Abstract
The (6S,8R,11S) 1,N2-HNE-dG adduct of trans-4-hydroxynonenal (HNE) was incorporated into the duplex 5′-d(GCTAGCXAGTCC)-3′•5′-d(GGACTAGCTAGC)-3′ [X=(6S,8R,11S) HNE-dG], in which the lesion was mismatched opposite dA. The (6S,8R,11S) adduct maintained the ring-closed 1,N2-HNE-dG structure. This was in contrast to when this adduct was correctly paired with dC, conditions under which it underwent ring opening and re-arrangement to diastereomeric minor groove hemiacetals [Huang, H., Wang, H., Qi, N., Lloyd, R.S., Harris, T.M., Rizzo, C.J., & Stone, M.P. (2008) J. Am. Chem. Soc. 130, 10898–10906]. The (6S,8R,11S) adduct exhibited a syn/anti conformational equilibrium about the glycosyl bond. The syn conformation was predominant in acidic solution. Structural analysis of the syn conformation revealed that X7 formed a distorted base pair with the complementary protonated A18. The HNE moiety was located in the major groove. Structural perturbations were observed at the neighbor C6•G19 and A8•T17 base pairs. At basic pH, the anti conformation of X7 was the major species. At X7 the 1,N2-HNE-dG intercalated and displaced the complementary A18 in the 5′-direction, resulting in a bulge at the X7•A18 base pair. The HNE aliphatic chain was oriented towards the minor groove. The Watson-Crick hydrogen bonding of the neighboring A8•T17 base pair was also disrupted.
Introduction
trans-4-Hydroxynonenal (HNE) is produced from the metabolism of membrane lipids (1), and it is the major in vivo peroxidation product of ω-6 polyunsaturated fatty acids (2, 3). Several routes for the formation of HNE from ω-6 polyunsaturated fatty acids have been described (4–6). HNE forms Michael addition adducts with protein Cys, His, and Lys residues, which can further re-arrange to cyclic hemiacetals (2, 7–11). Many of the cytotoxic effects attributed to HNE involve alteration in gene expression and cell signaling to cell proliferation and apoptosis (12–18), and these are associated with the etiology of human disease arising as a result of oxidative stress, e.g., Alzheimer’s disease (19), Parkinson’s disease (20), arteriosclerosis (21), and hepatic ischemia reperfusion injury (22).
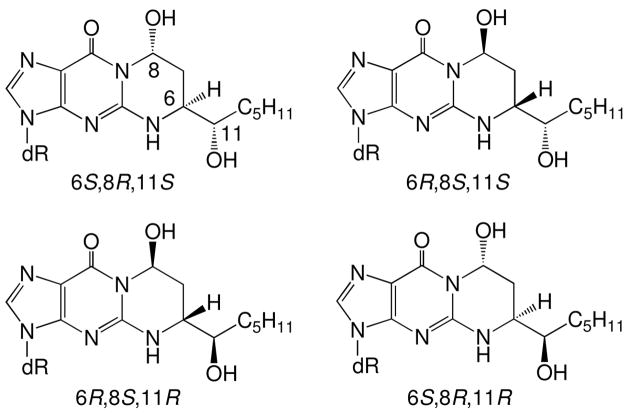
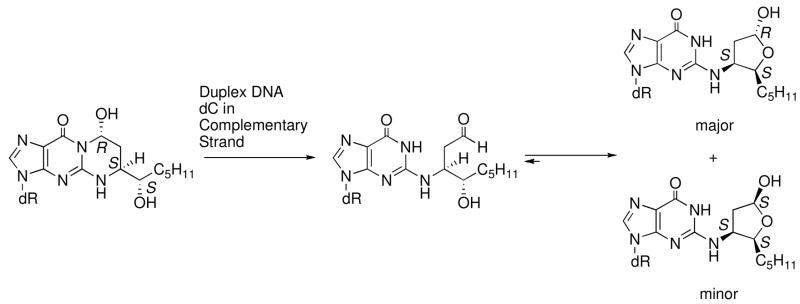
Michael addition of the N2-amino group of deoxyguanosine to HNE gives four diastereomeric 1,N2-HNE-dG adducts (Figure 1) (23–26), which have been detected in cellular DNA (27–33). Wang et al. (34, 35) synthesized the four stereoisomers of the exocyclic 1,N2-HNE dG adduct and incorporated them into 5′-d(GCTAGCXAGTCC)-3′•5′-d(GGACTCGCTAGC)-3′, in which X denotes the 1,N2-HNE-dG adduct. When placed opposite dC in duplex DNA, the exocyclic 1,N2-HNE-dG adducts underwent ring-opening at the N1 imine nitrogen of dG, thus exposing the Watson-Crick base pairing face of the adducted dG. The diastereomeric (6S,8R,11S) and (6R,8S,11R) 1,N2-HNE-dG adducts in fact existed primarily as minor groove cyclic hemiacetals when placed into this duplex (Scheme 1) (36). The initial ring-opening of the 1,N2-HNE-dG adducts likely occurs via a mechanism similar to that proposed for the related 3-(2′-deoxy-β-D-erythro-pentofuranosyl)pyrimido[1,2-α]purin-10(3H)-one (M1dG) adduct (37, 38). It should also be noted that an alternative pathway to the formation of HNE-derived DNA adducts involves oxidation of HNE to 2,3-epoxy-4-hydroxynonanal, yielding etheno adducts (39–43).
Figure 1.
Four stereoisomers of HNE derived exocyclic 1,N2-deoxyguanosine adducts.
Scheme 1.
Ring-chain tautomerization of HNE derived diastereomeric (6S,8R,11S) 1,N2-dG adduct when placed opposite dC.
The potential to form DNA adducts (Figure 1) suggests that HNE may be genotoxic. HNE induces the SOS response in Escherichia coli (44). Chromosomal aberrations are observed upon exposures to HNE in rodent (45, 46), mammalian (47, 48), and human (49) cells. In mammalian cells, the genotoxicity of HNE depends upon glutathione levels, which modulate the formation of HNE-DNA adducts (50–52). The mutational spectrum induced by HNE adduct in the lacZ gene of the single-stranded M13 phage transfected into wild type Escherichia coli revealed recombination events, C→T transitions, followed by G→C and A→C transversions, and frameshift mutations (25). HNE is mutagenic (53) and carcinogenic in rodent cells (54). Hussain et al. (55) reported that HNE caused G→T transversions at codon 249 of wild type p53 in lymphosblastoid cells. Hu et al. (56) reported that HNE-DNA adducts were preferentially formed with guanine at the third base of codon 249 in the p53 gene. The mutational spectrum induced by HNE adducts in the supF gene of shuttle vector pSP189 replicated in human cells showed that HNE induced primarily G→T transversions, accompanied by lower levels of G→A transitions (57). Fernandes et al. (58) conducted site-specific mutagenesis studies and observed that the (6S,8R,11S) and (6R,8S,11R) 1,N2-HNE-dG adducts were mutagenic, inducing low levels of G→T transversions and G→A transitions. The nucleotide excision repair pathway is involved in the excision of HNE-dG lesions (57, 59, 60).
The propensity of the 1,N2-HNE-dG adducts to undergo ring-opening when placed opposite dC in duplex DNA, potentially facilitating successful lesion bypass by Y-family polymerases, may account for the low levels of mutations associated with these lesions (58). Wolfle et al. (61) reported that the sequential activity of pols ι and κ bypassed the (6S,8R,11S) and (6R,8S,11R) 1,N2-HNE-dG adducts. Significantly, pol ι correctly inserted dCTP and to a lesser extent dTTP opposite the HNE adduct. Further extension was achieved in the presence of pol κ, which elongated from a C•HNE-dG template-primer terminous when T was opposite the adducts (61).
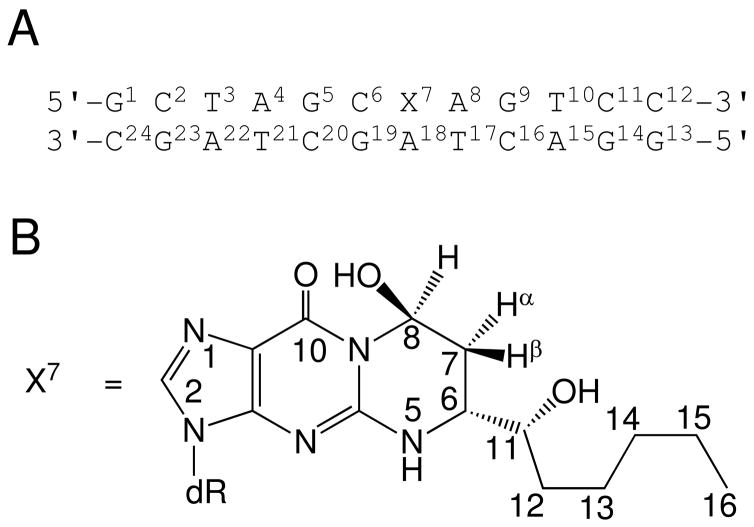
In the present work, the (6S,8R,11S) 1,N2-HNE-dG adduct (36) has been examined as to structure in 5′-d(GCTAGCXAGTCC)-3′•5′-d(GGACTAGCTAGC)-3′, containing the X•A mismatch sequence [X = (6S,8R,11S) 1,N2-HNE-dG], using NMR (Scheme 2). This duplex mimics the situation following incorrect incorporation of dATP opposite the (6S,8R,11S) 1,N2-HNE-dG adduct, and leading to HNE-induced G→T transversions (55, 57, 58). Solution structures of the oligodeoxynucleotide duplex have been refined from NMR data collected as a function of pH. The (6S,8R,11S) 1,N2-HNE-dG adduct maintains the exocyclic structure when placed complementary to dA. The adduct undergoes a syn/anti conformational equilibrium about the glycosyl bond, as was predicted by Xing et al. (26). The syn conformation predominates in acidic solution. Structural analysis reveals that X7 forms a distorted base pair with the complementary protonated A18. The HNE moiety is located in the major groove. Structural perturbations are observed at the neighbor C6•G19 and A8•T17 base pairs. At basic pH, the anti conformation of X7 is the major species. At X7 the 1,N2-HNE-dG intercalates and displaces the complementary A18 in the 5′-direction, resulting in a bulge at the X7•A18 base pair. The HNE aliphatic chain is oriented towards the minor groove. The Watson-Crick hydrogen bonding of the neighboring A8•T17 base pair is also disrupted.
Scheme 2.
A. The numbering scheme of the mismatched 5′-CpX-3′ duplex. B. The numbering scheme of the stereospecific HNE derived 1,N2-deoxyguanosine adduct.
Materials and Methods
Materials
The oligodeoxynucleotide 5′-d(GGACTAGCTAGC)-3′ was synthesized and purified by anion-exchange chromatography by the Midland Certified Reagent Co. (Midland, TX). The 1,N2-HNE-dG adduct of (6S,8R,11S) configuration was incorporated into 5′-d(GCTAGCXAGTCC)-3′ [X = (6S,8R,11S) 1,N2-HNE-dG] as reported (34, 35). The oligodeoxynucleotides were characterized by MALDI-TOF mass spectrometry. Capillary gel electrophoresis and C-18 HPLC were utilized to assess their purities. The oligodeoxynucleotides were desalted by chromatography on Sephadex G-25 (Sigma-Aldrich, St. Louis, MO). Oligodeoxynucleotide concentrations were determined by UV absorption at 260 nm, using calculated extinction coefficients for both sequences of 1.1 × 105 L·mol−1·cm−1 (62). The strands were annealed at 1:1 stoichiometry in 10 mM NaH2PO4, 100 mM NaCl, and 50 μM Na2EDTA (pH 7.0). The solutions were heated to 95 °C for 10 min and cooled to room temperature. The duplex DNA was purified using DNA Grade hydroxylapatite chromatography, with a gradient from 10 to 200 mM NaH2PO4 in 100 mM NaCl, and 50 μM EDTA (pH 7.0), and desalted using Sephadex G-25.
Melting Temperature
UV thermal melting profiles were collected as a function of pH with the oligodeoxynucleotides containing either the X7•C18 or X7•A18 base pair in 100 mM NaCl buffer. The strand concentration was 10 nM. Data were collected on a Varian Cary 4E spectrometer. The temperature was increased at a rate of 1 °C/min. The temperature and the absorbance at 260 nm were read and stored at 1-min intervals from 10–70 °C.
NMR
NMR experiments were performed at 1H frequencies of 600 MHz and 800 MHz; the data at 800 MHz were collected using a cryogenic probe. Samples were at 1.0 mM strand concentration. Samples for the non-exchangeable protons were dissolved in 10 mM NaH2PO4, 100 mM NaCl, and 50 μM Na2EDTA (pH 7.0) to a volume of 280 μL. They were exchanged with D2O and suspended in 280 μL 99.996% D2O. The pH was adjusted using dilute DCl or NaOD. The temperature was 15 °C. Samples for the observation of exchangeable protons were dissolved in 280 μL of the same buffer containing 9:1 H2O:D2O (v/v). The temperature was 5 °C. The 1H chemical shifts were referenced to water. Data were processed using FELIX 2000 (Accelrys Inc., San Diego, CA) on UNIX workstations (Dell Inc., Austin, TX). For all experiments, a relaxation delay of 1.5 s was used. The NOESY spectra were recorded with 512 real data in the t2 dimension and 2048 real data in the t1 dimension. For assignment of exchangeable protons, NOESY experiments used the Watergate solvent suppression scheme (63). The mixing time was 250 ms. The spectrum was zero-filled during processing to create a matrix of 1024 × 512 real points. For assignment of non-exchangeable protons and the derivation of distance restraints, NOESY experiments used TPPI quadrature detection and mixing times of 60, 150, 200 and 250 ms were used. The spectra were zero-filled during processing to create a matrix of 1024 × 1024 real points. The DQF-COSY experiments were performed with TPPI quadrature detection and pre-saturation of the residual water during the relaxation delay. 1H-31P HMBC spectra (64, 65) were obtained at 25 °C. The data matrix was 96 (t1) × 1024 (t2) complex points. The data were Fourier transformed after zero filling in the t1 dimension, resulting in a matrix size of 128 (D1) × 512 (D2) real points. The 31P chemical shifts were not calibrated.
