. 7.
ChIP analysis of binding proteins. ChIP analysis was performed using chromatin prepared from cells treated with 100 nM Dex for 1 h. The cross-linked DNA protein complexes were subjected to immunoprecipitation using antibodies to GR (monoclonal antibodies: anti-GR mAB or rabbit polyclonal antibodies: anti-GR pAb) or to the AP1 family including c-Fos, Fos B, c-Jun, Jun D, and JunB, as well as phospho-CREB. The purified DNA after reversal of cross-links was used as template in PCR along with primers for the region that spanned TH-GRE/AP-1 sequence shown in A: F -5772, 5′-CATGATTGCTGTCACATCACC, and R -5638, 5′- CAACCTGTGCACCAGTGAGT. B, semiquantitative PCR was performed for 35 cycles of amplification; the reactions were run on 2% agarose gel, and DNA bands were visualized by staining with Biotium GelRed DNA dye. The DNA ladder is on the left. Lane 1, no template control (NTC); lanes 2 and 3, control IgG; lanes 4 and 5, anti-GR mAB; lanes 6 and 7, anti-GR pAB; lanes 8 and 9, anti-cFos; lanes 10 and 11, anti-Fos B; lanes 12 and 13, anti-cJun; lanes 14 and 15, anti-Jun D; lanes 16 and 17, anti-JunB; and lanes 18 and 19, anti-P-CREB. C, quantitative PCR was performed using the same primer set along with SyBr Green master mix in a standard 40-cycle PCR using ABI Prism 7900 instrument. Amplification of specific products was seen between 13 and 32 cycles, and a single peak was seen in the dissociation curve analysis (data not shown). -Fold enrichment was calculated from the Ct values. The results are expressed relative to control IgG. Mean data from two similar experiments are shown.
Discussion
We have clearly identified a seven-nucleotide sequence, TGACTAA, in the rat TH promoter as critical to the stimulatory glucocorticoid response of the TH gene. This sequence of TH promoter is located nearly 5.7 kb from the transcription start site from -5734 to -5728. This glucocorticoid response element (TGACTAA) has no semblance to the classic GRE (5′-AGAACANNNTGTTCT) but closely resembles the signature motif (5′-TGACTCA) for the AP-1 family of proteins. Deleting just these seven base pairs from the 9-kb TH promoter completely abolished the glucocorticoid response. Conversely, complete GC responsiveness was retained after deletion of most of the 9-kb promoter, except for the 100 bp around the newly identified element at -5.7 kb. Moreover, results from EMSA and ChIP assays provided clear evidence for binding of GR and AP-1 to this putative GRE in the rat TH promoter. Therefore, we refer to this AP-1-like sequence as the TH-GRE/AP-1 site.
Although it is well known that glucocorticoids stimulate TH mRNA levels in adrenal chromaffin cells through transcription (Fossom et al., 1992), the mechanism by which glucocorticoids mediate this effect has remained elusive. In previous studies from our laboratory aimed at identifying a functional TH GRE, we used the TH3.6 reporter construct, reported to contain 3.6 kb of 5′ promoter sequences of the mouse TH gene (National Center for Biotechnology Information accession number X53503) (Morgan et al., 1996). Using this construct, we identified an imperfect GRE (5′-GGCACAGTGTGGTCT-3′) approximately -2.4 kb from the transcription start site (Hagerty et al., 2001b), that seemed to confer responsiveness to glucocorticoid. However, we have since re-evaluated our conclusion that this sequence is a biologically relevant GRE for several reasons. First, upon stimulation with dexamethasone, the magnitude of the increase in promoter activity of the TH3.6 promoter construct used in our previous studies was relatively weak, only 1.5- to 2-fold. Second, a recent comparison of the DNA sequence of this promoter with the 5′-flanking genomic sequence of the mouse TH gene in the ENSEMBL database revealed discrepancies. Thus, 1.2 kb of the region upstream of the TH-GRE identified previously in the TH3.6 construct corresponded partly to those on two different mouse chromosomes. Third, when the errant 1.2-kb sequence was removed and the remaining 2.4 kb containing the putative GRE was retested in the context of a different plasmid, responsiveness to glucocorticoid receptor stimulation was lost. It is noteworthy that a comparison of nucleotide sequences between the rat and the mouse TH gene revealed considerable homology in 5′-flanking sequences at approximately -5.7 kb. Moreover, we identified a sequence, TGACTAA, approximately -5.7 kb of the mouse TH promoter, which may be the mouse TH-GRE/AP-1 site. Further studies are underway to determine whether this sequence is also a functional TH-GRE/AP-1 site in the mouse gene.
