Abstract
Cryptophane-A has generated considerable interest based on its high affinity for xenon and potential for creating biosensors for 129Xe nuclear magnetic resonance (NMR) spectroscopy. Here, we report the cellular delivery of three peptide-functionalized cryptophane biosensors. Cryptophanes were delivered using two cationic cell penetrating peptides into several human cancer and normal cell lines. An RGD peptide targeting αvβ3 integrin receptor was shown to increase specificity of cryptophane cell uptake. Labeling the peptides with Cy3 made it possible to monitor cellular delivery using confocal laser scanning microscopy. The peptido-cryptophanes were determined to be relatively non-toxic by MTT assay at the micromolar cryptophane concentrations that are required for 129Xe NMR biosensing experiments.
Introduction
Magnetic resonance imaging (MRI) identifies molecules at high spatial resolution within deep tissue, and can facilitate the diagnosis of many diseases. However, MRI suffers from low sensitivity and high background signals due to intrinsic 1H signals from water and fat. In MRI procedures involving human subjects, signals are typically enhanced using gadolinium- or iron-oxide-based agents at approximately 0.1 mmol/kg. However, it has been challenging using these contrast agents to identify biomolecular targets, such as proteins, which are typically present in cells at low concentrations (1, 2). Moreover, U.S. and European agencies recently issued advisories regarding the risks posed by gadolinium agents in patients with impaired renal function. These findings motivate studies of non-proton-based, hyperpolarized MRI agents such as 129Xe, 13C, and 3He, which enable physiological assays, and provide potentially more sensitive methods for studying proteins and metabolites (3-5). 129Xe is particularly attractive due to its large chemical shift window and laser polarization of nuclear spins, which enhances 129Xe NMR signals by more than 10,000-fold. Importantly, the environmental sensitivity of the xenon chemical shift allows the simultaneous detection of multiple species in solution. 129Xe NMR biosensing has been elegantly demonstrated using a cryptophane organic host molecule that binds xenon and can be targeted to specific analytes in solution (6).
Cryptophanes are versatile host molecules whose size can be tuned to bind xenon and a variety of organic solvents and cationic species (7). In cryptophane-A, two cyclotriveratrylenes are joined by three ethylene linkers to form a 1-nm diameter pseudo-spherical cage with internal volume of ∼95 Å3. Hill et al. showed that a tricarboxylate-modified cryptophane-A binds xenon with comparable affinity in buffer (KA = 30,000 M-1) and human plasma (KA = 22,000 M-1) at 310 K (8). This avidity for xenon has motivated the design of cryptophane-A biosensors for detecting protein binding (9) or enzyme-related activity (10) by laser-polarized 129Xe nuclear magnetic resonance (NMR) spectroscopy. Biotin-functionalized cryptophanes produced a 1-4 ppm change in 129Xe NMR chemical shift upon streptavidin binding (11-14). Similarly, peptide-decorated cryptophane-A reported cleavage by matrix metalloproteinase-7 (MMP-7), which is secreted from many human tumors (10). Importantly, the cryptophane did little to affect MMP-7 activity, with peptide cleavage producing a 0.5 ppm change in 129Xe NMR chemical shift. These results motivate the development of cryptophane biosensors for in vivo 129Xe NMR studies. However, very few water-soluble cryptophanes (15) have been synthesized, and little is known about their biological activity. We are not aware of any cell or animal studies involving cryptophanes that have been reported to this time. In the current study, we investigated the uptake of peptide-modified cryptophane-A in human cancer, fibroblasts, and red blood cells.
Cryptophane-A has poor (low micromolar) water solubility and a molecular weight of 895 g/mol, which limits spontaneous intracellular diffusion, as can occur with smaller drug-like compounds. Thus, in order to test cryptophane cellular compatibility, one possible delivery method involves the use of polycationic peptides that help to solubilize the cryptophane and are rich in the basic amino acids arginine and lysine. These peptides, known generally as cell penetrating peptides (CPPs), can transport through cell membranes covalently attached cargo such as therapeutics, nucleic acids, and imaging agents (16-19). In one example, porphyrins useful for photodynamic therapy were delivered to cellular compartments by conjugating a segment of the human immunodeficiency transcriptional activator (HIV-1 TAT) or short peptides containing 6-9 consecutive arginines (20). Buckminsterfullerene, which is similar to cryptophane-A in molecular weight and chemical formula, was delivered to HEK-293 cells by attaching oligomers of lysine (21). These studies highlight the utility of CPPs for improving cellular uptake and bioavailability of large organic molecules.
Another cell delivery method involves the specific targeting of cell surface receptors, such as integrins (22-24), somatostatin (25-27), or folate (28). Integrin receptors are overexpressed in a variety of cancer cells and linked to tumor metastasis and invasion (29, 30). It is found that the αvβ3 integrin receptor in particular is substantially up-regulated in fast growing tumor cells, compared to minimum expression in most normal tissues (31, 32). A cyclic tripeptide RGD sequence is known to target the αvβ3 integrin receptor (ABIR) (33-36). More recently, it was demonstrated that linear multimeric (RGD)n sequences also target the ABIR, and promote the uptake of near infrared-emitting cypate in A549 non-small cell carcinoma cells (37). This strategy of selectively delivering biologically active molecules is particularly useful for imaging tumors, which overexpress a large number of cell surface receptors.
In the present study, we report the synthesis of cryptophane-A conjugated to HIV-1 TAT (residues 48-60), a nonamer of D-arginine, or the (RGD)4 dodecamer. Cell penetrating peptides were chosen to examine initially whether cryptophanes could be delivered to cells and to measure the effects of nonspecific delivery on cell viability. The (RGD)4 peptide was chosen for its ability to target cells overexpressing ABIR (37). Cysteine incorporation within these peptides provided a route for subsequent labeling with Cy3. Cell uptake and cytotoxicity were measured for AsPC-1 and CAPAN-2 pancreatic cancer cells, HFL-1 human lung fibroblasts, and human red blood cells.