Restraints
Footprints were drawn around cross peaks obtained at a mixing time of 250 ms using FELIX2000. Identical footprints were transferred and fit to the corresponding cross peaks obtained at the other two mixing times. Cross peak intensities were determined by volume integrations. These were combined as necessary with intensities generated from complete relaxation matrix analysis of a starting structure to generate a hybrid intensity matrix (66, 67). MARDIGRAS (68–70) iteratively refined the hybrid intensity matrix and optimized agreement between calculated and experimental NOE intensities. The RANDMARDI algorithm carried out 50 iterations for each set of data, randomizing peak volumes within limits specified by the input noise level (70). Calculations were initiated using isotropic correlation times of 2, 3, and 4 ns, and with both A-form and B-form starting structures and the three mixing times, yielding eighteen sets of distances. Analysis of these data yielded experimental distance restraints used in subsequent rMD calculations, and the corresponding standard deviations for the distance restraints.
Deoxyribose pseudorotational angles (P) were estimated by examining the 3JHH of sugar protons (71). The J1′-2′ and J1′-2″ couplings were measured from ECOSY spectra, while the intensities of H2″-H3′ and H3′-H4′ cross peaks were determined from DQF-COSY spectra. The data were fit to curves relating the coupling constants to the deoxyribose pseudorotation (P), sugar pucker amplitude (φ), and the percentage S-type conformation. The pseudorotation and amplitude ranges were converted to the five dihedral angles ν0 to ν4. Coupling constants measured from 1H-31P HMBC spectra were applied (72, 73) to the Karplus relationship (74) to determine the backbone dihedral angle ε (C4′-C3′-O3′-P), related to the H3′-C3′-O3′-P angle by a 120° shift. The ζ (C3′-O3′-P-O5′) backbone angles were calculated from the correlation between ε and ζ in B-DNA.
rMD Calculations
The HNE-adducted duplexes, either in A-form or B-form DNA helical coordinates, were constructed by bonding the stereospecific HNE C1 and C3 to G7 N1 and G7 N2, respectively using Insight II. The partial charges on the HNE atoms were obtained from density function theory (DFT) calculations using a neutral total charge, utilizing B3LYP/6-31G* basis set and the program GAUSSIAN (75). To obtain the A-form and B-form starting structures that were used for subsequent restrained molecular dynamics (rMD) calculations, these A-form or B-form modified duplexes were energy minimized using 200 iterations with the conjugate gradients algorithm, in the absence of experimental restraints.
Distance restraints were divided into classes weighted according to the error assessed in their measurements. Class 1, class 2, class 3, class 4 and class 5 were calculated from completely resolved, somewhat overlapped, slightly overlapped, medium overlapped, or heavily overlapped cross-peaks, respectively, which were at least 0.5 ppm from the water resonance or the diagonal line of the spectrum. Class 5 also included all other cross peaks. NOEs that did not have a distance calculated by MARDIGRAS were estimated by the relative peak intensities. The spectroscopic data indicated that the duplexes conserved Watson-Crick base pairing, so empirical restraints preserving Watson-Crick hydrogen bonding and preventing propeller twisting between base pairs were used (76). Empirical backbone and deoxyribose torsion angle restraints derived from B-DNA were used (77). The potential energy wells associated with the dihedral angle restraints were ± 30°. The force constants of the restraints were scaled from 3.2 kcal mol−1 Å−2 to 32 kcal mol−1 Å−2 during the first 10 ps and were maintained at 32 kcal mol−1 Å−2 for the remainder of the simulations.
Ten sets of randomly seeded rMD calculations (5 from A- and and 5 from B-type DNA starting structures) were conducted using the program AMBER (v 8.0) (78) and the parm99 force field. The Hawkins, Cramer, Truhlar pairwise generalized Born (GB) model (79, 80) was used to simulate implicit waters. The parameters developed by Tsui and Case (81) were used. The cutoff radius for nonbonding interactions was 18 Å. The restraint energy function contained terms describing distance and torsion angle restraints, both in the form of square well potentials. Bond lengths involving hydrogens were fixed with the SHAKE algorithm (82). A 1,000-step energy minimization was performed with an integrator time of 1 fs without experimental restraints, followed by a 100,000-iteration simulated annealing protocol with an integrator time step of 1 fs. The system was heated to 600 K in 5,000 iterations and kept at 600 K for 5,000 iterations, then cooled to 100 K with a time constant of 4.0 ps over 80,000 iterations. A final cooling was applied to relax the system to 0 K with a time constant of 1.0 ps over 10,000 iterations.
Convergence was assessed for structures having the lowest number of deviations from the experimental distance and dihedral restraints, lowest van der Waals energies, and the lowest overall energies. Finally, the ten refined structures were energy minimized for 250 iterations without restraints to obtain average structures. The program CORMA (67) was utilized to calculate the predicted NOE intensities from the structures refined from rMD calculations. Input volumes (intensities) were normalized from the intensities of protons with fixed intranuclear distances (i.e. cytosine H5-H6, and thymine CH3-H6 distances). Random noise was added to all intensities to simulate spectral noise. An isotropic correlation time (τc) of 3 ns was used. The rotation of thymidine CH3 groups was modeled using a 3-jump site model (83). A sixth root residual (R1x) factor (84) was calculated for each structure. Helicoidal analysis was carried out with the program 3DNA (85).
Results
Characterization of the Mismatched Duplex
The 5′-d(GCTAGCXAGTCC)-3′•5′-d(GGACTAGCTAGC)-3′ (X = 1,N2-HNE-dG) oligodeoxynucleotide was characterized by MALDI-TOF mass spectrometry, capillary gel electrophoresis, and C-18 HPLC. It was obtained at > 95% purity. However, at pH 7.3, two sets of NMR resonances that exhibited exchange cross-peaks on the NMR time scale were observed. The data suggested that the mismatched duplex adopted two conformations. The COSY spectra exhibited seven cytosine H5→H6 correlations in acidic, neutral, or basic solutions. The COSY spectra at neutral or basic conditions were similar, suggesting that the major conformation in neutral solution was similar to that in basic solution. Furthermore, the C6 H5→H6 correlation was broad in acidic and neutral solutions whereas it was sharp in basic solution, suggesting in the acidic and neutral solutions the mismatched duplex underwent a slow conformational exchange involving the adduct region. Shifting the pH to 5.5 or to 8.9 yielded spectra suitable for structural refinement. The resolution of the NMR spectra in acidic solution remained somewhat compromised by resonance broadening, but resonance assignments could be made. The resolutions of NMR spectra obtained in basic solution were outstanding. Hence, subsequent NMR experiments were performed at either acidic or basic pH, in an effort to characterize the two conformational species that were present in equilibrium at neutral pH.
The Mismatched X•A Duplex at pH 5.5
(a) Thermal Melting (Tm) Experiments
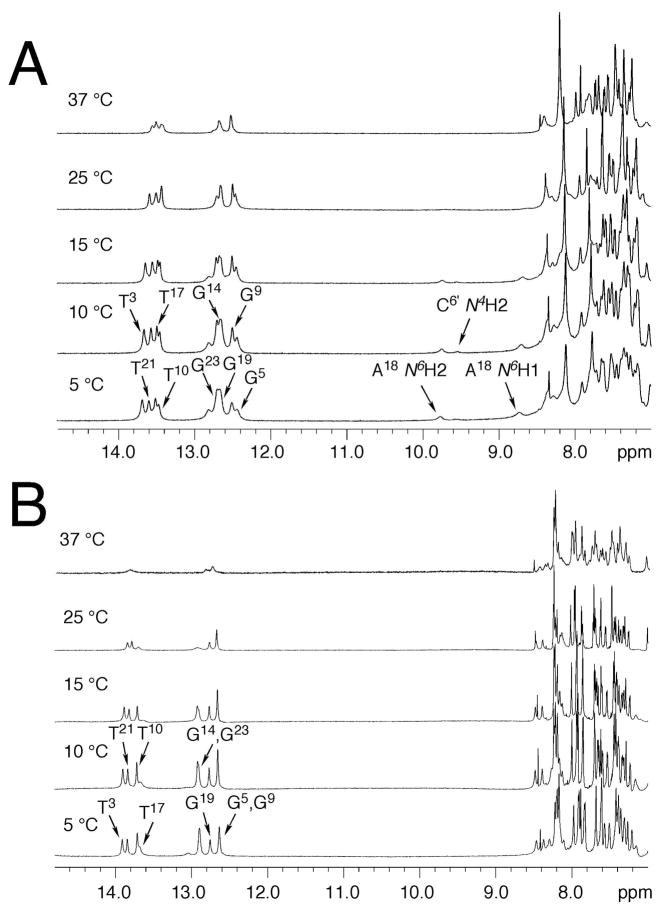
The Tm of the mismatched duplex at pH 5.5 was 37 °C, lower than that of the corresponding duplex containing X7•C18 base pair, which was 40 °C. 1H NMR spectra of the mismatched duplex at different temperatures in acidic solution are shown in Figure 2A. No imino resonance was assigned to X7. The imino resonances of the neighboring C6•G19 and A8•T17 base pairs broadened more rapidly than did the imino resonances of the nucleotides located in the middle of the duplex.
Figure 2.
A. 1H NMR spectra of the mismatched duplex at different temperatures at pH 5.5. Two small resonances at ~9.8 and 8.8 ppm at low temperature were assigned to the hydrogen bonded and non-hydrogen bonded amino protons of the protonated A18. The small peak at ~9.6 ppm was assigned to the partially protonated C6 hydrogen bonded amino proton. (B) 1H NMR of the mismatched duplex at different temperatures at pH 8.9. The broad resonance at ~13.7 ppm was tentatively assigned to the T17 imine proton.
(b) Non-Exchangeable Protons
The sequential NOE assignment of the non-exchangeable protons was accomplished using standard protocols (86, 87). The sequential NOEs between the aromatic (note that the X7 aromatic proton is designated as H2) and anomeric protons are displayed in Figures 3A and 3B. Complete sequential NOESY connectivities without an interruption were observed for both modified and complementary strands. Notably, A18 H8 was the most downfield among the adenine aromatic protons and X7 H2 was the most upfield among the guanine aromatic protons. The A18 H8 and A8 H2 resonances were observed at 8.42 ppm and 7.03 ppm. These peaks become weaker at pH 7.3 and disappeared at pH 8.9. The T17 H6 resonance was observed at 7.37 ppm.
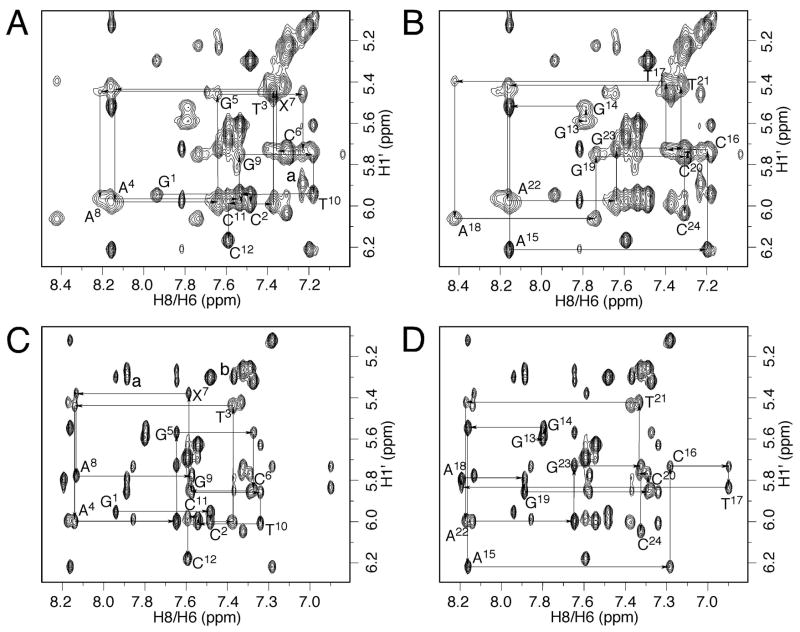
Figure 3.