Although deletion of the TH-GRE/AP-1 completely abolished the response to dexamethasone, it did not alter the response to forskolin. The lack of requirement for the cAMP/PKA pathway in the response to glucocorticoid receptor stimulation was corroborated by the intact response of the 9-kb promoter to Dex in PKA-deficient PC12 cells. These results clearly demonstrate that Dex and forskolin stimulate TH promoter activity via distinct pathways, corroborating our earlier observations (Hagerty et al., 2001a). Although there is one clearly identified consensus CRE sequence (-45/-38) in the rat TH promoter, there are several AP-1 sites. Mutation or deletion of the CRE or AP-1 in the proximal part of the -9-kb TH promoter had no influence on the Dex response but affected the responses to other agonists. Thus, mutation of the CRE and to a lesser extent the AP-1 (-207/-201), reduced the forskolin response. Our data are consistent with earlier reports that, in addition to the canonical sequence at -45/-38, other elements also contribute to the cAMP/forskolin response. (Lewis et al., 1987; Fung et al., 1992; Best et al., 1995; Nagamoto-Combs et al., 1997; Yang et al., 1998; Lewis-Tuffin et al., 2004).
Besides abolishing the Dex response, deletion of the distal 7-bp TH-GRE/AP-1 sequence also significantly reduced the response to phorbol ester (Fig. 4B), a stimulator of PKC, known to act via AP-1 sites (Icard-Liepkalns et al., 1992). However, this response was not completely abolished, unlike that seen for Dex. This may not be surprising because several AP-1 sites exist in the rat TH promoter that can perhaps collectively contribute to the response seen with the AP-1 agonists. Conversely, unlike the 40% reduction seen with the deletion of the distal 7-bp GRE/AP-1, the response to TPA was totally unaffected by the deletion/mutation of the proximal AP-1/CRE sites. This was surprising, because other studies have shown the importance of the proximal AP-1 and CRE sites in TPA response (Icard-Liepkalns et al., 1992; Best and Tank, 1998; Piech-Dumas et al., 2001). A major difference among the various studies seems to be the length of the promoter constructs used. We tested deletion/mutation of just the proximal 7/8-bp AP-1/CRE sites in the context of the entire 9-kb promoter, whereas others have used deletions made in shorter constructs containing varying lengths of the proximal part of the TH promoter cloned in other reporter systems. Indeed, when we used a short -775 bp TH-Luc construct, we also saw a response to TPA (Fig. 5B), similar to that published previously using shorter constructs. More recently, an interaction between AP-1 and other factors such as Egr1 has been proposed to be important for TPA up-regulation of TH gene (Nakashima et al., 2003; Stefano et al., 2006). Nevertheless, our results show that in the context of the 9-kb long promoter, deletion of just the proximal AP-1 or CRE sites minimally affects the response to AP-1 agonist. On the other hand, in a construct that deleted the entire proximal region but retained only 100 bp around the distal TH-GRE/AP-1 sequence (the 130-bp promoter, Fig. 5), the TPA response was only slightly (∼20%) diminished, implying that a major contribution to the TPA response comes from this distal TH-GRE/AP-1 site. Because the Dex response, mediated via the glucocorticoid receptor, was entirely dependent on the 7-bp site, this fits the description of a composite GRE/AP-1 element in the rat TH promoter.
The interaction of nuclear receptors with other transcription factors in regulating promoter function is well known for various genes (Pfahl, 1993). Indeed, the term “composite element” was coined to describe such a GRE in the mouse proliferin promoter in which both GR and AP-1 proteins, c-Jun and c-Fos, bound to overlapping sites, invoking a model involving DNA binding and protein-protein interactions of receptor and nonreceptor factors (Mordacq and Linzer, 1989). Using a synthetic promoter containing juxtaposed AP-1 and GR binding sites, GR was shown to negatively or positively modulate transcription, depending on the AP-1 subunits. It was thus hypothesized that expression of genes containing “composite elements” is activated or repressed in a hormone-dependent manner, depending on the ratio of positively and negatively acting AP-1 subunits (Diamond et al., 1990; Miner and Yamamoto, 1992).