Experimental Procedures
Reagents
Organic reagents and solvents were used as purchased from the following commercial sources: Sigma-Aldrich: dimethyl sulfoxide (DMSO), dimethylformamide (DMF), methanol, triisopropylsilane (TIS), 2,6-lutidine, piperidine, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT); Fisher: sodium chloride, copper(II) sulfate, trifluoroacetic acid (TFA), diethyl ether (Et2O), glutathione; Alfa Aesar: cesium carbonate, L-glutathione; Novabiochem: 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU), N-methylmorpholine (0.4 M), O-(2-azidoethyl)-O′-(N-diglycolyl-2-aminoethyl)heptaethyleneglycol ((PEG)7 linker), Rink amide resin, Fmoc-protected amino acids including Fmoc-L-Lys(Boc)-OH, Fmoc-Gly-OH, Fmoc-L-Arg(Pbf)-OH, Fmoc-Gln(Trt)-OH, Fmoc-Cys(tButhio)-OH, Fmoc-L-Pro-OH); GE Healthcare: Cy3 mono-reactive dye pack, Sephadex G-25 size exclusion column; Calbiochem: tris(2-carboxyethyl)phosphine hydrochloride (TCEP-HCl), cyclo-Arg-Gly-Asp-D-Phe-Val (cyclo-RGDfV); Invitrogen: RPMI medium 1640, Dulbecco's phosphate buffered saline (DPBS), anti-CD51 mouse antibody; Pierce: Ellman's reagent. For biological assays, all solutions were prepared using deionized water purified by Mar Cor Premium Grade Mixed Bed Service Deionization. The standard buffer is defined herein as 50 mM Tris, 5 mM CaCl2, 300 mM NaCl, pH 7.2.
General Methods
All air- and moisture-sensitive reactions were performed under dinitrogen with glassware oven-dried and then flamed under partial vacuum. Peptides were generated using a Protein Technologies PS3 Peptide Synthesizer. HPLC analysis was performed on an Agilent 1100 system equipped with a quaternary pump and diode array detector using a Zorbax Rx-C8 semi-preparative (9.4 × 250 mm, 5 microns) or analytical column (4.6 × 150 mm, 5 microns). The gradient eluent was composed of two solvents: 0.1% aqueous TFA (solvent A) and a 0.1% solution of TFA in CH3CN (solvent B). Mass identification of all peptide-containing compounds was performed by the Wistar Institute Proteomics Facility using an Applied Biosystems Voyager 6030 MALDI-TOF mass spectrometer. UV-visible spectra for peptides and peptide-cryptophane-A conjugates were measured using an Agilent 89090A spectrophotometer. Fluorescence spectra were collected for Cy3-labeled peptides dissolved in standard buffer (λex = 552 nm) using a Varian Cary Eclipse fluorescence spectrophotometer in small volume, 1-cm pathlength quartz cuvettes at rt (1 nm steps, 5 nm excitation and emission slits). The 96 well fluorescence measurements were made using a Labsystems Fluoroskan II microplate reader.
Peptide Synthesis and Purification
Peptides 1-3 (Scheme 1) were prepared by solid-phase synthesis using standard Fmoc amino acid protection chemistry on Rink Amide resin (0.1 mmol scale). Couplings of Fmoc-protected amino acids to the resin were carried out with HBTU and N-methylmorpholine to generate the activated ester. The resin was swelled in DMF (10 min) prior to synthesis. Amino acids were then added sequentially until 3-azidoproponic acid was attached at the N-terminus as the final step. All residues were coupled onto resin by the following procedure: removal of Fmoc group (20% piperidine solution in DMF, 2×5 min), wash (DMF, 6×30 sec), activation (amino acid/HBTU/N-methylmorpholine, 1×30 sec) coupling (amino acid/HBTU/N-methylmorpholine, 1×20 min), rinse (DMF, 3×30 sec). Cleavage from the resin was accomplished with a mixture of TFA, TIS, and water (90/5/5) at rt for 3 h. The cleavage cocktail removed side chain protecting groups from all amino acids except for cysteine, which was protected by t-butylthiol. Semi-preparative HPLC purification of all Cys-protected peptides was accomplished with a gradient: time 0, A/B = 100/0; 0-25 min, linear increase to A/B = 75/25; 25-27 min, linear change to A/B = 20/80; 27-37 min, A/B = 20/80. Selective cysteine deprotection was later performed by treating with TCEP (1 equiv) for 30 min, and running the reaction mixture through a NAP-10 gel filtration to remove TCEP.
Scheme 1.
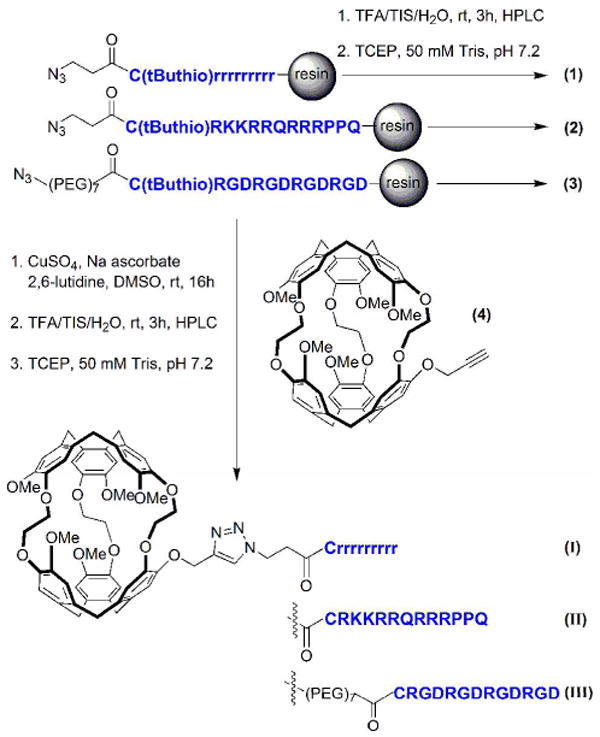
Synthesis of peptides 1-3 and peptide-cryptophane conjugates I-III. Azidopeptide on solid support was reacted with monopropargyl-cryptophane-A via a Cu(I)-mediated [3 + 2] cycloaddition reaction. (PEG)7 = -(CH2)2-O-(CH2CH2O)7-CH2CH2NHCOCH2OCH2.
The peptide sequences prepared were: N3-CH2CH2-CONH-Crrrrrrrrr (D-Arg9, 1), N3-CH2CH2-CONH-CGRKKRRQRRRPPQ (TAT, 2), and N3-CH2CH2O(PEG)7-CONH-CRGDRGDRGDRGD (RGD4, 3). MALDI MS calculated for D-Arg9 peptide, C60H119N40O12S1 (M + H+): 1623.97; found: 1624.09. MALDI MS calculated for TAT peptide, C76H139N39O18S1 (M + H+): 1918.09; found: 1919.15. MALDI MS calculated for RGD4 peptide C73H128N29O33S1 (M + H+): 1970.90; found: 1971.07. Pure peptides 1-3 were studied as-is, or subsequently conjugated with Cy3.
Synthesis of Peptide-Cryptophane Conjugates, I-III
[3+2] Cycloaddition reactions between azide-terminated peptides (still attached to solid support) and monopropargylated cryptophane-A 4 were performed to generate the peptide-cryptophane-A conjugates (Scheme 1) following established protocols (10). Briefly, an aqueous solution of copper (II) sulfate (0.006 mmol, 0.5 equiv) was added to the resin containing azidopeptide (20 mg, maximum 0.0124 mmol azidopeptide, 1 equiv) followed by 2,6-lutidine (2.78 ml, 0.024 mmol, 2 equiv), sodium ascorbate (0.018 mmol, 1.5 equiv) and 4 (21 mg, 0.024, 2 equiv). Monopropargylated cryptophane-A was synthesized as previously described (10). The suspension was degassed with N2 and stirred at rt for 16 h. The resin was then transferred to a fritted reaction vessel and washed sequentially with CH2Cl2, MeOH, water, and 1:1 MeOH:CH2Cl2 before drying under vacuum.