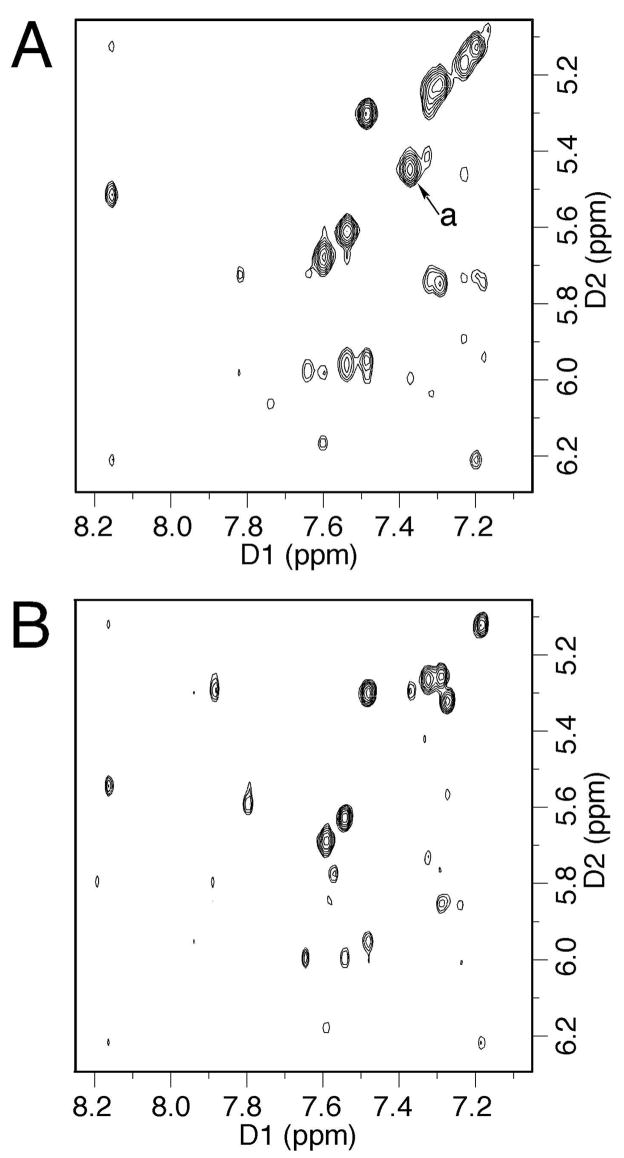
Expansions of NOESY spectra (250 ms) of the mismatched duplex showing the sequential connectivity of the base aromatic protons with sugar H1′ protons. A. Modified strand at pH 5.5. The strong cross peak designated as peak “a” was assigned to the C6 H6→X7 H8 correlation. B. Complementary strand at pH 5.5. C. Modified strand at pH 8.9. Extra cross peaks designated as peaks “a” and “b” were assigned to the A8 H2→X7 H8 and A18 H2→X7 H8 correlations, respectively. D. Complementary strand at pH 8.9.
The C6 H1′→X7 H2, X7 H1′→A8 H8, and T17 H1′→A18 H8 NOEs were weaker compared to other internucleotide deoxyribose H1′ →purine H8 NOEs. In contrast, the X7 H2→X7 H1′ NOE, which overlapped with the T3 H6→T3 H1′ NOE, was very strong. The overlapped NOE cross peak was comparable in intensity to the cytosine H5→H6 correlations in the spectrum with 60 ms mixing time (Figure 4A). The deoxyribose sugar proton resonances were assigned by utilizing a combination of DQF-COSY and NOESY spectra. Compared with the other H2′ protons, X7 H2′ shifted downfield. The assignments of the non-exchangeable protons are provided in Table S1 in the Supporting Information.
Figure 4.
Expansions of NOESY spectra (60 ms) of the mismatched duplex. A. At pH 5.5 the strong X7 H2→X7 H1′ correlation (peak “a”) suggested that X7 adopted the syn conformation about the glycosyl bond. B. The corresponding spectrum at pH 8.9.
(c) Exchangeable Protons
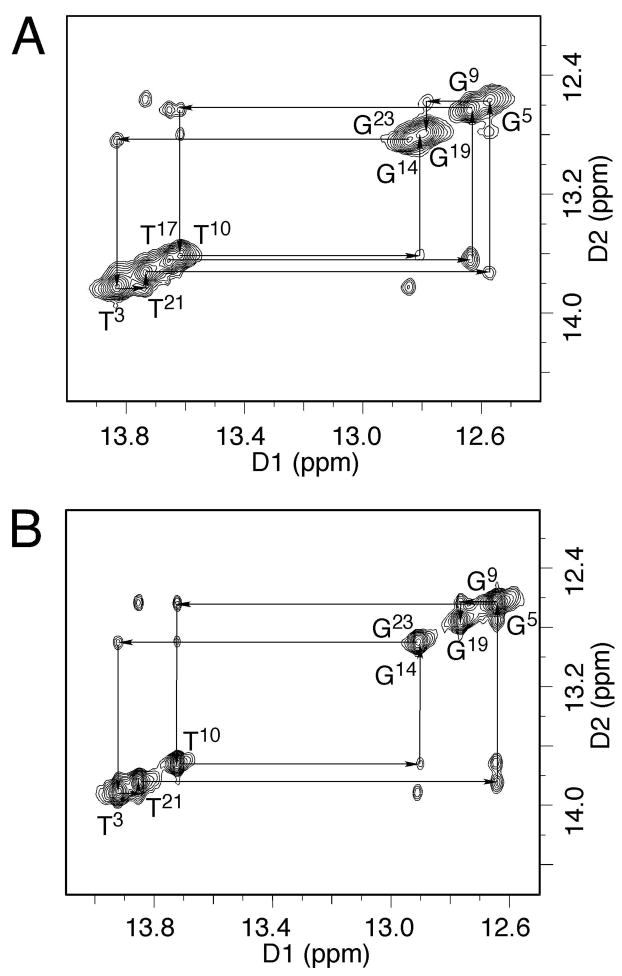
The base imino protons were assigned based on their sequential connectivities in NOESY spectra (Figure 5A) and these assignments were supported by their NOE cross peaks to Watson-Crick base paired amino protons (88). Since the X7 imino resonance was missing, no NOEs arising from the X7 imino proton were observed. The mismatched duplex exhibited two broad resonances at ~9.8 and ~8.8 ppm at low temperatures. They broadened further at higher temperature and disappeared at 25°C (Figure 2A). These resonances had NOE correlations with T17 CH3, T17 N3H, G19 N1H, and G19 N2H, and were assigned to hydrogen-bonded and non-bonded amino protons of the protonated A18, respectively (76). Another weak resonance was observed at ~9.6 ppm at low temperature (Figure 2A). It was also temperature-dependent and exhibited a weak NOE correlation with the resonance at 8.55 ppm at 5 °C. These two resonances were assigned to the amino protons of the partially protonated C6 (89). The NOE cross peaks of the imino protons arising from Watson-Crick base pairing for C2•G23, T3•A22, A4•T21, G5•C20, C6•G19, A8•T17, G9•C16, T10•A15, and C11•G14 base pairs were observed.
Figure 5.
NOE connectivity of the base imino protons. A. The NOE spectrum at pH 5.5. B. The NOE spectrum at pH 8.9. No imino resonance was observed for X7 in either spectrum, indicating X7 maintained the 1,N2-HNE-dG structure. The T17 imino proton was also missing at pH 8.9.
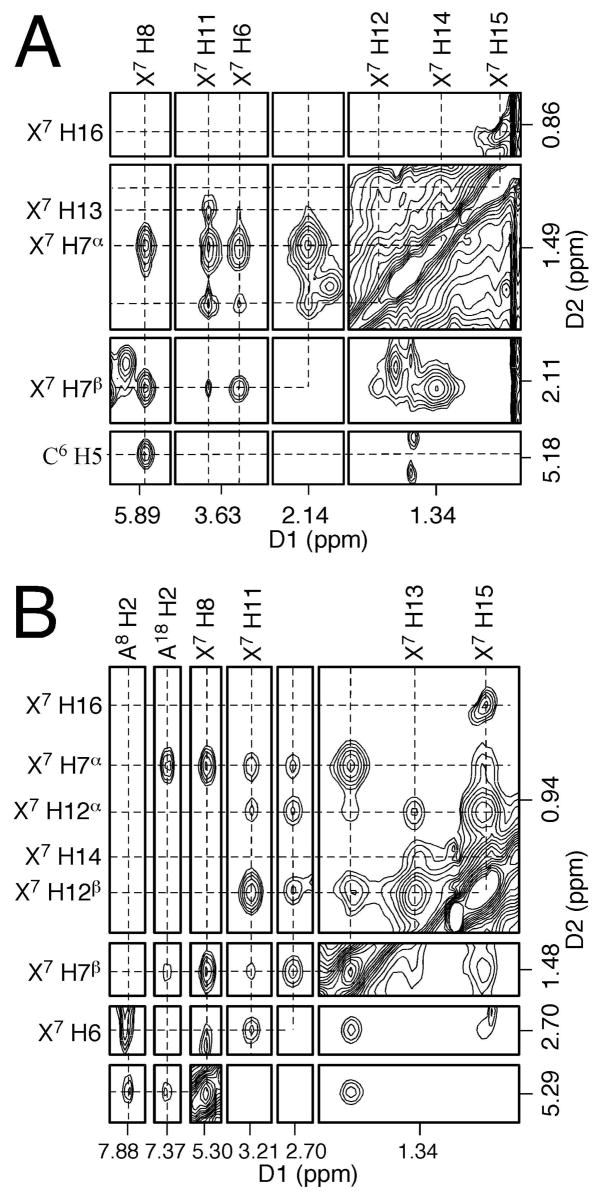
(d) HNE Protons
X7 H8 exhibited strong NOE correlations with C6 H6 (Figure 3A) and C6 H5 (Figure 6A). Other HNE protons were assigned based on the NOE correlations with 60 ms mixing time (Figure 6A). The resonances of X7 H7 geminal protons were well-resolved. X7 H7α, which was in the cis-configuration with respect to X7 H8, exhibited a relatively stronger NOE with X7 H8. X7 H7β, which was in the trans-configuration with respect to X7 H8, had a relatively weaker correlation with X7 H8. X7 H6 exhibited strong NOEs with the X7 H7 and X7 H11 protons. The X7 H11 germinal protons had strong NOE correlationswith X7 H12 and relatively weaker correlations with X7 H13. X7 H16 was the most upfield and exhibited a strong NOE cross peak with X7 H15. X7 H14 was overlapped with X7 H7α. It exhibited strong NOE correlations with X7 H13 and X7 H15. These assignments were supported by COSY, DQF-COSY, TOCSY, and NOE correlations with nucleotide protons. The chemical shifts of the HNE protons are summarized in Table 1. Notably, the HNE protons exhibited NOE correlations with the C6 H5 and C6 H6 protons in the mismatched duplex.
Figure 6.
Assignments of HNE protons based on the NOE correlations (60 ms). A. The NOE spectrum at pH 5.5. B. The NOE spectrum at pH 8.9.
Table 1.
Chemical Shifts of HNE Protons and Related NOEs at pH 5.5 Converted to rMD Distance Restraints.
| Proton | δ (ppm) | NOEs a |
|---|---|---|
| H6 | 3.59 | C6 H6(w), C6 H3′ (w), X7 H7α(s), X7 H7β(s), X7 H8(s), X7 H12(m), X7 H13(m), X7 H14(m), X7 H15(m) |
| H7α | 1.49 | C6 H5(m), C6 H6(m), C6 H1′ (w), C6 H3′ (w), X7 H7β(s), X7 H8(s), X7 H11(m) |
| H7β | 2.13 | C6 H5(m), C6 H6(m), C6 H1′ (w), C6 H3′ (w), X7 H8(s), X7 H11(m), X7 H12(m), X7 H13(w) |
| H8 | 5.89 | C6 H5(s), C6 H6(m), C6 H2′ (m), C6 H5′ (w), X7 H11(m), X7 H12(m), X7 H13(w), X7 H15(w) |
| H11 | 3.66 | C6 H6(w), C6 H3′ (w), X7 H12(m), X7 H13(m), X7 H14(m), X7 H15(m) |
| H12 | 1.65 | C6 H6(w), C6 H3′ (w), X7 H13(s), X7 H14(s), X7 H15(m) |
| H13 | 1.40 | C6 H3′ (w), X7 H14(s), X7 H16(m) |
| H14 | 1.48 | C6 H3′ (m), X7 H15(s) |
| H15 | 1.33 | C6 H3′ (m), X7 H16(s) |
| H16 | 0.88 |
Letters in brackets indicate peak intensity, s: strong, m: medium, w: weak.
(e) Deoxyribose and Backbone Angle Conformations
Deoxyribose and backbone angle conformations were determined spectroscopically by DQF-COSY and 31P-H3′ HMBC correlations. Evaluation of the DQF-COSY spectrum revealed that the pseudorotation of the sugar rings of all nucleotides except X7 and A18 were either C1′-exo or C2′-endo.
(f) Structural Refinement
A total of 414 distance restraints, including 239 intranucleotide and 175 internucleotide restraints, were calculated from the intensities of NOE cross peaks by MARDIGRAS. A total of 50 NOEs were assigned to HNE protons (Table 1). In addition, 50 empirical distance restraints defining Watson-Crick base pairing were used; their use was predicated upon inspection of the NMR data, which indicated that Watson-Crick base pairing was intact throughout the duplex except at the X7•A18 base pair. Finally, an additional 180 empirical backbone torsion angle restraints were used; these were based upon inspection of the NMR data, which suggested that the adducted duplex maintained the B-type architecture. The A18 imino nitrogen N1 was protonated to allow formation of a hydrogen bond with X7, and empirical distance restraints were used to position the hydrogen bonds of the protonated X7•A18 base pair. Torsion angle restraints were not used at the protonated X7•A18 base pair (Table 2).