To our knowledge, this is the first study of any TH promoter that describes such an indirect mechanism for GR action involving AP-1 sites. Our competition studies using EMSA clearly showed that the sequence identified as TH-GRE/AP-1 was able to bind both AP-1 and GRE binding proteins in nuclear extracts. Subsequent ChIP assays provided clear evidence for the presence of both GR and AP-1 family of proteins in these DNA-binding complexes. It is noteworthy that, using the ChIP assay, we detected several proteins from the AP-1 family, but not c-Jun. Such differential effects of glucocorticoid on Fos and Jun proteins have also been described for other genes (Autelitano, 1994). Given their importance as transcriptional regulators, it is not surprising that AP-1 activity is tightly controlled at the level of fos and jun gene transcription and by posttranslational modification of AP-1 proteins (Angel and Karin, 1991; Teurich and Angel, 1995). Further studies are underway to establish the effects of glucocorticoids on the relative expression of AP-1 family members and their interactions with GR in our cells. Nevertheless, our studies show that the TH gene joins a growing list of genes containing atypical GREs, in which the glucocorticoid receptor influences gene activation in collaboration with other transcription factors.
Acknowledgments
We thank Lianna Martinez and Michelle Carreon for technical assistance. We are grateful to Dr. James Lechleiter, Professor, Department of Cellular and Structural Biology, University of Texas Health Science Center at San Antonio, for critical reading of the manuscript.
This work was supported in part by the National Institutes of Health National Institute on Aging [Grant AG022307] and by the Merit Review Medical Research Program of the Department of Veterans Affairs.
ABBREVIATIONS: GR, glucocorticoid receptor; TH, tyrosine hydroxylase; PC12, pheochromocytoma cell line; GRE, glucocorticoid response element; AP-1, activator protein 1; CRE, cAMP-response element; PKA, cAMP-dependent protein kinase A; Dex, dexamethasone; TPA, tetradecanoyl-12,13-phorbol acetate; bp, base pair(s); Luc, luciferase; EMSA, electrophoretic mobility shift assay; ChIP, chromatin immunoprecipitation; kb, kilobase(s); PCR, polymerase chain reaction; WT, wild type; NBM, Neurobasal A medium; CREB, cAMP response element-binding protein; F, forward; R, reverse; NE, nuclear extract; RU486, mifepristone.
References
- Angel P and Karin M (1991) The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta 1072 129-157. [DOI] [PubMed] [Google Scholar]
- Autelitano DJ (1994) Glucocorticoid regulation of c-Fos, c-Jun and transcription factor AP-1 in the AtT-20 corticotrope cell. J Neuroendocrinol 6 627-637. [DOI] [PubMed] [Google Scholar]
- Best JA, Chen Y, Piech KM, and Tank AW (1995) The response of the tyrosine hydroxylase gene to cyclic AMP is mediated by two cyclic AMP response elements. J Neurochem 65 1934-1943. [DOI] [PubMed] [Google Scholar]
- Best JA and Tank AW (1998) The THCRE2 site in the rat tyrosine hydroxylase gene promoter is responsive to phorbol ester. Neurosci Lett 258 131-134. [DOI] [PubMed] [Google Scholar]
- Chan GC, Hess P, Meenakshi T, Carlstedt-Duke J, Gustafsson JA, and Payvar F (1991) Delayed secondary glucocorticoid response elements. J Biol Chem 266 22634-22644. [PubMed] [Google Scholar]
- De Bosscher K, Vanden Berghe W, and Haegeman G (2003) The interplay between the glucocorticoid receptor and nuclear factor-{kappa}B or activator protein-1: molecular mechanisms for gene repression. Endocr Rev 24 488-522. [DOI] [PubMed] [Google Scholar]
- Del Monaco M, Covello SP, Kennedy SH, Gilinger G, Litwack G, and Uitto J (1997) Identification of novel glucocorticoid response elements in human elastin promoter and demonstration of nucleotide sequence specificity of the receptor binding. J Invest Dermatol 108 938-942. [DOI] [PubMed] [Google Scholar]