Cleavage from the resin was accomplished with a mixture of TFA, TIS, and water (90/5/5) at rt for 3 h. The Cys-protected peptide-cryptophane conjugates were precipitated and washed with anhydrous Et2O before drying under vacuum. Analytical HPLC was performed using a gradient: time 0, A/B = 75/25; 0-25 min, linear increase to A/B = 50/50; time 25-27 min, linear change to A/B = 20/80; 27-37 min, A/B = 20/80. HPLC traces and retention times are provided in the Supporting Information. The cysteine was subsequently deprotected with TCEP in order to generate conjugates I-III (Scheme 1). MALDI MS calculated for C116H173N40O24S1 (D-Arg9-cryptophane, I) (M + H+): 2542.33; found: 2542.08. MALDI MS calculated for C132H193N39O30S1 (TAT-cryptophane, II) (M + H+): 2836.45; found: 2836.96. MALDI MS calculated for C129H182N29O45S1 ((RGD)4-cryptophane, III) (M + H+): 2889.26; found: 2889.07. An extinction coefficient for I–III, 9,300 M-1 cm-1 at 280 nm in water, was determined from solutions containing a weighed sample, as described previously (10).
Cysteine Labeling with Cy3
The pure Cys-protected peptides and Cys-protected peptide-cryptophane-A conjugates were dissolved in Tris buffer (50 mM, pH 7.2) at a concentration of 60 μM. Cysteine labeling with the Cy3-maleimide construct was performed according to the manufacturer's protocol. As described previously, cysteine was deprotected with TCEP, which was then removed by NAP-10 column. Cy3 dye was added dropwise and reacted for 3 h. The reaction was quenched by addition of glutathione (1 equiv). The reaction mixture was run through a NAP-10 gel filtration column before analytical HPLC purification.
For Cy3-labeled I-III, HPLC elution was achieved with the following gradient: time 0, A/B = 75/25; 0-25 min, linear increase to A/B = 50/50; 25-27 min, linear change to A/B = 20/80; 27-37 min, A/B = 20/80. MALDI MS calculated for C153H215N44O33S3 (Cy3-I) (M+H+): 3304.57; MALDI found: 3304.82. MALDI MS calculated for C170H235N43O39S3 (Cy3-II) (M+H+): 3586.69; MALDI found: 3587.84. MALDI MS calculated for C167H224N33O54S3 (Cy3-III) (M+H+): 3639.50; MALDI found: 3640.07. Extinction coefficients used to determine solution concentrations of Cy3-labeled I–III were ε280 = 12,300 M-1cm-1 and ε552 = 150,000 M-1cm-1 in water.
For Cy3-labeled 1-3, HPLC elution was achieved with the following gradient: time 0, A/B = 100/0; 0-25 min, linear increase to A/B = 75/25; 25-27 min, linear change to A/B = 20/80; 27-37 min, A/B = 20/80. HPLC traces and retention times are provided in the Supporting Information. MALDI MS calculated for C97H161N44O21S3 (Cy3-1) (M + H+ - H2O): 2356.30; MALDI found: 2357.68. MALDI MS calculated for C113H181N43O27S3 (Cy3-2) (M + H+ - 2 H2O): 2632.29; MALDI found: 2632.12. MALDI MS calculated for C110H170N33O42S3 (Cy3-3) (M + H+ - N2): 2697.12; MALDI found: 2696.37. The concentration of Cy3-labeled peptides 1-3 in solution was determined using ε552 = 150,000 M-1cm-1, and accounting for the fraction labeled. Cy3 labeling efficiency was determined using Ellman's reagent according to manufacturer's protocol.
Cell Culture
HFL-1 human diploid lung fibroblasts, and AsPC-1 and CAPAN-2 human pancreatic carcinoma cell lines were obtained from the Cell Culture Core of the Center for Molecular Studies in Liver and Digestive Diseases (University of Pennsylvania Medical School, Philadelphia, PA). NCI-H1975 cells were from Dr. Intae Lee and red blood cells were from Dr. Don Siegel (University of Pennsylvania Medical School, Philadelphia, PA). All cells were grown in 25 cm2 tissue culture flasks in RPMI-1640 with 25 mM HEPES supplemented with 2 mM L-glutamine, 15% fetal calf serum, 100 units of penicillin and 100 units of streptomycin. Cells were subcultured weekly, or more frequently, as needed.
Cytotoxicity Assays
In 96 well plates, 25,000 cells were plated per well and allowed to grow overnight. Non-fluorescently labeled peptide-cryptophane conjugates I-III were added from a stock solution in DPBS to wells in triplicate at final concentrations of 2, 10, 25, 50, 80 and 100 μM and incubated for 24 h in the dark. The medium was removed and the cells were washed thrice with DPBS before being treated with 20 μL of MTT for 3 h. The medium was removed once more and the resulting crystals were solubilized in DMSO. Absorbance at 540 nm was recorded in each well using the plate reader. Absorbance readings were subtracted from the value of wells containing untreated cells, and the reduction in cell growth was calculated as a percentage of control absorbance in the absence of any treatment. Data show the mean of at least three independent experiments +/- SD.
Cell Uptake Studies
Cells were grown to confluence on LabTek 8-well microscope slides with glass coverslip bottoms. For uptake studies, cells were incubated with 2 μM solutions of Cy3-labeled I, II, or III for 15 min. For blocking studies, cells were pretreated for 45 min with an 100 μM solution of a cyclic RGDfV peptide containing D-phenylalanine that was previously identified to bind tightly to the integrin αvβ3 receptor (37, 38). In the antibody blocking studies, cells were pretreated for 30 min with 10 μM blocking anti-α antibody. For visualizing the results of both the uptake and blocking studies, the medium was removed and the cells were washed three times with DPBS. Cells were visualized using an Olympus FV1000 confocal laser scanning microscope with 543 nm (HeNe) laser excitation and Cy3 emission filter under 40× magnification (Olympus UApo/340 40×, 1.15 W).
