Abstract
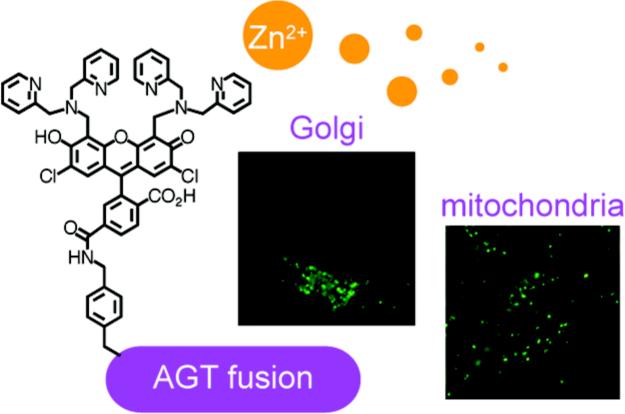
A protein labeling approach is employed for the localization of a zinc-responsive fluorescent probe in the mitochondria and in the Golgi apparatus of living cells. ZP1, a zinc sensor of the Zinpyr family, was functionalized with a benzylguanine moiety and thus converted into a substrate (ZP1BG) for the human DNA repair enzyme alkylguaninetransferase (AGT or SNAP-Tag). The labeling reaction of purified glutathione S-transferase tagged AGT with ZP1BG and the zinc response of the resulting protein-bound sensor were confirmed in vitro. The new detection system, which combines a protein labeling methodology with a zinc fluorescent sensor, was tested in live HeLa cells expressing AGT in specific locations. The enzyme was genetically fused to site-directing proteins that anchor the probe onto targeted organelles. Localization of the zinc sensors in the Golgi apparatus and in the mitochondria was demonstrated by fluorescence microscopy. The protein-bound fluroescence detection system is zinc-responsive in living cells.
Zinc-selective fluorescent sensors are valuable tools for the investigation of zinc physiology.1-3 Their applications will help to construct a detailed network map of biological zinc and its transporter proteins and to elucidate how zinc influences processes that include neurotransmission4 and proliferative signaling.5 The high-fidelity tracking of zinc mobilization in live cells requires sensors that monitor specific subcellular compartments without relying on the spontaneous distribution of the probes. To address this need, we present a genetically targeted fluorescent detection system for monitoring zinc flux in selected organelles in live cells.
Fluorescence bioimaging has greatly benefited from methodologies for the labeling of proteins by genetic fusion techniques.6-9 For instance, in applications aimed at biological calcium detection, the topological control of the probe has been accomplished by genetic targeting of protein-based indicators10,11 and, more recently, small organic dyes.12,13
A recent labeling approach14 devised by Johnsson and coworkers employs the alkylation of Cys145 of human O6-alkylguanine transferase (AGT) with various benzylguanine derivatives. For bioimaging studies in live cells, AGT was fused to partners that naturally localize to specific subcellular compartments (e.g., nucleus, cytosol, plasma membrane), and then labeled with fluorescent substrates carrying a benzylguanine moiety.14,15 We therefore evaluated the AGT labeling methodology for the immobilization of a fluorescent zinc sensor of the Zinpyr family2 onto targeted organelles.
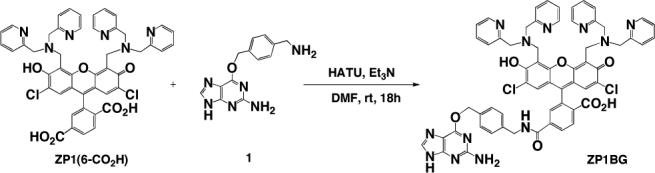
The design of ZP1BG (Scheme 1) combines the Zn(II)-sensitive ZP1 sensor and the AGT substrate benzylguanine. Reaction of ZP1(6-CO2H)16 with amine 1 in the presence of the coupling reagent HATU gave the target compound ZP1BG, which was isolated in high purity by reverse-phase preparative HPLC in 43% yield.
Scheme 1.
The ability of AGT to accept ZP1BG as a substrate was assayed in vitro. A glutathione S-transferase-tagged AGT fusion was overexpressed in E. coli and purified by affinity chromatography (Supporting Information). After incubation of an ∼1:1 ratio of the AGT fusion protein and ZP1BG for 10 min at room temperature, electrospray mass spectrometry confirmed the formation of the covalently labeled product ZP1-AGT (Figure S3 and Table S1, Supporting Information).
The fluorescence properties and Zn(II) response of ZP1-AGT were next examined in aqueous solution buffered at pH 7.0. ZP1-AGT displayed maximum fluorescence emission at 532 nm (λex = 490 nm, Figure 1). Addition of 2 equiv of ZnCl2 caused an ∼2.7-fold increase in integrated emission, which is in good agreement with the ∼3.1-fold turn-on previously reported for the parent sensor ZP1.17 Subsequent addition of the high-affinity zinc chelator N,N,N’,N’-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN) reverted the emission intensity to that of the zinc-free system. The presence of the covalently attached AGT protein did not perturb the emission properties of the small-molecule sensor, which therefore functions as a reversible zinc-responsive probe.