- Diamond MI, Miner JN, Yoshinaga SK, and Yamamoto KR (1990) Transcription factor interactions: selectors of positive or negative regulation from a single DNA element. Science 249 1266-1272. [DOI] [PubMed] [Google Scholar]
- Drouin J, Trifiro MA, Plante RK, Nemer M, Eriksson P, and Wrange O (1989) Glucocorticoid receptor binding to a specific DNA sequence is required for hormone-dependent repression of pro-opiomelanocortin gene transcription. Mol Cell Biol 9 5305-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossom LH, Sterling CR, and Tank AW (1992) Regulation of tyrosine hydroxylase gene transcription rate and tyrosine hydroxylase mRNA stability by cyclic AMP and glucocorticoid. Mol Pharmacol 42 898-908. [PubMed] [Google Scholar]
- Fung BP, Yoon SO, and Chikaraishi DM (1992) Sequences that direct rat tyrosine hydroxylase expression. J Neurochem 58 2044-2052. [DOI] [PubMed] [Google Scholar]
- Hagerty T, Fernandez E, Lynch K, Wang SS, Morgan WW, and Strong R (2001a) Interaction of a glucocorticoid-responsive element with regulatory sequences in the promoter region of the mouse tyrosine hydroxylase gene. J Neurochem 78 1379-1388. [DOI] [PubMed] [Google Scholar]
- Hagerty T, Morgan WW, Elango N, and Strong R (2001b) Identification of a glucocorticoid-responsive element in the promoter region of the mouse tyrosine hydroxylase gene. J Neurochem 76 825-834. [DOI] [PubMed] [Google Scholar]
- Icard-Liepkalns C, Biguet NF, Vyas S, Robert JJ, Sassone-Corsi P, and Mallet J (1992) AP-1 complex and c-fos transcription are involved in TPA provoked and trans-synaptic inductions of tyrosine hydroxylase gene: Insights into long-term regulatory mechanisms. J Neurosci Res 32 290-298. [DOI] [PubMed] [Google Scholar]
- Iwawaki T, Kohno K, and Kobayashi K (2000) Identification of a potential Nurr1 response element that activates the tyrosine hydroxylase gene promoter in cultured cells. Biochem Biophys Res Commun 274 590-595. [DOI] [PubMed] [Google Scholar]
- Jonat C, Rahmsdorf HJ, Park KK, Cato AC, Gebel S, Ponta H, and Herrlich P (1990) Antitumor promotion and antiinflammation: down-modulation of AP-1 (Fos/Jun) activity by glucocorticoid hormone. Cell 62 1189-1204. [DOI] [PubMed] [Google Scholar]
- Kumer SC and Vrana KE (1996) Intricate regulation of tyrosine hydroxylase activity and gene expression. J Neurochem 67 443-462. [DOI] [PubMed] [Google Scholar]
- Lai M, Seckl J, and Macleod M (2005) Overexpression of the mineralocorticoid receptor protects against injury in PC12 cells. Mol Brain Res 135 276-279. [DOI] [PubMed] [Google Scholar]
- Lewis EJ, Harrington CA, and Chikaraishi DM (1987) Transcriptional regulation of the tyrosine hydroxylase gene by glucocorticoid and cyclic AMP. Proc Natl Acad Sci U S A 84 3550-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis-Tuffin LJ, Quinn PG, and Chikaraishi DM (2004) Tyrosine hydroxylase transcription depends primarily on cAMP response element activity, regardless of the type of inducing stimulus. Mol Cell Neurosci 25 536-547. [DOI] [PubMed] [Google Scholar]
- Min N, Joh TH, Kim KS, Peng C, and Son JH (1994) 5′ upstream DNA sequence of the rat tyrosine hydroxylase gene directs high level tissue-specific expression to catecholaminergic neurons in the central nervous system of transgenic mice. Mol Brain Res 27 281-289. [DOI] [PubMed] [Google Scholar]
- Miner JN and Yamamoto KR (1992) The basic region of AP-1 specifies glucocorticoid receptor activity at a composite response element. Genes Dev 6 2491-2501. [DOI] [PubMed] [Google Scholar]
- Mordacq JC and Linzer DI (1989) Co-localization of elements required for phorbol ester stimulation and glucocorticoid repression of proliferin gene expression. Genes Dev 3 760-769. [DOI] [PubMed] [Google Scholar]
- Morgan WW, Walter CA, Windle JJ, and Sharp ZD (1996) 3.6 kb of the 5′ Flanking DNA Activates the Mouse Tyrosine Hydroxylase Gene Promoter Without Catecholaminergic-Specific Expression. J Neurochem 66 20-25. [DOI] [PubMed] [Google Scholar]