Table 2.
rMD Restraints and Statistical Analysis of rMD Converged Structures of the Mismatched Duplexes in Acidic and Basic Solutions.
| Solution pH | pH 5.5 | pH 8.9 |
|---|---|---|
| Total restraints for rMD calculation | 644 | 671 |
| Experimental NOE distance restraints | 414 | 467 |
| Intraresidue NOE restraints | 239 | 251 |
| Interresidue NOE restraints | 175 | 216 |
| Restraints of HNE unit | 49 | 83 |
| Empirical base pair restraints | 50 | 44 |
| Empirical torsion angle restraints | 180 | 160 |
| Backbone torsion angles restraints | 90 | 80 |
| Sugar torsion angles restraints | 90 | 80 |
|
| ||
| Structure Statistics a | ||
| NMR R-factor (R1x) (×10−2) b | 7.83 | 8.77 |
| Intraresidue NOEs | 7.66 | 8.08 |
| Interresidue NOEs | 8.08 | 9.69 |
| RMSD deviation of refined structures | 0.50 | 0.52 |
HNE unit considered to be an single residue attached to guanine G7 in the rMD calculation and the statistical analysis;
Mixing time used to calculate R1x was 250ms. , where (a0) and (ac) are the intensities of observed (nonzero) and calculated NOE cross peaks, respectively
The randomly seeded rMD calculations were performed starting with initial structures, which were created either with A- or B-form geometries. Pairwise rmsd analysis of emergent structures indicated that the calculations converged, irrespective of starting structure (Table 2). The accuracies of the emergent structures were evaluated by comparison of theoretical NOE intensities calculated by complete relaxation analysis for the refined structure, to the experimental NOE intensities, to yield sixth root residuals (R1x). This residual was less 0.1 for the modified duplex (Table 2), indicating that the refined structures provided an accurate depiction of the data.
(g) Analysis of rMD Structures
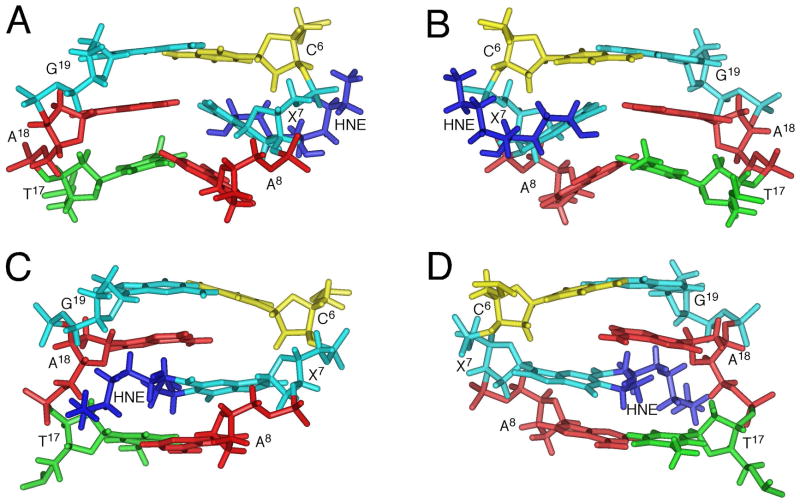
The backbone torsion angles of the refined structures showed the oligodeoxynucleotide remained in the B-type geometry except for the adducted region. Expanded views of the average structure around the adducted region are shown in Figures 7A and 7B, and the base stacking at the modified region is demonstrated in Figures 8A and 8B. All nucleosides except X7 maintained the anti conformation about the glycosyl bond. The neighboring C6•G19 and A8•T17 base pairs were distorted but Watson-Crick hydrogen bonding was conserved. X7 adopted the syn conformation about the glycosyl bond and the χ torsion angle (O4′-C1′-N3-C2) was 106°. The protonated A18 adopted the anti conformation about the glycosyl bond and formed hydrogen bonds with X7. The HNE was located in the major groove and exposed to the solvent.
Figure 7.
Expanded views of average structures of the mismatched duplex. A. View from the minor groove at pH 5.5. B. View from the major groove at pH 5.5. X7 adopts the syn conformation about the glycosyl bond allowing formation of hydrogen bonds with protonated A18. C. View from the minor groove at pH 8.9. D. View from the major groove at pH 8.9. X7 is intercalated and displaces A18 in the 5′-direction.
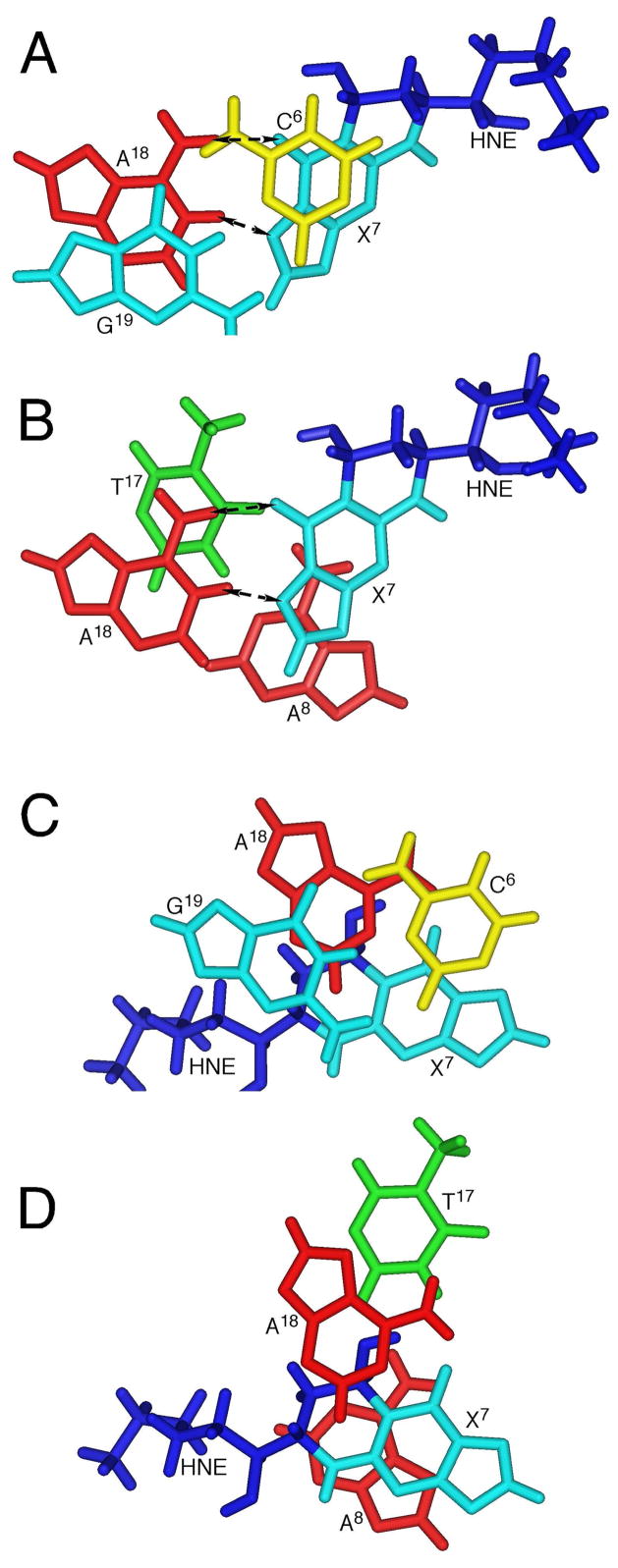
Figure 8.
A, B. Base stacking of the mismatched duplex at pH 5.5. The dashed arrows indicate the potential hydrogen bonds of the X7•A18 mismatched base pair. C, D. Base stacking of the mismatched duplex at pH 8.9. No hydrogen bond is formed for the mismatched X7•A18 or A8•T17 base pairs.
The Mismatched Duplex at pH 8.9
(a) Thermal Melting Experiments
The Tm of the mismatched duplex at pH 8.9 was 32 °C, lower than that of the correctly paired duplex containing the X7•C18 base pair, which was 37 °C. Figure 2B shows the temperature dependence of 1H NMR of the mismatched duplex. No imino resonance was assigned to X7. The G19 imino resonance from the 5′-neighbor C6•G19 base pair broadened more rapidly than the imino resonances of the nucleotides located in the middle of the sequence. A resonance tentatively assigned as the T17 imino resonance from the 3′-neighbor A8•T17 base pair was broad even at 5 °C.
(b) Non-Exchangeable Protons
The sequential NOE assignment of the non-exchangeable protons was also accomplished using standard protocols (86, 87). The sequential NOEs between the aromatic and anomeric protons are displayed in Figures 3C and 3D. Complete sequential NOESY connectivities without interruption or peak intensity differences were observed for both the modified and complementary strands. The deoxyribose sugar proton resonances were assigned by utilizing a combination of DQF-COSY and NOESY spectra. The resonances of A18 H8 and A8 H2 were found at 8.20 ppm and 7.89 ppm, respectively. In addition, a singlet at 6.91 ppm was assigned to T17 H6. It became smaller at pH 7.3 and almost disappeared at pH 5.5. The assignments of the non-exchangeable protons are provided in Table S2 in the Supporting Information.
The chemical shifts of the mismatched duplex in basic solution were compared with the corresponding unmodified G7•A18 mismatched duplex. Large chemical shift perturbations were located at the adducted X7•A18 base pair and the neighboring C6•G19 and A8•T17 base pairs, indicating some perturbation at the adduct region. The chemical shifts were also compared with those in acidic solution. Large differences were also observed at the modified X7•A18 base pair and the neighboring C6•G19 and A8•T17 base pairs, suggesting large conformational differences between in acidic and basic solutions were located at the adduct region.
(c) Exchangeable Protons
The resonances of the base imino protons were assigned based on their sequential connectivty in NOESY spectra (Figure 5B) and were supported by their NOE cross peaks to Watson-Crick hydrogen bonded amino protons (88). X7 was not assigned an imino resonance. The T17 N3H resonance could not be assigned, although a broad resonance at ~13.7 ppm was observed (Figure 2B), and located in the thymine imino region of the spectrum. The NOE cross peaks of the imino protons arising from Watson-Crick base pairing for C2•G23, T3•A22, A4•T21, G5•C20, C6•G19, G9•C16, T10•A15, and C11•G14 base pairs were observed.
(d) HNE Protons
X7 H8 exhibited strong NOEs with A8 H2 and A18 H2 (Figure 3C). Other HNE protons were also assigned based on the NOE correlations and the peak intensities at 60 ms mixing time (Figure 6B). In addition to the resonances of the geminal X7 H7 protons, the X7 H12 geminal protons were also resolved. The assignments were supported by COSY, DQF-COSY, TOCSY, and NOE correlations with nucleotide protons. The chemical shifts of the HNE protons are summarized in Table 3. Compared with those at pH 5.5, the resonances of the HNE protons were shifted upfield.85 NOE cross peaks were assigned to HNE protons (Table 3). HNE protons were found to exhibit NOE interactions with minor groove protons, including A8 H2, A18 H2, and A18 H1′.
Table 3.
Chemical Shifts of HNE Protons and Related NOEs at pH 8.9 Converted to rMD Distance Restraints.
| Proton | δ (ppm) | NOEs a |
|---|---|---|
| H6 | 2.70 | X7 H7α(m), X7 H7β(s), X7 H8(m), X7 H11(s), X7 H12α(m), X7 H12β(s), X7 H13(w), X7 H14(w), A18 H2(m) |
| H7α | 0.95 | X7 H7β(s), X7 H8(s), X7 H11(m), X7 H12β(m), X7 H13(m), A8 H2(w), T17 H1′ (w), A18 H2(m) A18 H1′ (w), A18 H4′ (w) |
| H7β | 1.48 | X7 H8(s), X7 H11(m), X7 H12α(s), X7 H12β(s), X7 H13(m), X7 H14(w), A8 H2(w), A18 H2(m), A18 H1′ (w), A18 H4′ (w) |
| H8 | 5.30 | X7 H11(m), X7 H12α(m), X7 H12β(m), X7 H13(w), A8 H2(m), T17 H1′ (w), A18 H2(m), A18 H1′ (w) |
| H11 | 3.21 | X7 H12α(m), X7 H12β(s), X7 H13(m), X7 H14(m), A18 H2(m), A18 H4′ (w) |
| H12α | 1.04 | X7 H12β(s), X7 H13(s), A18 H2(w), A18 H1′ (w), A18 H4′ (m), A18 H5′ (w), G19 H4′ (w), G19 H5′ (w) |
| H12β | 1.21 | X7 H13(s), A8 H2(w), A18 H2(m), A18 H8(w), A18 H1′ (w), A18 H4′ (m), A18 H5′ (w), G19 H4′ (w), G19 H5′ (w) |
| H13 | 1.36 | X7 H14(s), X7 H15(m), X7 H16(w), A18 H2(w), A18 H4′ (m), A18 H5′ (w), G19 H4′ (m) |
| H14 | 1.14 | X7 H16(m), A18 H2(w), A18 H4′ (m), A18 H5′ (w), A18 H5″ (w), G19 H4′ (w), G19 H5′ (w) |
| H15 | 1.21 | X7 H16(s), A18 H4′ (m), A18 H5′ (m), A18 H5″ (m), G19 H4′ (m), G19 H5′ (m) |
| H16 | 0.82 | G9 H5′ (w), A18 H4′ (w), G19 H1′ (w), G19 H4′ (w), G19 H5′ (w) |
Letters in brackets indicate peak intensity, s: strong, m: medium, w: weak.
(e) Deoxyribose and Backbone Angle Conformations
Deoxyribose and backbone angle conformations were determined spectroscopically from DQF-COSY and 31P-H3′ HMBC correlations. Evaluation of the DQF-COSY spectrum revealed that the pseudorotation of the sugar rings of all nucleotides except X7 and A18 was either C1′-exo or C2′-endo. The T17 phosphate resonance shifted downfield.