Figure 1.
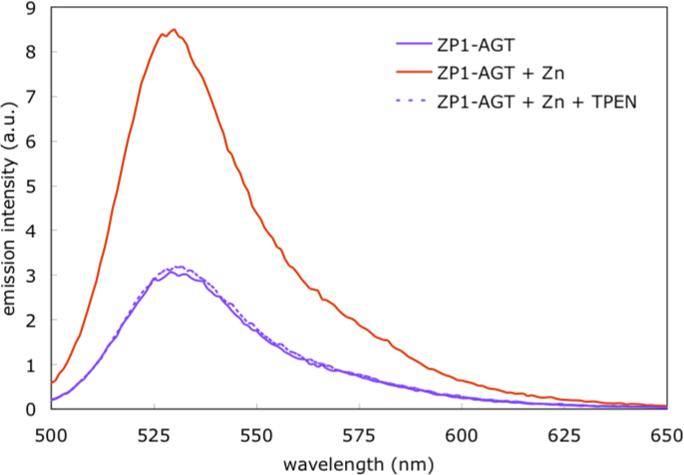
Fluorescence emission spectrum (λex 490 nm) of ZP1-AGT (1 μM) in 50 mM PIPES buffer and 100 mM KCl at pH 7.0, and spectral changes after addition of 2 equiv of ZnCl2 and subsequent addition of 20 equiv of TPEN.
Conversely, the addition of ZnCl2 to a solution of the AGT substrate ZP1BG did not elicit a fluorescence emission turn-on (Figure S2). This behavior is ascribed to the presence of the guanine moiety, which complicates both the coordinative and physicochemical properties of the receptor. Guanine quenches the emission of several fluorophores, including fluorescein.18,19 The lack of zinc responsiveness of the ZP1BG “pro-sensor” is a favorable feature of this detection system because it ensures that only the immobilized and not the free probe affords a zinc-induced signal.
Preliminary fluorescent imaging studies using HeLa cells indicated that ZP1BG is cell permeable and provides a weak and sparse staining (Figure S4, Supporting Information).
Because of the role played by the Golgi apparatus and the mitochondria in intracellular zinc transport and storage,20,21 we prepared constructs pGolgi–AGT and pMito–AGT to express AGT in these organelles. The pGolgi–AGT construct directs AGT to the trans-Golgi cisternae by fusion with human β-1,4-galactosyltransferase,22 whereas pMito–AGT employs the targeting signal from subunit VIII of cytochrome c oxidase to direct AGT to the mitochondrial matrix.23
After transient transfection using standard liposome-mediated gene-transfer techniques, HeLa cells were incubated with ZP1BG (10 μM, 1 h, 37 °C) and colocalization studies were performed to assess the probe localization. Specifically, pGolgi–mCherry was used to label the Golgi apparatus with the red fluorescent protein mCherry24 and the commercially available dye Mitotracker Red was employed as a mitochondrial stain. Selective ZP labeling of the targeted organelles was confirmed by live cell fluorescence imaging (Figure 2). ZP1BG treatment of non-transfected cells, as well as cross-colocalization controls, confirmed that the staining patterns require targeted AGT expression (Figures S4-S6).
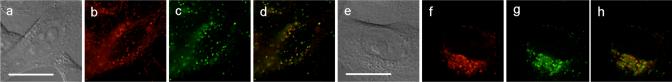
Figure 2.
HeLa cells expressing AGT in the mitochondria (a-d) and in the Golgi apparatus (e-h) were labeled with ZP1BG (10 μM, 1 h). The organelles were marked in the red by staining with 0.2 μM Mitotracker Red (b) or by expression of mCherry in the Golgi apparatus (f). For each series, from the left: (i) DIC image, (ii) emission from the organelle marker, (iii) emission from site-specifically localized probe, and (iv) overlay of the red and green fluorescence images. Scale bar: 25 μm. See Figure S5 for full-view images.
The fluorescence response of immobilized ZP1 in the Golgi and mitochondria of HeLa cells to exogenous Zn(II) was evaluated (Figure 3). In both cases, addition of 25−50 μM ZnCl2 and the ionophore pyrithione, 2-mercaptopyridine-N-oxide, caused a prompt increase of the green fluorescence emission in the labeled organelles. Consistent with a zinc-induced fluorescence response, the effect was reversed by TPEN.
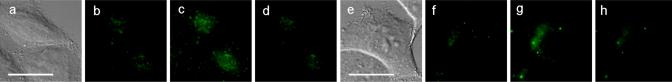
Figure 3.