- Nagamoto-Combs K, Piech KM, Best JA, Sun B, and Tank AW (1997) Tyrosine hydroxylase gene promoter activity is regulated by both cyclic AMP-responsive element and AP1 sites following calcium influx—evidence for cyclic AMP-responsive element binding protein-independent regulation. J Biol Chem 272 6051-6058. [DOI] [PubMed] [Google Scholar]
- Nakashima A, Ota A, and Sabban EL (2003) Interactions between Egr1 and AP1 factors in regulation of tyrosine hydroxylase transcription. Mol Brain Res 112 61-69. [DOI] [PubMed] [Google Scholar]
- Newton R and Holden NS (2007) Separating transrepression and transactivation: a distressing divorce for the glucocorticoid receptor? Mol Pharmacol 72 799-809. [DOI] [PubMed] [Google Scholar]
- Owen GI, Richer JK, Tung L, Takimoto G, and Horwitz KB (1998) Progesterone regulates transcription of the p21waf1 cyclin-dependent kinase inhibitor gene through Sp1 and CBP/p300. J Biol Chem 273 10696-10701. [DOI] [PubMed] [Google Scholar]
- Pfahl M (1993) Nuclear receptor/AP-1 interaction. Endocr Rev 14 651-658. [DOI] [PubMed] [Google Scholar]
- Piech-Dumas KM, Best JA, Chen Y, Nagamoto-Combs K, Osterhout CA, and Tank AW (2001) The cAMP responsive element and CREB partially mediate the response of the tyrosine hydroxylase gene to phorbol ester. J Neurochem 76 1376-1385. [DOI] [PubMed] [Google Scholar]
- Rhen T and Cidlowski JA (2005) Antiinflammatory action of glucocorticoids—new mechanisms for old drugs. N Engl J Med 353 1711-1723. [DOI] [PubMed] [Google Scholar]
- Rüdiger JJ, Roth M, Bihl MP, Cornelius BC, Johnson M, Ziesche R, and Block LH (2002) Interaction of C/EBPα and the glucocorticoid receptor in vivo and in non-transformed human cells. FASEB J 16 177-184. [DOI] [PubMed] [Google Scholar]
- Scheidereit C, Geisse S, Westphal HM, and Beato M (1983) The glucocorticoid receptor binds to defined nucleotide sequences near the promoter of mouse mammary tumor virus. Nature 304 749-752. [DOI] [PubMed] [Google Scholar]
- Schreiber E, Matthias P, Müller MM, and Schaffner W (1989) Rapid detection of octamer binding proteins with “mini-extracts”, prepared from a small number of cells. Nucleic Acids Res 17 6419-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano L, Al Sarraj J, Rössler OG, Vinson C, and Thiel G (2006) Up-regulation of tyrosine hydroxylase gene transcription by tetradecanoylphorbol acetate is mediated by the transcription factors Ets-like protein-1 (Elk-1) and Egr-1. J Neurochem 97 92-104. [DOI] [PubMed] [Google Scholar]
- Teurich S and Angel P (1995) The glucocorticoid receptor synergizes with Jun homodimers to activate AP-1-regulated promoters lacking GR binding sites. Chem Senses 20 251-255. [DOI] [PubMed] [Google Scholar]
- Truss M and Beato M (1993) Steroid hormone receptors: interaction with deoxyribonucleic acid and transcription factors. Endocr Rev 14 459-479. [DOI] [PubMed] [Google Scholar]
- Uht RM, Anderson CM, Webb P, and Kushner PJ (1997) Transcriptional activities of estrogen and glucocorticoid receptors are functionally integrated at the AP-1 response element. Endocrinology 138 2900-2908. [DOI] [PubMed] [Google Scholar]
- Van Buskirk R, Corcoran T, and Wagner JA (1985) Clonal variants of PC12 pheochromocytoma cells with defects in cAMP-dependent protein kinases induce ornithine decarboxylase in response to nerve growth factor but not to adenosine agonists. Mol Cell Biol 5 1984-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Kim HS, Seo H, and Kim KS (1998) Identification and characterization of potential cis-regulatory elements governing transcriptional activation of the rat tyrosine hydroxylase gene. J Neurochem 71 1358-1368. [DOI] [PubMed] [Google Scholar]
- Yang-Yen HF, Chambard JC, Sun YL, Smeal T, Schmidt TJ, Drouin J, and Karin M (1990) Transcriptional interference between c-Jun and the glucocorticoid receptor: Mutual inhibition of DNA binding due to direct protein-protein interaction. Cell 62 1205-1215. [DOI] [PubMed] [Google Scholar]