(f) Structural Refinement
A total of 467 distance restraints, including 251 intraresidue and 216 interresidue restraints, were calculated from the intensities of NOE cross peaks by MARDIGRAS. In addition, 44 empirical distance restraints defining Watson-Crick base pairing were used to refine the structure of the duplex; their use was predicated upon inspection of the NMR data, which indicated that Watson-Crick base pairing was intact throughout the duplex except for the X7•A18 and A8•T17 base pairs. Finally, an additional 160 empirical backbone torsion angle restraints were also used for structure refinement; these were based upon inspection of the NMR data, which suggested that the adducted duplex maintained the B-type architecture. Hydrogen bonding and torsion angle restraints were not used for the X7•A18 and A8•T17 base pairs (Table 2). The randomly seeded rMD calculations were performed starting with initial structures, which were created either with A- or B-form geometries. Pairwise rmsd analysis of emergent structures indicated that the calculations converged, irrespective of starting structure (Table 2). The accuracies of the emergent structures were evaluated by comparison of theoretical NOE intensities calculated by complete relaxation analysis for the refined structure, to the experimental NOE intensities, to yield sixth root residuals (R1x). This residual was less 0.1 for the modified duplex (Table 2), indicating that the refined structures provided an accurate depiction of the data.
(g) Analysis of the rMD Structure
The backbone torsion angles of the refined structures showed the oligodeoxynucleotide remained in the B-type geometry except at the adduct region. Expanded views of the average structure around the adduct region are shown in Figures 7C and 7D, and the base stacking around the modified region is shown in Figures 8C and 8D. All nucleosides including X7 maintained the anti conformation about the glycosyl bond. However, the duplex was highly perturbed. X7 was intercalated into the duplex and the complementary A18 was displaced in the 5′-direction. No hydrogen bond was observed between them. The neighboring C6•G19 base pair maintained Watson-Crick hydrogen bonding with minimal distortion. The A8•T17 base pair was also highly perturbed, and no hydrogen bond was formed between these nucleotides. The aliphatic HNE chain was oriented towards the minor groove.
Discussion
The (6S,8R,11S) 1,N2-HNE-dG Adduct Does Not Undergo Ring-Opening When Placed Opposite dA in Duplex DNA
The ring-closed (6S,8R,11S) 1,N2-HNE-dG adduct in duplex DNA opposite a mismatched dA contrasts with the situation when the same adduct is placed opposite the correct complementary nucleotide dC in this sequence. In the latter instance, the exocyclic ring of the (6S,8R,11S) 1,N2-HNE-dG adduct undergoes ring-opening and exists primarily as a minor groove cyclic hemiacetal (36). The conclusion that the (6S,8R,11S) exocyclic 1,N2-dG adduct does not undergo ring-opening when placed opposite dA derives, in part, from the failure to observe the X7 N1H imino resonance in acidic, neutral, or basic solutions (Figure 5). The alternative possibility that ring-opening had occurred, but that the resulting X7 N1H imino resonance was in rapid exchange with solvent, and hence was not observed, was considered. However, no spectroscopic evidence for a ring-opened aldehydic proton is observed, under acidic, neutral, or basic solution conditions. An aldehydic 1H resonance was observed when this adduct was placed opposite the correct complementary nucleotide dC nucleotide in this sequence, even though it existed primarily as a minor groove cyclic hemiacetal (36). Moreover, the data of the present case suggest that the bulky ring-closed (6S,8R,11S) 1,N2-HNE-dG adduct rotates about the glycosyl bond, into the syn conformation under acidic conditions; this is supported by the observation of NOEs with major groove protons, which would not be anticipated if ring opening of the bulky lesion to the corresponding minor groove cyclic hemiacetal form were present (36). Additionally, the chemical shifts of X7 H6-H8 and H11 are similar to those observed for the exocyclic 1,N2-dG nucleotide (35) (Table 4), but differ from the chemical shifts of X7 H6-H8 and H11 when the adduct exists as diastereomeric N2-dG cyclic hemiacetals when placed complementary to dC in duplex DNA (36).
Table 4.
Comparison of the Chemical Shifts of the HNE Protons.
| sequence | X7 a | X7•C18 b | X7•A18 | |
|---|---|---|---|---|
| solution | methanol | pH 7.0 | pH 5.5 | pH 8.9 |
| H6 | 3.61 | 4.55 | 3.59 | 2.70 |
| H7α | 1.60 | 2.17 | 1.49 | 0.95 |
| H7β | 2.19 | 2.17 | 2.13 | 1.48 |
| H8 | 6.40 | 5.43 | 5.89 | 5.30 |
| H11 | 3.47 | 4.23 | 3.66 | 3.21 |
The observation that the (6S,8R,11S) 1,N2-HNE-dG adduct maintains the ring-closed structure in DNA when mismatched with dA is also consistent with the notion that placement of enal-derived 1,N2-hydroxypropano-dG adducts opposite dC in duplex DNA facilitates the ring-opening reaction to aldehydic products. Riggins et al. (37, 38) reported mechanistic studies of the ring-opening and closing of the related malondialdehyde- derived adduct 3-(2′-deoxy-β-D-erythro-pentofuranosyl)pyrimido- [1,2-α]purin-10(3H)-one (M1dG). They concluded that ring-opening of M1dG as a nucleoside or in oligodeoxynucleotides occurs via a reversible second-order reaction with hydroxide, and is catalyzed by the complementary dC in duplex DNA. The closure of the resulting N2-(3-oxo-1-propenyl)-dG anion is pH-dependent and under neutral and acidic conditions ring-closure is biphasic, leading to the rapid formation of intermediates that slowly convert to M1dG in a general-acid-catalyzed reaction, in the presence of dC in the complementary strand. It should be noted that the ring-opened N2-(3-oxo-1-propenyl)-dG adduct has a perturbed pKa (~6.9) relative to the extended conjugation offered by the N2-(3-oxo-1-propenyl) group, which is likely to play a significant role in the mechanism of ring-opening and closing (90). The enal-dG adducts are saturated and more likely to have a pKa similar to dG.
Structure of the (6S,8R,11S) 1,N2-HNE-dG Adduct Mismatched with dA
(a) Acidic Solution
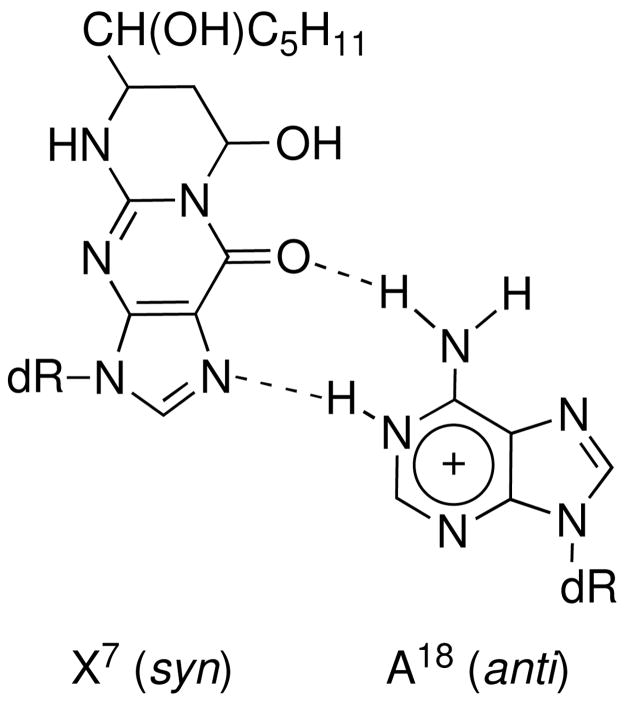
The (6S,8R,11S) 1,N2-HNE-dG adduct maintains a ring-closed form and rotates about the glycosyl bond into the syn conformation with a predicted χ torsion angle at X7 of 106° when mismatched with dA at low pH. The strong X7 H2→X7 H1′ NOE correlation, the downfield chemical shift of the X7 H2′ resonance (91–94), the upfield chemical shift of X7 H2 compared to other guanine H8 resonances, and the observation of X7 H2→A8 H2 and X7 H2→A18 H2 NOEs, are each consistent with this conclusion. The downfield chemical shifts of A18 N1H, A18 N2H(s), and A18 H8 as compared with the other adenines is consistent with the conclusion that A18 is protonated (76, 89, 95, 96). Similar to the PdG•dA base pair (76), the anticipated far downfield resonance of the protonated A18 N1H is not observed, suggesting that A18 N1H undergoes rapid exchange with solvent, and is probably only weakly hydrogen bonded (Figures 8A and 8B). The presence of the X7 syn conformation and A18 protonation facilitates base pairing through X7 O10→A18 N6H and X7 N1→A18 N1H hydrogen bonds (note that X7 N1 corresponds to N7 of unmodified guanine and X7 O10 corresponds to O6 of unmodified guanine) (Chart 1). The syn conformation of the glycosyl torsion angle places the HNE moiety in the major groove (Figures 7A and 7B), and consistent with the NOE correlations with major groove protons, notably C6 H5 and C6 H6 (Table 1). The 5′-neighbor C6•G19 and 3′-neighbor A8•T17 base pairs maintain Watson-Crick base pairing, but are distorted (Figures 7A and 7B). This conclusion is supported by the observation that the T17 and G19 imino resonances broaden more rapidly as compared to the other imino resonances (Figure 2A).
Chart 1.
Hydrogen bonding of X7•A18 base pair in acidic solution.
(b) Basic Solution
In basic solution, all nucleotides including X7 adopt the anti conformation about the glycosyl bond. Thus, the HNE moiety is oriented towards the minor groove (Figures 7C and 7D). The presence of the anti conformation about the glycosyl bond at X7 disrupts Watson Crick hydrogen bonding at the adducted base pair (Figures 8C and 8D), and greatly perturbs the DNA duplex at the lesion site. The rMD calculations predict that the complementary A18 is displaced in the 5′-direction, resulting in a bulge at the X7•A18 base pair. The presence of Watson-Crick hydrogen bonding at the 5′-neighboring C6•G19 base pair is supported by the observation of the G19 imino resonance and NOEs between the G19 imino proton and the exocyclic amino protons of C6 (Figure 5B). However, these disappear at lower temperatures as compared to other imino resonances (Figure 2B) consistent with the prediction from the rMD calculations that base pair C6•G19 is distorted (Figure 8C). In the 3′-direction, the broad T17 imino resonance (Figure 2B) and lack of NOEs to the A8 N6H(s) and A8 H2 resonances lead to the conclusion that base pair A8•T17 is also significantly disrupted (Figure 8D). The downfield shift of the associated 31P resonance suggests that that the phosphodiester backbone is distorted at the lesion site.
Conformational Equilibrium of the Mismatched Duplex in Neutral Solution
At pH 7.3, both the syn and anti conformations of X7 about the glycosyl bond are populated. This conclusion is supported by the observation that two sets of 1H NMR resonances, matching those in acidic and basic solutions respectively, are obtained at pH 7.3. Exchange NOE crosspeaks are observed for X7 H2, A8 H2, T17 H6, A18 H2, A18 H8, and G19 H8, indicating a slow exchange between the acidic and basic conformations of the HNE adduct, on the NMR timescale (Chart 2) (Figure S3 in the Supporting Information). The steric bulk of the HNE aliphatic chain presumably slows the conformational exchange rate. At pH 7.3, the set of resonances corresponding to those observed in basic solution are stronger than are those observed in acidic solution, suggesting the major conformer, which constitutes 60–70% of the overall conformations based on the integration of the 1H resonances, is that in which X7 adopts the anti conformation about the glycosyl bond. This is also supported by the similarity of COSY spectra at pH 7.3 and pH 8.9. The pKa of adenosine is ~7.6 (97), therefore, A18 is largely protonated at pH 5.5, X7 adopts the syn conformation about the glycosyl bond to form hydrogen bonds with A18; A18 is weakly protonated at pH 7.3, and the anti conformation of the glycosyl bond is the major specie present; A18 is not protonated at pH 8.9, and the anti conformation of the glycosyl bond predominates.
Chart 2.
pH-dependent syn/anti conformational equilibrium of the ring-closed 1,N2-HNE-dG adduct when mismatched with dA in duplex DNA.
Structure-Activity Relationships
The inability of the (6S,8R,11S) 1,N2-HNE-dG adduct to undergo ring-opening when placed opposite dA in duplex DNA suggests that it does not undergo further chemistry. In contrast, when placed complementary to dC in this 5′-CpG-3′ sequence, the (6S,8R,11S) adduct slowly forms an interstrand cross-link (35). The slow rate of interstrand cross-link formation is attributed to the fact that the 6S,8R,11S 1,N2-HNE-dG adduct exists primarily as a set of diastereomeric cyclic hemiacetals when placed into this duplex (36); these cyclic hemiacetals mask the aldehyde necessary for cross-link formation. In contrast, the corresponding 1,N2-dG adducts of acrolein (98, 99) and crotonaldehyde (99, 100), which exist predominantly as aldehydes and do not rearrange to cyclic hemiacetals, form inter-chain cross-links more rapidly when paired opposite dC in this sequence context (99, 101, 102). In the case of the crotonaldehyde adduct, the 6R stereoisomer forms cross-links more efficiently than does the 6S stereoisomer (102, 103). This is attributed to the relative orientations of the reactive aldehyde species within the minor groove for the two diasteromeric adducts, such that interstrand cross-linking is favored for the 6R stereoisomer (100). Significantly, the (6S,8R,11S) diastereomeric adduct derived from HNE possesses the same relative stereochemistry as does the 6R crotonaldehyde adduct, and likewise, favorably orients the reactive aldehyde species to facilitate interstrand cross-link formation (104).