Time-Dependent Cellular Uptake
Cells were plated at 10,000 cells per well in a CoStar 96-well plate and allowed to grow overnight. Cy3-labeled I, II, or III was added to the wells at a concentration of 10 μM and incubated for 0, 1, 2, 4, 8, and 24 h at 37 °C. At the end of the incubation time the loading medium was removed, and the cells were washed thrice with DPBS. The cells were solubilized by the addition of 100 μL of 0.25% Triton X-100 in DPBS. To determine conjugate concentration, samples were excited at 544 nm and fluorescence at 590 nm was recorded using a plate reader. The cell numbers were then quantified using the CyQuant reagent (Molecular Probes) according to manufacturer protocol.
Results and Discussion
Synthesis and Characterization of Peptide-Cryptophane Conjugates
Monopropargylated cryptophane-A 4 was synthesized in 12 nonlinear steps and 3% overall yield (10). The yields of purified peptides (1-3) were 80-85% for all peptide coupling and cleavage steps, based on the maximum possible yield from starting resin. Azidopeptides, while still attached to the solid support, were reacted with 4 by a copper (I)-catalyzed [3+2] cycloaddition in 80-89% yield (39, 40). Peptide-cryptophane conjugates (with Cys still protected) were cleaved from the resin, and purified by reverse-phase HPLC (Figures S1-S3, Supporting Information). The cysteine was deprotected before use with TCEP to yield I-III. The deprotected cysteines showed no evidence of disulfide formation leading to cryptophane dimerization on the timescale of hours when purified by HPLC. The free thiols, however, may interact with endogenous sulfhydryls when introduced into cells.
The (RGD)4 linear peptide was chosen to target ABIR because it was readily synthesized and attached to cryptophane using established protocols (10). Furthermore, polypeptides of similar length were found to confer excellent cryptophane water solubility. In constrast, the cryptophane-(RGD)4 conjugate presented solubility problems, most likely due to charge interactions between the repeating sequence of arginine and glutamic acid residues. A prior version of conjugate III was synthesized without the PEG7 linker separating the cryptophane from the RGD tetramer. This prototype exhibited poor water solubility, which prevented its usefulness for in vitro cell studies. Addition of the PEG7 linker increased water solubility, allowing concentrations up to 120 μM when DMSO (10% final concentration) was added to the buffer solutions.
The cysteine sulfhydryl group on the peptides and peptide-cryptophane conjugates was deprotected and reacted with maleimide-functionalized Cy3 to generate Cy3-labeled 1-3 and I-III. Mixtures of fluorescently labeled and unlabeled conjugates were obtained after reverse-phase HPLC purification (Figures S4-S6, Supporting Information). For the cryptophane-peptide conjugates I-III, Cy3 labeling efficiencies of 50-69% were determined from the ratio of dye absorbance at 552 nm to the cryptophane absorbance at 280 nm. Cy3 labeling efficiencies were initially lower than expected. To improve yields, the reducing agent TCEP was removed from the solution by gel filtration chromatography before the addition of the Cy3 mono-reactive dye pack. Removal of TCEP improved Cy3 conjugation yields by about 10%.
Ellman's reagent was used to determine the Cy3 labeling efficiencies for 1-3, and confirmed the previous measurements determined for Cy3-labeled I-III. The 5,5′-dithiobis(2-nitrobenzoic acid) reacted with any free (previously unreacted) sulfhydryl groups to yield 2-nitro-5-thiobenzoic acid, which was quantified by measuring absorbance at 412 nm (ε412 =14,150 M-1cm-1) (41). This assay found slightly higher Cy3 labeling efficiencies for 1-3 (60-72%) than for I-III (47-70%), which was in good agreement with the previously described ratiometric (A552/A280) UV-Vis measurements for Cy3-labeled I-III.
Cell Toxcity Assays
The cytotoxicity of the cryptophane conjugates was evaluated in AsPC-1, CAPAN-2, and HFL-1 cell lines by exposing cells to increasing concentrations of I, II, or III as shown in Figure 1. The (RGD)4-cryptophane conjugate III showed 25-30% proliferation inhibition of the various cell lines at the highest concentration studied (100 μM). Slightly higher amounts of cytotoxicity were also found for the D-Arg9- and TAT-cryptophane conjugates, I and II, in both the AsPC-1 and CAPAN-2 cell lines. In general, the three conjugates appeared to be more toxic to HFL-1 fibroblast cells than either AsPC-1 or CAPAN-2 cancer cells. The toxicity profile is comparable to that of the similarly sized fullerene C60 molecule, which decreased viability of HEK cells by 10-50% over a similar range of concentrations (42). For compound III, 50% inhibition of proliferation was not reached in any of the cell lines at the concentrations tested. Compounds I and II showed 50% inhibition at 60 and 75 μM in HFL-1 fibroblasts. I also showed IC50 of 75 μM in AsPC-1 cells and more than 100 μM in CAPAN-2. II had IC50 values of 100 μM in AsPC-1 cells while 50% inhibition was not reached in CAPAN-2 cells at 100 μM. Increased toxicity for compounds I and II as compared to III is thought to be a result of cryptophane delivery to the cell nucleus by TAT or D-Arg9 nuclear localization peptides. For compound III, 10% DMSO was required to work at the higher concentrations. Cell viability assays for this compound were performed using 10% DMSO at all sample concentrations and these viability data were normalized to cells treated with 10% DMSO without cryptophane. In absolute terms, 10% DMSO alone reduced cell counts by 2000-3500, corresponding to a 8-14% decrease compared to untreated cells.
Figure 1.
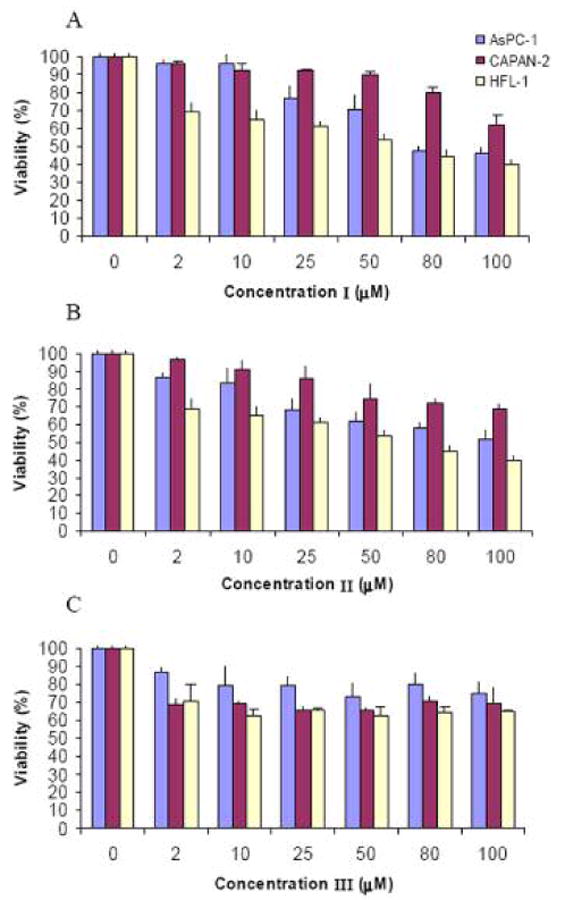
Cytotoxicity assay for conjugates I (A), II (B), and III (C) in AsPC-1 (blue), CAPAN-2 (maroon) and HFL-1 (off-white) cells. %Viability was determined via MTT assay after 24 h incubation with increasing concentrations of each conjugate, as compared to untreated cells.