Zinc-induced fluorescence response in HeLa cells expressing AGT in the mitochondria (a-d) and in the Golgi apparatus (e-h). For each series, from the left: (i) DIC image, (ii) labeling with ZP1BG (10 μM, 1 h), (iii) addition of Zn(II)/pyrithione (1:1, 50 μM, 5 min), and (iv) subsequent addition of TPEN (100 μM, 5 min). Scale bar: 25 μm. See Figure S7 for full-view images.
The AGT labeling methodology therefore provides a way to direct the intracellular distribution of a Zn(II) sensor and to visualize qualitatively a turn-on emission response to addition of zinc. Current work on this approach focuses on improvement of the system sensitivity by use of probes displaying a greater dynamic range, on localization of the sensor(s) in other cellular compartments, and on detection of endogenous zinc. In addition, we continue to evaluate opportunities offered by several protein-labeling techniques that, combined with zinc-selective and ratiometric sensors, could provide valuable tools to illuminate the complex zinc trafficking network in living cells and to address contemporary issues in zinc biology.
Supplementary Material
Acknowledgement
This work was supported by grant GM65519 from the NIGMS. We thank Prof. Kai Johnsson and Dr. Simone Schmitt (EPFL, Lausanne, Switzerland) for generously providing the AGT cDNA used for cloning and the AGT plasmid used for protein expression, and for helpful discussions.
Footnotes
Supporting Information Available: Experimental procedures, characterization data, Table S1, and Figures S1-S7. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Que EL, Domaille DW, Chang CJ. Chem. Rev. 2008;108:1517–1549. doi: 10.1021/cr078203u. [DOI] [PubMed] [Google Scholar]
- 2.Chang CJ, Lippard SJ. Metal Ions Life Sci. 2006;1:321–370. [Google Scholar]
- 3.Kikuchi K, Komatsu K, Nagano T. Curr. Opin. Chem. Biol. 2004;8:182–191. doi: 10.1016/j.cbpa.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Frederickson CJ, Koh JY, Bush AI. Nat. Rev. Neurosci. 2005;6:449–462. doi: 10.1038/nrn1671. [DOI] [PubMed] [Google Scholar]
- 5.MacDonald RS. J. Nutr. 2000;130:1500S–1508S. doi: 10.1093/jn/130.5.1500S. [DOI] [PubMed] [Google Scholar]
- 6.Miyawaki A, Sawano A, Kogure T. Nat. Cell Biol. 2003:S1–S7. [PubMed] [Google Scholar]
- 7.Marks KM, Nolan GP. Nature Methods. 2006;3:591–596. doi: 10.1038/nmeth906. [DOI] [PubMed] [Google Scholar]
- 8.Johnsson N, Johnsson K. ChemBioChem. 2003;4:803–810. doi: 10.1002/cbic.200200603. [DOI] [PubMed] [Google Scholar]
- 9.Chen I, Ting AY. Curr. Opin. Biotechnol. 2005;16:35–40. doi: 10.1016/j.copbio.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Rizzuto R, Simpson AWM, Brini M, Pozzan T. Nature. 1992;358:325–327. doi: 10.1038/358325a0. [DOI] [PubMed] [Google Scholar]
- 11.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 12.Marks KM, Rosinov M, Nolan GP. Chem. Biol. 2004;11:347–356. doi: 10.1016/j.chembiol.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Tour O, Adams SR, Kerr RA, Meijer RM, Sejnowski TJ, Tsien RW, Tsien RY. Nat. Chem. Biol. 2007;3:423–431. doi: 10.1038/nchembio.2007.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. Nat. Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 15.Keppler A, Pick H, Arrivoli C, Vogel H, Johnsson K. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9955–9959. doi: 10.1073/pnas.0401923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodroofe CC, Masalha R, Barnes KR, Frederickson CJ, Lippard SJ. Chem. Biol. 2004;11:1659–1666. doi: 10.1016/j.chembiol.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 17.Burdette SC, Walkup GK, Spingler B, Tsien RY, Lippard SJ. J. Am. Chem. Soc. 2001;123:7831–7841. doi: 10.1021/ja010059l. [DOI] [PubMed] [Google Scholar]
- 18.Crockett AO, Wittwer CT. Anal. Biochem. 2001;290:89–97. doi: 10.1006/abio.2000.4957. [DOI] [PubMed] [Google Scholar]
- 19.Misra A, Kumar P, Gupta KC. Anal. Biochem. 2007;364:86–88. doi: 10.1016/j.ab.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 20.Sekler I, Sensi SL, Hershfinkel M, Silverman WF. Mol. Med. 2007;13:337–343. doi: 10.2119/2007-00037.Sekler. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pierrel F, Cobine PA, Winge DR. BioMetals. 2007;20:675–682. doi: 10.1007/s10534-006-9052-9. [DOI] [PubMed] [Google Scholar]
- 22.Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, Tsien RY. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizzuto R, Brini M, Pizzo P, Murgia M, Pozzan T. Curr. Biol. 1995;5:635–642. doi: 10.1016/s0960-9822(95)00128-x. [DOI] [PubMed] [Google Scholar]
- 24.Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.