Biological Implications
The low levels of mutations induced by the (6S,8R,11S) 1,N2-HNE-dG adduct when present in duplex DNA opposite cytosine are likely related to the observation that it undergoes ring-opening to expose the Watson-Crick hydrogen bonding face of the adducted dG (58), which facilitate the correct incorporation of dCTP opposite the lesion during lesion bypass. A similar explanation has been advanced to explain the low levels of mutations induced by the acrolein (105, 106)- and crotonaldehyde-derived exocyclic 1,N2-dG adducts (107). Wolfle et al. (61) reported that the sequential activity of pols ι and κ bypassed the (6S,8R,11S) and (6R,8S,11R) 1,N2-HNE-dG adducts. Significantly, pol ι correctly inserted dCTP and to a lesser extent dTTP opposite the HNE adduct. Further extension was achieved in the presence of pol κ, which elongated from a C opposite the HNE adducts much more efficiently than when T was opposite the adducts (61).
The source of the G→T transversions induced by the (6S,8R,11S) 1,N2-HNE-dG adduct in this sequence remains obscure. Xing et al. (26) attributed the low levels of G→T transversions to the re-orientation of the adduct into the syn conformation about the glycosyl bond, thus allowing misincorporation of dATP opposite the lesion. This might allow for subsequent extension from the mismatched template primer, e.g., by polymerase ζ, which efficiently extends from primer-terminal base pairs containing mismatches or lesions (108). The present structural data confirm that such a re-orientation about the glycosyl bond does occur when the (6S,8R,11S) 1,N2-HNE-dG adduct is mismatched with dA in duplex DNA, and that the syn conformation of the adduct is present in equilibrium with the anti conformation at physiological pH values. Xing et al. (26) subsequently invoked the transient presence of the rare imine tautomer of dATP during trans-lesion bypass as a potential source of the G→T transversions. In contrast, the present results suggest that a pH-mediated transient protonation of the N1 imine of dA occurs in duplex DNA. The (6S,8R,11S) 1,N2-HNE-dG adduct also induces G→A transitions in the human p53 gene (55, 57) and in the sequence used in the present study (58). Thus, the extent to which the HNE-dG•T mismatched sequence perturbs the DNA duplex will also be of interest. The (6S,8R,11S) HNE-induced adduct is anticipated to also maintain the exocyclic 1,N2-dG structure when mismatched with T in the complementary strand.
The saturated 1,N2-propanodeoxyguanosine adduct (PdG), which is stable in the exocyclic 1,N2-dG configuration, provides a surrogate for the chemically unstable enal-derived exocyclic 1,N2-dG adducts (76, 89, 95, 109–115). The conformation of PdG in either PdG•dC (89, 95) or PdG•dA (76, 109, 111, 112) base pairs is also pH-dependent. PdG adopts the syn conformation about the glycosyl bond with complementary dC or dA in acidic solution whereas it adopts the anti conformation without hydrogen bonds with the opposite base in basic solution. At neutral pH or a pKa value of 7.6, the two conformations exist in equilibrium. Plum et al. (116) showed that the PdG lesion alters the differential thermal stability (ΔTm) but not the differential thermodynamic stability (ΔΔG) of duplexes with correctly paired dC or mismatched dA cross-strand partners. This has led to the suggestion that the observed preference for insertion of dATP over dCTP across from the PdG lesion should not be rationalized in terms of thermodynamic differences between the final duplex states, and probably instead reflects properties of the DNA polymerase and the replication fork. Likewise, the present data indicate that the Tm for the X7•A18 base pair in either basic or acidic solution is lower than is the Tm for the X7•C18 base pair. In light of these findings, the structure of the (6S,8R,11S) 1,N2-HNE-dG adduct, particularly in complexes with the bypass polymerases ι and κ (61) will be of interest.
A similar pH-dependent syn/anti conformational exchange has also been observed for the 1,N2-ethenodG (1,N2-εdG)•dC (117–119) base pair. Similar to the PdG adduct, and differing from the (6S,8R,11S) 1,N2-HNE-dG adduct, 1,N2-εdG is stable in the exocyclic ring-closed configuration. Under basic conditions, both 1,N2-εdG and the complementary dC adopted the anti conformation about the glycosyl bonds (117, 118). In contrast, under acidic conditions, the 1,N2-εdG adduct formed a Hoogsteen pair with the complementary cytosine, characterized by downfield shifts of the amino protons of the cytosine complementary to the exocyclic adduct (119).
Summary
The HNE-dG adduct with the (6S,8R,11S) configuration maintains the exocyclic 1,N2-dG structure when mismatched with dA in this duplex. It undergoes syn/anti conformational equilibrium with the anti conformation being the major species in neutral solution. The syn conformation favors base pairing with the complementary protonated A18 in acidic solution. In basic solution, X7 adopts the anti conformation about the glycosyl bond without hydrogen bonding with A18. The X7•A18 mismatch greatly perturbs the neighboring C6•G19 and A8•T17 base pairs in both acidic and basic solutions.
Supplementary Material
Table S1, showing chemical shifts of the mismatched duplex at pH 5.5; Table S2, showing chemical shifts of the mismatched duplex at pH 8.9; Table S3, showing NOE restraints utilized in the rMD calculation at pH 5.5; Table S4, showing NOE restraints utilized in the rMD calculation at pH 8.9, Table S5, showing torsion angles of the rMD structure at pH 5.5; Table S6, showing torsion angles of the rMD structure at pH 8.9; Figure S1, showing force field parameters for the HNE-dG adduct; Figure S2, showing 1H NMRs of the mismatched duplex at pH 5.5, 7.3, and 8.9; Figure S3, showing the exchange NOE correlations of the two sets of resonances at pH 7.3; Figure S4, showing NOE crosspeaks of the imino protons at pH 5.5 and 8.9; Figure S5, showing NOE crosspeaks of the protonated A18 and C6 amino protons at pH 5.5; Figure S6, showing NOE crosspeaks associated with HNE protons at pH 5.5 and 8.9; Figure S7, showing chemical shift perturbations of the mismatched duplex at pH 8.9; Figure S8, showing chemical shift comparisons of the mismatched duplex at pH 5.5 and 8.9; Figure S9, showing residue-by-residue sixth root residuals (R1x) at pH 5.5 and 8.9; and Figure S10, showing CPK models of average structures of the mismatched duplex at pH 5.5 and 8.9. This material is available free of charge via the internet at http://pubs.acs.org.
Acknowledgments
Dr. Markus Voehler assisted with NMR experiments and Ms. Albena Kozekova assisted with the synthesis of the (6S,8R,11S) HNE-modified oligodeoxynucleotide. This work was supported by NIH Grant PO1 ES-05355 (R.S.L, C.J.R., & M.P.S.). Funding for the NMR spectrometers was supplied by Vanderbilt University; by NIH grant RR-05805, and the Vanderbilt Center in Molecular Toxicology, ES-00267. The Vanderbilt-Ingram Cancer Center is supported by NIH grant CA-68485.
Abbreviations
- HNE
trans-4-hydroxynonenal
- 1N2-HNE-dG
HNE derived 1,N2-2′-deoxyguanosine adduct
- PdG
1,N2-propano-2′-deoxyguanosine
- M1dG
3-(2′-deoxy-β-D-erythro-pentofuranosyl)pyrimido[1,2-α]purin-10(3H)-one
- NOESY
nuclear Overhauser effect spectroscopy
- COSY
correlation spectroscopy
- TOCSY
total correlation spectroscopy
- DQF-COSY
double-quantum filtered COSY
- NOE
nuclear Overhauser effect
- rMD
restrained molecular dynamics
Contributor Information
Hai Huang, Department of Chemistry, Center in Molecular Toxicology, Center for Structural Biology and Vanderbilt-Ingram Cancer Center, Vanderbilt University, Nashville, Tennessee 37235.
Hao Wang, Department of Chemistry, Center in Molecular Toxicology, Center for Structural Biology and Vanderbilt-Ingram Cancer Center, Vanderbilt University, Nashville, Tennessee 37235.
R. Stephen Lloyd, Center for Research in Occupational and Environmental Toxicology, Oregon Health and Science University, 3181 SW Sam Jackson Park Road, L606, Portland, Oregon 97239-3098.
Carmelo J. Rizzo, Department of Chemistry, Center in Molecular Toxicology, Center for Structural Biology and Vanderbilt-Ingram Cancer Center, Vanderbilt University, Nashville, Tennessee 37235
Michael P. Stone, Department of Chemistry, Center in Molecular Toxicology, Center for Structural Biology and Vanderbilt-Ingram Cancer Center, Vanderbilt University, Nashville, Tennessee 37235
References
- 1.Benedetti A, Comporti M, Esterbauer H. Identification of 4-hydroxynonenal as a cytotoxic product originating from the peroxidation of liver microsomal lipids. Biochim Biophys Acta. 1980;620:281–296. doi: 10.1016/0005-2760(80)90209-x. [DOI] [PubMed] [Google Scholar]
- 2.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radical Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 3.Burcham PC. Genotoxic lipid peroxidation products: Their DNA damaging properties and role in formation of endogenous DNA adducts. Mutagenesis. 1998;13:287–305. doi: 10.1093/mutage/13.3.287. [DOI] [PubMed] [Google Scholar]
- 4.Lee SH, Blair IA. Characterization of 4-oxo-2-nonenal as a novel product of lipid peroxidation. Chem Res Toxicol. 2000;13:698–702. doi: 10.1021/tx000101a. [DOI] [PubMed] [Google Scholar]
- 5.Schneider C, Tallman KA, Porter NA, Brash AR. Two distinct pathways of formation of 4-hydroxynonenal. Mechanisms of nonenzymatic transformation of the 9- and 13-hydroperoxides of linoleic acid to 4-hydroxyalkenals. J Biol Chem. 2001;276:20831–20838. doi: 10.1074/jbc.M101821200. [DOI] [PubMed] [Google Scholar]
- 6.Schneider C, Porter NA, Brash AR. Routes to 4-hydroxynonenal: Fundamental issues in the mechanisms of lipid peroxidation. J Biol Chem. 2008;283:15539–15543. doi: 10.1074/jbc.R800001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nadkarni DV, Sayre LM. Structural definition of early lysine and histidine adduction chemistry of 4-hydroxynonenal. Chem Res Toxicol. 1995;8:284–291. doi: 10.1021/tx00044a014. [DOI] [PubMed] [Google Scholar]
- 8.Amarnath V, Valentine WM, Montine TJ, Patterson WH, Amarnath K, Bassett CN, Graham DG. Reactions of 4-hydroxy-2(E)-nonenal and related aldehydes with proteins studied by carbon-13 nuclear magnetic resonance spectroscopy. Chem Res Toxicol. 1998;11:317–328. doi: 10.1021/tx970176n. [DOI] [PubMed] [Google Scholar]
- 9.Doorn JA, Petersen DR. Covalent modification of amino acid nucleophiles by the lipid peroxidation products 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Chem Res Toxicol. 2002;15:1445–1450. doi: 10.1021/tx025590o. [DOI] [PubMed] [Google Scholar]
- 10.Zhang WH, Liu J, Xu G, Yuan Q, Sayre LM. Model studies on protein side chain modification by 4-oxo-2-nonenal. Chem Res Toxicol. 2003;16:512–523. doi: 10.1021/tx020105a. [DOI] [PubMed] [Google Scholar]
- 11.Sayre LM, Lin D, Yuan Q, Zhu X, Tang X. Protein adducts generated from products of lipid oxidation: Focus on HNE and ONE. Drug Metab Rev. 2006;38:651–675. doi: 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- 12.Parola M, Bellomo G, Robino G, Barrera G, Dianzani MU. 4-Hydroxynonenal as a biological signal: Molecular basis and pathophysiological implications. Antioxid Redox Signal. 1999;1:255–284. doi: 10.1089/ars.1999.1.3-255. [DOI] [PubMed] [Google Scholar]