Cell Uptake Studies
The time-dependent uptake of Cy3-labeled peptide-cryptophane conjugates I-III and peptides 1-3 at 10 μM was investigated in CAPAN-2 cells (Figure 2). All conjugates showed similar uptake kinetics with a rapid accumulation of compounds within the first hour. Maximum uptake was achieved by 4 h and a plateau was observed for the remaining 20 h. In order to quantify cell uptake for each time point, 10,000 cells containing each fluorescent compound were placed in 100 μL lysis buffer. The concentrations of fluorescent peptides and conjugates were quantified by fluorometry after generating a fluorescence standard curve for Cy3 in the same lysis buffer, and correcting for the Cy3-labeling efficiency of each compound. Fluorescent peptides 1-3 were measured in the cell lysate solution at concentrations of 1.0-1.4 μM, whereas fluorescent cryptophane conjugates I-III were determined to be 500-800 nM. Within the error of the measurements, we cannot draw distinctions regarding the uptake efficiency of the three peptide motifs.
Figure 2.
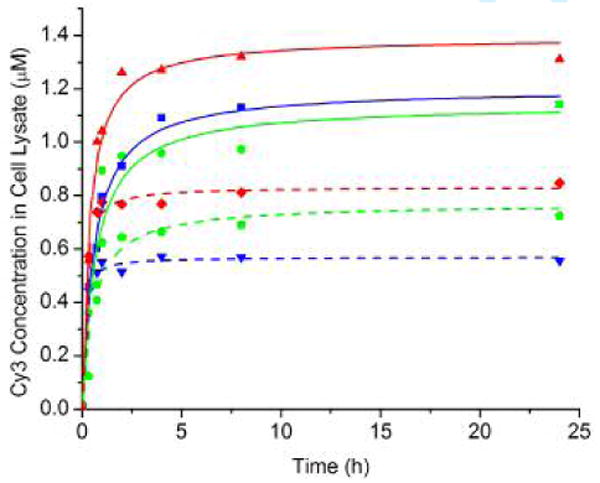
Uptake of peptides (solid lines) was measured for 1 (blue squares), 2 (red triangles) and 3 (green circles) and peptide-cryptophane conjugates (dashed lines) for I (blue inverted triangles), II (red diamonds) and III (green pentagons) by CAPAN-2 cells over 24 h.
A procedure was developed to estimate the intracellular concentration of each compound from the lysate data. When detached from the surface, the CAPAN-2 cells were spherical and measured with a microscope to be ∼35 μm in diameter. The average cell volume was calculated, and the fraction of the 100 μL cell lysate originally occupied by the cells was determined to be 0.0050. Thus, the measured cell lysate concentrations of 500-800 nM gave an approximate range of intracellular concentrations of 100-160 μM for I-III after the 24-h incubation period. This corresponds to 10-16-fold enrichment of I-III inside the cell (originally present in the media at 10 μM). Support for this finding comes from ratiometric measurements (Figures S7-S9, Supporting Information) showing a range of fluorescence intensities that is 4-20 times greater inside the CAPAN-2 cells than the surrounding cell media. Importantly, these intracellular concentrations are within the limits of detection for laser-polarized 129Xe NMR experiments (43).
Confocal laser scanning microscopy (CLSM) confirmed cellular delivery of cryptophane with the D-Arg9- and TAT-cryptophane conjugates, I and II (Figure 3). After 1-h exposure to Cy3-labeled I and II, fluorescence was seen evenly distributed throughout most AsPC-1, CAPAN-2 and HFL-1 cells. Cy3-labeled I and II translocated into the cell nucleus, as was expected for conjugates of these peptides (44-46). After 1-h incubation at 4 °C with Cy3-labeled I and II, no fluorescence was observed (Figures 3D, 3H) by CLSM, which confirmed that uptake occurred via an energy dependent endocytic pathway.
Figure 3.
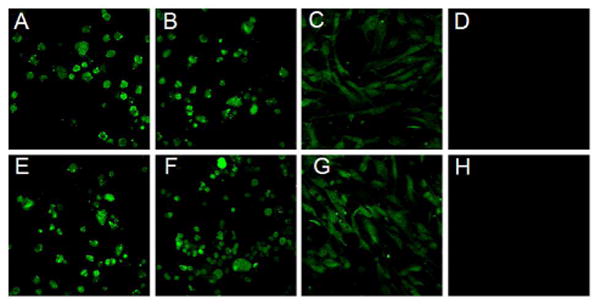
Cellular uptake of 2 μM Cy3-labeled I (top row) and II (bottom row) in AsPC-1 (A, E), CAPAN-2 (B, F) and HFL-1 cells (C, G) after 1 h incubation at 37 °C. Uptake of I and II was inhibited after 1 h incubation at 4 °C (D, H).
Cell uptake of Cy3-labeled III was also confirmed by CLSM. After 30-min incubation and rigorous cell washing, fluorescence was observed in NCI-H1975, CAPAN-2, and AsPC-1 cells (Figures 4A-C). Translocation of Cy3-labeled III was not observed in red blood cells, as expected, as these cells do not express αvβ3 integrin receptor (Figure 4D). In contrast to Cy3-labeled I and II, Cy3-labeled III was not observed to cross the nuclear membrane. In general, nuclear internalization of imaging agents is undesirable because of potential mutagenic effects on healthy cells. To determine whether uptake of III was mediated by ABIR, cells were preincubated with 100 μM cyclic RGDfV peptide, which is known to bind αvβ3 integrin receptor with high affinity (47). Figure 4D shows that the cyclic RGDfV peptide inhibited uptake of (RGD)4-cryptophane as indicated by reduction in fluorescence. Internalization was also inhibited by co-incubation with 10 μM anti-αv mouse antibody (Figure 4F). Cyclic RGDfV and anti-αv mouse antibody blocking groups also inhibited uptake in HFL-1 and CAPAN-2 cells (data not shown). These blocking data suggest that the (RGD)4-labeled conjugate was specifically recognized by the αvβ3 integrin receptor and the αv subunit is critical for cellular uptake of III.
Figure 4.
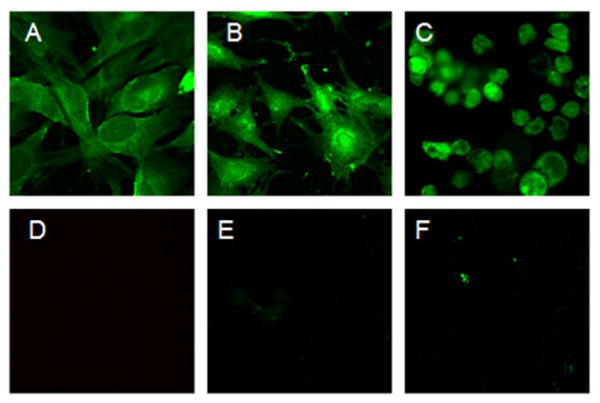
Uptake of 2 μM Cy3-labeled III targeting αvβ3 integrin in NCI-H1975 (A), HFL-1 (B), and CAPAN-2 (C) cells after 30 min incubation at 37 °C. Red blood cells (D) did not uptake this compound. Targeting in NCI-H1975 was successfully blocked with 100 μM cyclic RGDfV peptide (E) and 10 μM anti-αv antibody (F).
Attempts at performing hyperpolarized 129Xe NMR spectroscopy were made with compound III in isolated cells in culture. However, these experiments were unsuccessful due to the limited solubility of III and inability to achieve sufficient cell density within the detection volume. The requirement for incorporating DMSO in the buffers used to solubilize III further complicated the NMR data collection, as 129Xe NMR chemical shift is very sensitive to solvent composition. The requirement for DMSO also made it difficult to achieve the high density of viable cells required for these NMR experiments.
Conjugates I-III have molecular weights ∼3500 Da and their transport and clearance properties in vivo are not yet known. Fullerenes, which are similar in size and chemical composition, have been found to localize to a variety of tissues in rats with large accumulations in the liver, spleen, and kidneys, suggesting that these compounds are cleared rapidly (48, 49). PEG coatings may be used to improve bioavailability if the biodistribution of cryptophane is found to be similar to that of fullerene (50).
Conclusion
Cryptophane-A was conjugated to (D-Arg)9, TAT, and (RGD)4 peptides using a modified [3 + 2] cycloaddition reaction. This reaction provides a facile route for synthesizing chemically stable peptide-cryptophane conjugates (39, 51). All compounds were examined for cell toxicity, and fluorescently labeled with Cy3 to facilitate cellular imaging studies by CLSM.
Conjugates I and II were delivered nonspecifically to all cell types tested and showed little cytotoxicity in concentration ranges that are relevant for solution-based hyperpolarized 129Xe NMR experiments. Conjugate III, which contained an (RGD)4 αvβ3 integrin receptor binding motif, was selectively delivered to cells expressing this receptor and excluded from human red blood cells. III was similarly nontoxic as I and II. Importantly for laser-polarized 129Xe NMR studies, cryptophane concentrations in excess of 100 μM were achieved in CAPAN-2 cells using the three peptides.
An important goal of these studies is to identify cell delivery motifs that can be attached to cryptophane and effectively discriminate between cancer and normal cells. The results of this initial study are very promising, and motivate the search for peptides and small molecules that promote the cellular delivery of even higher concentrations of cryptophanes, with reduced toxicity, in order to facilitate in vivo NMR studies. As examples, the transportan and penetratin CPP sequences were shown previously to deliver higher concentrations of attached cargo than TAT and (D-Arg)9 peptides (52, 53). Additional cell targeting motifs will be explored for use with other cancer biomarkers. Finally, protocols must be developed to perform 129Xe MRI experiments using xenon biosensors in tissue samples, where higher cellular density (and associated cryptophane per unit volume) will improve 129Xe MR signals. This work demonstrates that due to their limited cytotoxicity and ready delivery to cells expressing specific cell surface receptors, cryptophanes offer considerable promise for in vivo xenon biosensing experiments.
Supplementary Material
HPLC analysis of peptide-cryptophane conjugates I-III and Cy3-labeled peptides 1-3, and CLSM images of Cy3-labeled peptide 3 in CAPAN-2 cells. This material is available free of charge via the Internet at http://pubs.acs.org/BC.
Acknowledgments
We thank Intae Lee for providing cells and helpful discussions; Jeffery Saven, Ronen Marmorstein, and Scott Diamond for access to instrumentation; the Cell Culture Core of the University of Pennsylvania Center for Molecular Studies in Liver and Digestive Diseases for cells; Tobias Baumgart and Eric Meggers for access to cell culture facilities. This work was supported by the DOD (W81XWH-04-1-0657), NIH (1R21CA110104, 1R33CA110104, 1S10RR021113-01), and Camille and Henry Dreyfus Foundation.
References
- 1.Degani H, Gusis V, Weinstein D, Feilds S, Strano S. Mapping pathophysiological features of breast tumors by MRI at high spatial resolution. Nat Medicine. 1997;3:780–782. doi: 10.1038/nm0797-780. [DOI] [PubMed] [Google Scholar]
- 2.Foster-Gareau P, Heyn C, Alejski A, Rutt BK. Imaging single mammalian cells with a 1.5 T clinical MRI scanner. Mag Res Med. 2003;49:968–971. doi: 10.1002/mrm.10417. [DOI] [PubMed] [Google Scholar]
- 3.Golman K, Zandt RI, Lerche M, Pehrson R, Ardenkjaer-Larsen JH. Metabolic imaging by hyperpolarized 13C magnetic resonance imaging for in vivo tumor diagnosis. Cancer Res. 2006;66:10855–10860. doi: 10.1158/0008-5472.CAN-06-2564. [DOI] [PubMed] [Google Scholar]
- 4.Hopkins SR, Levin DL, Emami K, Kadlecek S, Yu J, Ishii M, Rizi RR. Advances in magnetic resonance imaging of lung physiology. J Appl Physiol. 2007;102:1244–1254. doi: 10.1152/japplphysiol.00738.2006. [DOI] [PubMed] [Google Scholar]
- 5.Mugler JP, Driehuys B, Brookeman JR, Cates GD, Berr SS, Bryant RG, Daniel TM, deLange EE, Downs JH, Erickson CJ, Happer W, Hinton DP, Kassel NF, Maier T, Phillips CD, Saam BT, Sauer KL, Wagshul ME. MR imaging and spectroscopy using hyperpolarized Xe-129 gas: Preliminary human results. Magn Reson Med. 1997;37:809–815. doi: 10.1002/mrm.1910370602. [DOI] [PubMed] [Google Scholar]