- 13.Poli G, Schaur RJ. 4-Hydroxynonenal in the pathomechanisms of oxidative stress. IUBMB life. 2000;50:315–321. doi: 10.1080/713803726. [DOI] [PubMed] [Google Scholar]
- 14.Nakashima I, Liu W, Akhand AA, Takeda K, Kawamoto Y, Kato M, Suzuki H. 4-hydroxynonenal triggers multistep signal transduction cascades for suppression of cellular functions. Mol Aspects Med. 2003;24:231–238. doi: 10.1016/s0098-2997(03)00018-9. [DOI] [PubMed] [Google Scholar]
- 15.West JD, Ji C, Duncan ST, Amarnath V, Schneider C, Rizzo CJ, Brash AR, Marnett LJ. Induction of apoptosis in colorectal carcinoma cells treated with 4-hydroxy-2-nonenal and structurally related aldehydic products of lipid peroxidation. Chem Res Toxicol. 2004;17:453–462. doi: 10.1021/tx034248o. [DOI] [PubMed] [Google Scholar]
- 16.West JD, Marnett LJ. Alterations in gene expression induced by the lipid peroxidation product, 4-hydroxy-2-nonenal. Chem Res Toxicol. 2005;18:1642–1653. doi: 10.1021/tx050211n. [DOI] [PubMed] [Google Scholar]
- 17.West JD, Marnett LJ. Endogenous reactive intermediates as modulators of cell signaling and cell death. Chem Res Toxicol. 2006;19:173–194. doi: 10.1021/tx050321u. [DOI] [PubMed] [Google Scholar]
- 18.Dwivedi S, Sharma A, Patrick B, Sharma R, Awasthi YC. Role of 4-hydroxynonenal and its metabolites in signaling. Redox Rep. 2007;12:4–10. doi: 10.1179/135100007X162211. [DOI] [PubMed] [Google Scholar]
- 19.Sayre LM, Zelasko DA, Harris PL, Perry G, Salomon RG, Smith MA. 4-Hydroxynonenal-derived advanced lipid peroxidation end products are increased in Alzheimer’s disease. J Neurochem. 1997;68:2092–2097. doi: 10.1046/j.1471-4159.1997.68052092.x. [DOI] [PubMed] [Google Scholar]
- 20.Yoritaka A, Hattori N, Uchida K, Tanaka M, Stadtman ER, Mizuno Y. Immunohistochemical detection of 4-hydroxynonenal protein adducts in Parkinson disease. Proc Natl Acad Sci USA. 1996;93:2696–2701. doi: 10.1073/pnas.93.7.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Napoli C, D’Armiento FP, Mancini FP, Postiglione A, Witztum JL, Palumbo G, Palinski W. Fatty streak formation occurs in human fetal aortas and is greatly enhanced by maternal hypercholesterolemia. Intimal accumulation of low density lipoprotein and its oxidation precede monocyte recruitment into early atherosclerotic lesions. J Clin Invest. 1997;100:2680–2690. doi: 10.1172/JCI119813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamagami K, Yamamoto Y, Kume M, Ishikawa Y, Yamaoka Y, Hiai H, Toyokuni S. Formation of 8-hydroxy-2′-deoxyguanosine and 4-hydroxy-2-nonenal-modified proteins in rat liver after ischemia-reperfusion: Distinct localization of the two oxidatively modified products. Antioxid Redox Signal. 2000;2:127–136. doi: 10.1089/ars.2000.2.1-127. [DOI] [PubMed] [Google Scholar]
- 23.Winter CK, Segall HJ, Haddon WF. Formation of cyclic adducts of deoxyguanosine with the aldehydes trans-4-hydroxy-2-hexenal and trans-4-hydroxy-2-nonenal in vitro. Cancer Res. 1986;46:5682–5686. [PubMed] [Google Scholar]
- 24.Douki T, Odin F, Caillat S, Favier A, Cadet J. Predominance of the 1,N2-propano 2′-deoxyguanosine adduct among 4-hydroxy-2-nonenal-induced DNA lesions. Free Radical Biol Med. 2004;37:62–70. doi: 10.1016/j.freeradbiomed.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 25.Kowalczyk P, Ciesla JM, Komisarski M, Kusmierek JT, Tudek B. Long-chain adducts of trans-4-hydroxy-2-nonenal to DNA bases cause recombination, base substitutions and frameshift mutations in M13 phage. Mutat Res. 2004;550:33–48. doi: 10.1016/j.mrfmmm.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Xing DX, Sun LX, Cukier RI, Bu YX. Theoretical prediction of the p53 gene mutagenic mechanism induced by trans-4-hydroxy-2-nonenal. J Phys Chem B. 2007;111:5362–5371. doi: 10.1021/jp0673922. [DOI] [PubMed] [Google Scholar]
- 27.Yi P, Zhan D, Samokyszyn VM, Doerge DR, Fu PP. Synthesis and 32P-postlabeling/high-performance liquid chromatography separation of diastereomeric 1,N2-(1,3-propano)-2′-deoxyguanosine 3′-phosphate adducts formed from 4-hydroxy-2-nonenal. Chem Res Toxicol. 1997;10:1259–1265. doi: 10.1021/tx970100r. [DOI] [PubMed] [Google Scholar]
- 28.Chung FL, Nath RG, Ocando J, Nishikawa A, Zhang L. Deoxyguanosine adducts of t-4-hydroxy-2-nonenal are endogenous DNA lesions in rodents and humans: Detection and potential sources. Cancer Res. 2000;60:1507–1511. [PubMed] [Google Scholar]
- 29.Wacker M, Schuler D, Wanek P, Eder E. Development of a 32P-postlabeling method for the detection of 1,N2-propanodeoxyguanosine adducts of trans-4-hydroxy-2-nonenal in vivo. Chem Res Toxicol. 2000;13:1165–1173. doi: 10.1021/tx000058r. [DOI] [PubMed] [Google Scholar]
- 30.Wacker M, Wanek P, Eder E. Detection of 1,N2-propanodeoxyguanosine adducts of trans-4-hydroxy-2-nonenal after gavage of trans-4-hydroxy-2-nonenal or induction of lipid peroxidation with carbon tetrachloride in F344 rats. Chem Biol Interact. 2001;137:269–283. doi: 10.1016/s0009-2797(01)00259-9. [DOI] [PubMed] [Google Scholar]
- 31.Chung FL, Zhang L. Deoxyguanosine adducts of tert-4-hydroxy-2-nonenal as markers of endogenous DNA lesions. Methods Enzymol. 2002;353:523–536. doi: 10.1016/s0076-6879(02)53074-3. [DOI] [PubMed] [Google Scholar]
- 32.Liu X, Lovell MA, Lynn BC. Detection and quantification of endogenous cyclic DNA adducts derived from trans-4-hydroxy-2-nonenal in human brain tissue by isotope dilution capillary liquid chromatography nanoelectrospray tandem mass spectrometry. Chem Res Toxicol. 2006;19:710–718. doi: 10.1021/tx0502903. [DOI] [PubMed] [Google Scholar]
- 33.Pan J, Davis W, Trushin N, Amin S, Nath RG, Salem N, Jr, Chung FL. A solid-phase extraction/high-performance liquid chromatography-based 32P-postlabeling method for detection of cyclic 1,N2-propanodeoxyguanosine adducts derived from enals. Anal Biochem. 2006;348:15–23. doi: 10.1016/j.ab.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 34.Wang H, Rizzo CJ. Stereocontrolled syntheses of all four stereoisomeric 1,N2-deoxyguanosine adducts of the lipid peroxidation product trans-4-hydroxynonenal. Org Lett. 2001;3:3603–3605. doi: 10.1021/ol016810c. [DOI] [PubMed] [Google Scholar]
- 35.Wang H, Kozekov ID, Harris TM, Rizzo CJ. Site-specific synthesis and reactivity of oligonucleotides containing stereochemically defined 1, N2-deoxyguanosine adducts of the lipid peroxidation product trans-4-hydroxynonenal. J Am Chem Soc. 2003;125:5687–5700. doi: 10.1021/ja0288800. [DOI] [PubMed] [Google Scholar]
- 36.Huang H, Wang H, Qi N, Kozekova A, Rizzo CJ, Stone MP. Rearrangement of the (6S,8R,11S) and (6R,8S,11R) exocyclic 1,N2-deoxyguanosine adducts of trans-4-hydroxynonenal to N2-deoxyguanosine cyclic hemiacetal adducts when placed complementary to cytosine in duplex DNA. J Am Chem Soc. 2008;130:10898–10906. doi: 10.1021/ja801824b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riggins JN, Pratt DA, Voehler M, Daniels JS, Marnett LJ. Kinetics and mechanism of the general-acid-catalyzed ring-closure of the malondialdehyde-DNA adduct, N2-(3-oxo-1-propenyl)deoxyguanosine (N2OPdG-), to 3-(2′-deoxy-beta-D-erythro-pentofuranosyl)pyrimido[1,2-alpha]purin-10(3H)-one (M1dG) J Am Chem Soc. 2004;126:10571–10581. doi: 10.1021/ja040010q. [DOI] [PubMed] [Google Scholar]
- 38.Riggins JN, Daniels JS, Rouzer CA, Marnett LJ. Kinetic and thermodynamic analysis of the hydrolytic ring-opening of the malondialdehyde-deoxyguanosine adduct, 3-(2′-deoxy-beta-D-erythro-pentofuranosyl)- pyrimido[1,2-alpha]purin-10(3H)-one. J Am Chem Soc. 2004;126:8237–8243. doi: 10.1021/ja040009r. [DOI] [PubMed] [Google Scholar]
- 39.Sodum RS, Chung FL. 1,N2-ethenodeoxyguanosine as a potential marker for DNA adduct formation by trans-4-hydroxy-2-nonenal. Cancer Res. 1988;48:320–323. [PubMed] [Google Scholar]
- 40.Sodum RS, Chung FL. Structural characterization of adducts formed in the reaction of 2,3-epoxy-4-hydroxynonanal with deoxyguanosine. Chem Res Toxicol. 1989;2:23–28. doi: 10.1021/tx00007a004. [DOI] [PubMed] [Google Scholar]
- 41.Sodum RS, Chung FL. Stereoselective formation of in vitro nucleic acid adducts by 2,3-epoxy-4-hydroxynonanal. Cancer Res. 1991;51:137–143. [PubMed] [Google Scholar]
- 42.Chen HJ, Chung FL. Formation of etheno adducts in reactions of enals via autoxidation. Chem Res Toxicol. 1994;7:857–860. doi: 10.1021/tx00042a021. [DOI] [PubMed] [Google Scholar]
- 43.el Ghissassi F, Barbin A, Nair J, Bartsch H. Formation of 1,N6-ethenoadenine and 3,N4-ethenocytosine by lipid peroxidation products and nucleic acid bases. Chem Res Toxicol. 1995;8:278–283. doi: 10.1021/tx00044a013. [DOI] [PubMed] [Google Scholar]
- 44.Benamira M, Marnett LJ. The lipid peroxidation product 4-hydroxynonenal is a potent inducer of the SOS response. Mutat Res. 1992;293:1–10. doi: 10.1016/0921-8777(92)90002-k. [DOI] [PubMed] [Google Scholar]
- 45.Esterbauer H, Eckl P, Ortner A. Possible mutagens derived from lipids and lipid precursors. Mutat Res. 1990;238:223–233. doi: 10.1016/0165-1110(90)90014-3. [DOI] [PubMed] [Google Scholar]
- 46.Eckl PM, Ortner A, Esterbauer H. Genotoxic properties of 4-hydroxyalkenals and analogous aldehydes. Mutat Res. 1993;290:183–192. doi: 10.1016/0027-5107(93)90158-c. [DOI] [PubMed] [Google Scholar]
- 47.Karlhuber GM, Bauer HC, Eckl PM. Cytotoxic and genotoxic effects of 4-hydroxynonenal in cerebral endothelial cells. Mutat Res. 1997;381:209–216. doi: 10.1016/s0027-5107(97)00170-x. [DOI] [PubMed] [Google Scholar]
- 48.Eckl PM. Genotoxicity of HNE. Mol Aspects Med. 2003;24:161–165. doi: 10.1016/s0098-2997(03)00010-4. [DOI] [PubMed] [Google Scholar]
- 49.Emerit I, Khan SH, Esterbauer H. Hydroxynonenal, a component of clastogenic factors? Free Radical Biol Med. 1991;10:371–377. doi: 10.1016/0891-5849(91)90045-5. [DOI] [PubMed] [Google Scholar]
- 50.Chung FL, Komninou D, Zhang L, Nath R, Pan J, Amin S, Richie J. Glutathione depletion enhances the formation of endogenous cyclic DNA adducts derived from t-4-hydroxy-2-nonenal in rat liver. Chem Res Toxicol. 2005;18:24–27. doi: 10.1021/tx049728+. [DOI] [PubMed] [Google Scholar]
- 51.Falletti O, Cadet J, Favier A, Douki T. Trapping of 4-hydroxynonenal by glutathione efficiently prevents formation of DNA adducts in human cells. Free Radical Biol Med. 2007;42:1258–1269. doi: 10.1016/j.freeradbiomed.2007.01.024. [DOI] [PubMed] [Google Scholar]
- 52.Yadav UC, Ramana KV, Awasthi YC, Srivastava SK. Glutathione level regulates HNE-induced genotoxicity in human erythroleukemia cells. Toxicol Appl Pharmacol. 2008;227:257–264. doi: 10.1016/j.taap.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cajelli E, Ferraris A, Brambilla G. Mutagenicity of 4-hydroxynonenal in V79 Chinese hamster cells. Mutat Res. 1987;190:169–171. doi: 10.1016/0165-7992(87)90050-9. [DOI] [PubMed] [Google Scholar]
- 54.Chung FL, Chen HJ, Guttenplan JB, Nishikawa A, Hard GC. 2,3-epoxy-4-hydroxynonanal as a potential tumor-initiating agent of lipid peroxidation. Carcinogenesis. 1993;14:2073–2077. doi: 10.1093/carcin/14.10.2073. [DOI] [PubMed] [Google Scholar]
- 55.Hussain SP, Raja K, Amstad PA, Sawyer M, Trudel LJ, Wogan GN, Hofseth LJ, Shields PG, Billiar TR, Trautwein C, Hohler T, Galle PR, Phillips DH, Markin R, Marrogi AJ, Harris CC. Increased p53 mutation load in nontumorous human liver of Wilson disease and hemochromatosis: Oxyradical overload diseases. Proc Natl Acad Sci USA. 2000;97:12770–12775. doi: 10.1073/pnas.220416097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu W, Feng Z, Eveleigh J, Iyer G, Pan J, Amin S, Chung FL, Tang MS. The major lipid peroxidation product, trans-4-hydroxy-2-nonenal, preferentially forms DNA adducts at codon 249 of human p53 gene, a unique mutational hotspot in hepatocellular carcinoma. Carcinogenesis. 2002;23:1781–1789. doi: 10.1093/carcin/23.11.1781. [DOI] [PubMed] [Google Scholar]