- 6.Spence MM, Rubin SM, Dimitrov IE, Ruiz EJ, Wemmer DE, Pines A, Yao SQ, Tian F, Schultz PG. Functionalized xenon as a biosensor. Proc Natl Acad Sci USA. 2001;98:10654–10657. doi: 10.1073/pnas.191368398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collet A. Cryptophanes. Comp Supramol Chem. 1996;2:325–365. [Google Scholar]
- 8.Hill PA, Wei Q, Eckenhoff RG, Dmochowski IJ. Thermodynamics of xenon binding to cryptophane in water and human plasma. J Am Chem Soc. 2007;129:9262–9263. doi: 10.1021/ja072965p. [DOI] [PubMed] [Google Scholar]
- 9.Spence MM, Ruiz EJ, Rubin SM, Lowery TJ, Winssinger N, Schultz PG, Wemmer DE, Pines A. Development of a functionalized xenon biosensor. J Am Chem Soc. 2004;126:15287–15294. doi: 10.1021/ja0483037. [DOI] [PubMed] [Google Scholar]
- 10.Wei Q, Seward GK, Hill PA, Patton B, Dimitrov IE, Kuzma NN, Dmochowski IJ. Designing Xe-129 NMR biosensors for matrix metalloproteinase detection. J Am Chem Soc. 2006;128:13274–13283. doi: 10.1021/ja0640501. [DOI] [PubMed] [Google Scholar]
- 11.Spence MM, Rubin SM, Dimitrov IE, Ruiz EJ, Wemmer DE, Pines A, Yao SQ, Tian F, Schultz PG. Functionalized xenon as a biosensor. Proc Natl Acad Sci USA. 2001;98:10654–10657. doi: 10.1073/pnas.191368398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowery TJ, Garcia S, Chavez L, Ruiz EJ, Wu T, Brotin T, Dutasta JP, King DS, Schultz PG, Pines A, Wemmer DE. Optimization of xenon biosensors for detection of protein interactions. Chembiochem. 2006;7:65–73. doi: 10.1002/cbic.200500327. [DOI] [PubMed] [Google Scholar]
- 13.Ruiz EJ, Sears DN, Pines A, Jameson CJ. Diastereomeric Xe chemical shifts in tethered cryptophane cages. J Am Chem Soc. 2006;128:16980–16988. doi: 10.1021/ja066661z. [DOI] [PubMed] [Google Scholar]
- 14.Hilty C, Lowery TJ, Wemmer DE, Pines A. Spectrally resolved magnetic resonance imaging of a xenon biosensor. Angew Chem, Int Ed. 2006;45:70–73. doi: 10.1002/anie.200502693. [DOI] [PubMed] [Google Scholar]
- 15.Huber G, Brotin T, Dubois L, Desvaux H, Dutasta JP, Berthault P. Water soluble cryptophanes showing unprecedented affinity for xenon: Candidates as NMR-based biosensors. J Am Chem Soc. 2006;128:6239–6246. doi: 10.1021/ja060266r. [DOI] [PubMed] [Google Scholar]
- 16.Gupta B, Levchenko TS, Torchilin VP. Intracellular delivery of large molecules and small particles by cell-penetrating proteins and peptides. Adv Drug Delivery Rev. 2005;57:637–651. doi: 10.1016/j.addr.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Wright LR, Rothbard JB, Wender PA. Guanidinium rich peptide transporters and drug delivery. Curr Protein Pept Sci. 2003;4:105–124. doi: 10.2174/1389203033487252. [DOI] [PubMed] [Google Scholar]
- 18.Goun EA, Pillow TH, Jones LR, Rothbard JB, Wender PA. Molecular transporters: Synthesis of oligoguanidinium transporters and their application to drug delivery and real-time imaging. Chembiochem. 2006;7:1497–1515. doi: 10.1002/cbic.200600171. [DOI] [PubMed] [Google Scholar]
- 19.Goun EA, Shinde R, Dehnert KW, Adams-Bond A, Wender PA, Contag CH, Franc BL. Intracellular cargo delivery by an octaarginine transporter adapted to target prostate cancer cells through cell surface protease activation. Bioconj Chem. 2006;17:787–796. doi: 10.1021/bc0503216. [DOI] [PubMed] [Google Scholar]
- 20.Sibrian-Vazquez M, Jensen TJ, Hammer RP, Vicente MGH. Peptide-mediated cell transport of water soluble porphyrin conjugates. J Med Chem. 2006;49:1364–1372. doi: 10.1021/jm050893b. [DOI] [PubMed] [Google Scholar]
- 21.Yang JH, Wang K, Driver J, Yang JH, Barron AR. The use of fullerene substituted phenylalanine amino acid as a passport for peptides through cell membranes. Org Biomol Chem. 2007;5:260–266. doi: 10.1039/b614298b. [DOI] [PubMed] [Google Scholar]
- 22.Belvisi L, Bernardi A, Colombo M, Manzoni L, Potenza D, Scolastico C, Giannini G, Marcellini M, Riccioni T, Castorina M, LoGiudice P, Pisano C. Targeting integrins: Insights into structure and activity of cyclic RGD pentapeptide mimics containing azabicycloalkane amino acids. Bioorg Med Chem. 2006;14:169–180. doi: 10.1016/j.bmc.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 23.Harris TD, Kalogeropoulos S, Nguyen T, Liu S, Bartis J, Ellars C, Edwards S, Onthank D, Silva P, Yalamanchili P, Robinson S, Lazewatsky J, Barrett J, Bozarth J. Design, synthesis, and evaluation of radiolabeled integrin alpha(v)beta(3) receptor antagonists for tumor imaging and radiotherapy. Cancer Biother Radiopharm. 2003;18:627–641. doi: 10.1089/108497803322287727. [DOI] [PubMed] [Google Scholar]
- 24.Temming K, Schiffelers RM, Molema G, Kok RJ. RGD-based strategies for selective delivery of therapeutics and imaging agents to the tumour vasculature. Drug Resist Updates. 2005;8:381–402. doi: 10.1016/j.drup.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Cescato R, Schulz S, Waser B, Eltschinger V, Rivier JE, Wester HJ, Culler M, Ginj M, Liu QS, Schonbrunn A, Reubi JC. Internalization of sst(2), sst(3), and sst(5) receptors: Effects of somatostatin agonists and antagonists. J Nucl Med. 2006;47:502–511. [PubMed] [Google Scholar]
- 26.Ginj M, Zhang HW, Waser B, Cescato R, Wild D, Wang XJ, Erchegyi J, Rivier J, Macke HR, Reubi JC. Radiolabeled somatostatin receptor antagonists are preferable to agonists for in vivo peptide receptor targeting of tumors. Proc Natl Acad Sci USA. 2006;103:16436–16441. doi: 10.1073/pnas.0607761103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reubi JC, Eisenwiener KP, Rink H, Waser B, Macke HR. A new peptidic somatostatin agonist with high affinity to all five somatostatin receptors. Eur J Pharmacol. 2002;456:45–49. doi: 10.1016/s0014-2999(02)02651-1. [DOI] [PubMed] [Google Scholar]
- 28.Hilgenbrink AR, Low PS. Folate receptor-mediated drug targeting: From therapeutics to diagnostics. J Pharm Sci. 2005;94:2135–2146. doi: 10.1002/jps.20457. [DOI] [PubMed] [Google Scholar]
- 29.Byzova TV, Goldman CK, Pampori N, Thomas KA, Bett A, Shattil SJ, Plow EF. A mechanism for modulation of cellular responses to VEGF: Activation of the integrins. Mol Cell. 2000;6:851–860. [PubMed] [Google Scholar]
- 30.Byzova TV, Kim W, Midura RJ, Plow EF. Activation of integrin alpha(v)beta(3) regulates cell adhesion and migration to bone sialoprotein. Exp Cell Res. 2000;254:299–308. doi: 10.1006/excr.1999.4765. [DOI] [PubMed] [Google Scholar]
- 31.Cairns RA, Khokha R, Hill RP. Molecular mechanisms of tumor invasion and metastasis: An integrated view. Curr Mol Med. 2003;3:659–671. doi: 10.2174/1566524033479447. [DOI] [PubMed] [Google Scholar]
- 32.Felding-Habermann B. Integrin adhesion receptors in tumor metastasis. Clin Exp Metastas. 2003;20:203–213. doi: 10.1023/a:1022983000355. [DOI] [PubMed] [Google Scholar]
- 33.Pierschbacher MD, Hayman EG, Ruoslahti E. The Cell Attachment Determinant in Fibronectin. J Cell Biochem. 1985;28:115–126. doi: 10.1002/jcb.240280205. [DOI] [PubMed] [Google Scholar]
- 34.Pierschbacher MD, Ruoslahti E. Cell Attachment Activity of Fibronectin Can Be Duplicated by Small Synthetic Fragments of the Molecule. Nature. 1984;309:30–33. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 35.Ruoslahti E. The RGD story: a personal account. Matrix Biol. 2003;22:459–465. doi: 10.1016/s0945-053x(03)00083-0. [DOI] [PubMed] [Google Scholar]
- 36.Ruoslahti E, Pierschbacher MD. Arg-Gly-Asp - a Versatile Cell Recognition Signal. Cell. 1986;44:517–518. doi: 10.1016/0092-8674(86)90259-x. [DOI] [PubMed] [Google Scholar]
- 37.Ye YP, Bloch S, Xu BG, Achilefu S. Design, synthesis, and evaluation of near infrared fluorescent multimeric RGD peptides for targeting tumors. J Med Chem. 2006;49:2268–2275. doi: 10.1021/jm050947h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gottschalk KE, Kessler H. The structures of Integrins and integrin-ligand complexes: Implications for drug design and signal transduction. Angew Chem, Int Ed. 2002;41:3767–3774. doi: 10.1002/1521-3773(20021018)41:20<3767::AID-ANIE3767>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 39.Punna S, Kuzelka J, Wang Q, Finn MG. Head-to-tail peptide cyclodimerization by copper-catalyzed azide-alkyne cycloaddition. Angew Chem, Int Ed. 2005;44:2215–2220. doi: 10.1002/anie.200461656. [DOI] [PubMed] [Google Scholar]
- 40.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise Huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem, Int Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 41.Riddles PW, Blakeley RL, Zerner B. Reassessment of Ellman's Reagent. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- 42.Rouse JG, Yang JZ, Barron AR, Monteiro-Riviere NA. Fullerene-based amino acid nanoparticle interactions with human epidermal keratinocytes. Toxicol In Vitro. 2006;20:1313–1320. doi: 10.1016/j.tiv.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 43.Lowery TJ, Rubin SM, Ruiz EJ, Spence MM, Winssinger N, Schultz PG, Pines A, Wemmer DE. Applications of laser-polarized Xe-129 to biomolecular assays. Magn Reson Imag. 2003;21:1235–1239. doi: 10.1016/j.mri.2003.08.025. [DOI] [PubMed] [Google Scholar]
- 44.Lindgren M, Gallet X, Soomets U, Hallbrink M, Brakenhielm E, Pooga M, Brasseur R, Langel U. Translocation properties of novel cell penetrating transportan and penetratin analogues. Bioconj Chem. 2000;11:619–626. doi: 10.1021/bc990156s. [DOI] [PubMed] [Google Scholar]
- 45.Lindgren M, Hallbrink M, Prochiantz A, Langel U. Cell-penetrating peptides. Trends Pharmacol Sci. 2000;21:99–103. doi: 10.1016/s0165-6147(00)01447-4. [DOI] [PubMed] [Google Scholar]
- 46.Zorko M, Langel U. Cell-penetrating peptides: mechanism and kinetics of cargo delivery. Adv Drug Delivery Rev. 2005;57:529–545. doi: 10.1016/j.addr.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 47.Haubner R, Wester HJ, Burkhart F, Senekowitsch-Schmidtke R, Weber W, Goodman SL, Kessler H, Schwaiger M. Glycosylated RGD-containing peptides, tracer for tumor targeting and angiogenesis imaging with improved biokinetics. J Nucl Med. 2001;42:326–336. [PubMed] [Google Scholar]
- 48.Bolskar RD, Benedetto AF, Husebo LO, Price RE, Jackson EF, Wallace S, Wilson LJ, Alford JM. First soluble M@C-60 derivatives provide enhanced access to metallofullerenes and permit in vivo evaluation of Gd@C-60[C(COOH)(2)](10) as a MRI contrast agent. J Am Chem Soc. 2003;125:5471–5478. doi: 10.1021/ja0340984. [DOI] [PubMed] [Google Scholar]
- 49.Xu JY, Li QN, Li JG, Ran TC, Wu SW, Song WM, Chen SL, Li WX. Biodistribution of Tc-99m-C-60(OH)(x) in Sprague-Dawley rats after intratracheal instillation. Carbon. 2007;45:1865–1870. [Google Scholar]
- 50.Tabata Y, Ikada Y. Biological functions of fullerene. Pure Appl Chem. 1999;71:2047–2053. [Google Scholar]
- 51.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise Huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem-Int Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 52.Bendifallah N, Rasmussen FW, Zachar V, Ebbesen P, Nielsen PE, Koppelhus U. Evaluation of cell-penetrating peptides (CPPs) as vehicles for intracellular delivery of antisense peptide nucleic acid (PNA) Bioconj Chem. 2006;17:750–758. doi: 10.1021/bc050283q. [DOI] [PubMed] [Google Scholar]
- 53.Myrberg H, Lindgren M, Langel U. Protein delivery by the cell-penetrating peptide YTA2. Bioconj Chem. 2007;18:170–174. doi: 10.1021/bc060266g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HPLC analysis of peptide-cryptophane conjugates I-III and Cy3-labeled peptides 1-3, and CLSM images of Cy3-labeled peptide 3 in CAPAN-2 cells. This material is available free of charge via the Internet at http://pubs.acs.org/BC.