- 57.Feng Z, Hu W, Amin S, Tang MS. Mutational spectrum and genotoxicity of the major lipid peroxidation product, trans-4-hydroxy-2-nonenal, induced DNA adducts in nucleotide excision repair-proficient and -deficient human cells. Biochemistry. 2003;42:7848–7854. doi: 10.1021/bi034431g. [DOI] [PubMed] [Google Scholar]
- 58.Fernandes PH, Wang H, Rizzo CJ, Lloyd RS. Site-specific mutagenicity of stereochemically defined 1,N2-deoxyguanosine adducts of trans-4-hydroxynonenal in mammalian cells. Environ Mol Mutagen. 2003;42:68–74. doi: 10.1002/em.10174. [DOI] [PubMed] [Google Scholar]
- 59.Chung FL, Pan J, Choudhury S, Roy R, Hu W, Tang MS. Formation of trans-4-hydroxy-2-nonenal- and other enal-derived cyclic DNA adducts from ω-3 and ω-6 polyunsaturated fatty acids and their roles in DNA repair and human p53 gene mutation. Mutat Res. 2003;531:25–36. doi: 10.1016/j.mrfmmm.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 60.Choudhury S, Pan J, Amin S, Chung FL, Roy R. Repair kinetics of trans-4-hydroxynonenal-induced cyclic 1,N2-propanodeoxyguanine DNA adducts by human cell nuclear extracts. Biochemistry. 2004;43:7514–7521. doi: 10.1021/bi049877r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolfle WT, Johnson RE, Minko IG, Lloyd RS, Prakash S, Prakash L. Replication past a trans-4-hydroxynonenal minor-groove adduct by the sequential action of human DNA polymerase l and k. Mol Cell Biol. 2006;26:381–386. doi: 10.1128/MCB.26.1.381-386.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cavaluzzi MJ, Borer PN. Revised UV extinction coefficients for nucleoside-5′-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004;32:e13. doi: 10.1093/nar/gnh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 64.Sklenar V, Bax A, Zon G. Assignment of Z DNA NMR spectra of poly d(Gm5C) by two-dimensional multinuclear spectroscopy. Febs Lett. 1986;208:94–98. doi: 10.1016/0014-5793(86)81539-3. [DOI] [PubMed] [Google Scholar]
- 65.Sklenar V, Miyashiro H, Zon G, Miles HT, Bax A. Assignment of the 31P and 1H resonances in oligonucleotides by two-dimensional NMR spectroscopy. Febs Lett. 1986;208:94–98. doi: 10.1016/0014-5793(86)81539-3. [DOI] [PubMed] [Google Scholar]
- 66.James TL. Relaxation matrix analysis of two-dimensional nuclear Overhauser effect spectra. Curr Opin Struct Biol. 1991;1:1042–1053. [Google Scholar]
- 67.Keepers JW, James TL. A theoretical study of distance determinations from NMR - Two-dimensional nuclear Overhauser effect spectra. J Magn Reson. 1984;57:404–426. [Google Scholar]
- 68.Borgias BA, James TL. Two-dimensional nuclear Overhauser effect: Complete relaxation matrix analysis. Methods Enzymol. 1989;176:169–183. doi: 10.1016/0076-6879(89)76011-0. [DOI] [PubMed] [Google Scholar]
- 69.Borgias BA, James TL. MARDIGRAS--a procedure for matrix analysis of relaxation for discerning geometry of an aqueous structure. J Magn Reson. 1990;87:475–487. [Google Scholar]
- 70.Liu H, Spielmann HP, Ulyanov NB, Wemmer DE, James TL. Interproton distance bounds from 2D NOE intensities: Effect of experimental noise and peak integration errors. J Biomol NMR. 1995;6:390–402. doi: 10.1007/BF00197638. [DOI] [PubMed] [Google Scholar]
- 71.Salazar M, Fedoroff OY, Miller JM, Ribeiro NS, Reid BR. The DNA strand in DNA:RNA hybrid duplexes is neither B-form nor A-form in solution. Biochemistry. 1993;32:4207–4215. doi: 10.1021/bi00067a007. [DOI] [PubMed] [Google Scholar]
- 72.Wang H, Zuiderweg ERP, Glick GD. Solution structure of a disulfide cross-linked DNA hairpin. J Am Chem Soc. 1995;117:2981–2991. [Google Scholar]
- 73.Geen H, Freeman R. Band-selective radiofrequency pulses. J Magn Reson. 1991;93:93–141. [Google Scholar]
- 74.Lankhorst PP, Haasnoot AG, Erkelens C, Altona C. Carbon-13 NMR in conformational analysis of nucleic acid fragments. 3 The magnitude of torsional angle in d(TpA) from CCOP and HCOP NMR coupling constants. Nucleic Acids Res. 1984;12:5419–5428. doi: 10.1093/nar/12.13.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Pomelli J, Ochterski W, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewska VG, Daniels AD, Farkas O, Rabuck AD, Raghavachari K, Ortiz JV. GAUSSIAN 03. Gaussian, Inc; Wallingford, CT: 2004. [Google Scholar]
- 76.Kouchakdjian M, Marinelli E, Gao X, Johnson F, Grollman A, Patel D. NMR studies of exocyclic 1,N2-propanodeoxyguanosine adducts (X) opposite purines in DNA duplexes: Protonated X(syn):A(anti) pairing (acidic pH) and X(syn):G(anti) pairing (neutral pH) at the lesion site. Biochemistry. 1989;28:5647–5657. doi: 10.1021/bi00439a047. [DOI] [PubMed] [Google Scholar]
- 77.Arnott S, Hukins DWL. Optimised parameters for A-DNA and B-DNA. Biochem Biophys Res Commun. 1972;47:1504–1509. doi: 10.1016/0006-291X(72)90243-4. [DOI] [PubMed] [Google Scholar]
- 78.Case DA, Pearlman DA, Caldwell JW, Cheatham TE, III, Wang J, Ross WS, Simmerling CL, Darden TA, Merz KM, Stanton RV, Cheng AL, Vincent JJ, Crowley M, Tsui V, Gohlke H, Radmer RJ, Duan Y, Pitera J, Massova I, Seibel GL, Singh UC, Weiner PK, Kollman PA. AMBER 8.0. University of California; San Francisco, CA: 2002. [Google Scholar]
- 79.Hawkins GD, Cramer CJ, Truhlar DG. Pairwise solute descreening of solute charges from a dielectric medium. Chem Phys Lett. 1995;246:122–129. [Google Scholar]
- 80.Hawkins GD, Cramer CJ, Truhlar DG. Parametrized models of aqueous free energies of solvation based on pairwise descreening of solute atomic charges from a dielectric medium. J Phys Chem. 1996;100:19824–19839. [Google Scholar]
- 81.Tsui V, Case DA. Theory and applications of the generalized Born solvation model in macromolecular simulations. Biopolymers. 2000;56:275–291. doi: 10.1002/1097-0282(2000)56:4<275::AID-BIP10024>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 82.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comp Phys. 1977;23:327–341. [Google Scholar]
- 83.Liu Y, Zhao D, Altman R, Jardetzky O. A systematic comparison of three structure determination methods from NMR data: Dependence upon quality and quantity of data. J Biomol NMR. 1992;2:373–388. doi: 10.1007/BF01874815. [DOI] [PubMed] [Google Scholar]
- 84.Thomas PD, Basus VJ, James TL. Protein solution structure determination using distances from two-dimensional nuclear Overhauser effect experiments: Effect of approximations on the accuracy of derived structures. Proc Natl Acad Sci USA. 1991;88:1237–1241. doi: 10.1073/pnas.88.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu XJ, Olson WK. 3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Patel DJ, Shapiro L, Hare D. DNA and RNA: NMR studies of conformations and dynamics in solution. Q Rev Biophys. 1987;20:35–112. doi: 10.1017/s0033583500004224. [DOI] [PubMed] [Google Scholar]
- 87.Reid BR. Sequence-specific assignments and their use in NMR studies of DNA structure. Q Rev Biophys. 1987;20:2–28. doi: 10.1017/s0033583500004212. [DOI] [PubMed] [Google Scholar]
- 88.Boelens R, Scheek RM, Dijkstra K, Kaptein R. Sequential assignment of imino- and amino-proton resonances in 1H NMR spectra of oligonucleotides by two-dimensional NMR spectroscopy. Application to a lac operator fragment. J Magn Reson. 1985;62:378–386. [Google Scholar]
- 89.Weisenseel JP, Reddy GR, Marnett LJ, Stone MP. Structure of an oligodeoxynucleotide containing a 1,N2-propanodeoxyguanosine adduct positioned in a palindrome derived from the Salmonella typhimurium hisD3052 gene: Hoogsteen pairing at pH 5.2. Chem Res Toxicol. 2002;15:127–139. doi: 10.1021/tx0101090. [DOI] [PubMed] [Google Scholar]
- 90.Szekely J, Rizzo CJ, Marnett LJ. Chemical properties of oxopropenyl adducts of purine and pyrimidine nucleosides and their reactivity toward amino acid cross-link formation. J Am Chem Soc. 2008;130:2195–2201. doi: 10.1021/ja074506u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Norman D, Abuaf P, Hingerty BE, Live D, Grunberger D, Broyde S, Patel DJ. NMR and computational characterization of the N-(deoxyguanosin-8-yl)aminofluorene adduct [(AF)G] opposite adenosine in DNA: (AF)G[syn]:A[anti]pair formation and its pH dependence. Biochemistry. 1989;28:7462–7476. doi: 10.1021/bi00444a046. [DOI] [PubMed] [Google Scholar]
- 92.Mao B, Cosman M, Hingerty BE, Broyde S, Patel DJ. Solution conformation of [AF]dG opposite a -1 deletion site in a DNA duplex: Intercalation of the covalently attached aminoflourene ring into the helix with base displacement of the C8-modified syn guanine into the major groove. Biochemistry. 1995;34:6226–6238. doi: 10.1021/bi00018a027. [DOI] [PubMed] [Google Scholar]
- 93.Cosman M, Hingerty B, Geacintov NE, Broyde S, Patel DJ. Structural alignments of (+)- and (−)-trans-anti-benzo[a]pyrene-dG adducts positioned at a DNA template/primer junction. Biochemistry. 1995;34:15334–15350. doi: 10.1021/bi00046a043. [DOI] [PubMed] [Google Scholar]
- 94.Mao B, Gorin A, Gu Z, Hingerty BE, Broyde S, Patel DJ. Solution structure of the aminofluorene-intercalated conformer of the syn [AF]-C8-dG adduct opposite a -2 deletion site in the NarI hot spot sequence context. Biochemistry. 1997;36:14479–14490. doi: 10.1021/bi972205z. [DOI] [PubMed] [Google Scholar]
- 95.Singh US, Moe JG, Reddy GR, Weisenseel JP, Marnett LJ, Stone MP. 1H NMR of an oligodeoxynucleotide containing a propanodeoxyguanosine adduct positioned in a (CG)3 frameshift hotspot of Salmonella typhimurium hisD3052: Hoogsteen base-pairing at pH 5.8. Chem Res Toxicol. 1993;6:825–836. doi: 10.1021/tx00036a012. [DOI] [PubMed] [Google Scholar]
- 96.Kim HY, Voehler M, Harris TM, Stone MP. Detection of an interchain carbinolamine cross-link formed in a CpG sequence by the acrolein DNA adduct γ-OH-1,N2-propano-2′-deoxyguanosine. J Am Chem Soc. 2002;124:9324–9325. doi: 10.1021/ja020333r. [DOI] [PubMed] [Google Scholar]
- 97.Muth GW, Ortoleva-Donnelly L, Strobel SA. A single adenosine with a neutral pKa in the ribosomal peptidyl transferase center. Science. 2000;289:947–950. doi: 10.1126/science.289.5481.947. [DOI] [PubMed] [Google Scholar]
- 98.de los Santos C, Zaliznyak T, Johnson F. NMR characterization of a DNA duplex containing the major acrolein-derived deoxyguanosine adduct γ-OH-1,-N2-propano-2′-deoxyguanosine. J Biol Chem. 2001;276:9077–9082. doi: 10.1074/jbc.M009028200. [DOI] [PubMed] [Google Scholar]
- 99.Stone MP, Cho YJ, Huang H, Kim HY, Kozekov ID, Kozekova A, Wang H, Lloyd RS, Harris TM, Rizzo CJ. Interstrand DNA cross-links induced by α,β-unsaturated aldehydes derived from lipid peroxidation and environmental sources. Acc Chem Res. 2008;41:793–804. doi: 10.1021/ar700246x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cho YJ, Wang H, Kozekov ID, Kurtz AJ, Jacob J, Voehler M, Smith J, Harris TM, Lloyd RS, Rizzo CJ, Stone MP. Stereospecific formation of interstrand carbinolamine DNA cross-links by crotonaldehyde- and acetaldehyde-derived α-CH3-γ-OH-1,N2-propano-2′-deoxyguanosine adducts in the 5′-CpG-3′ sequence. Chem Res Toxicol. 2006;19:195–208. doi: 10.1021/tx050239z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kozekov ID, Nechev LV, Sanchez A, Harris CM, Lloyd RS, Harris TM. Interchain cross-linking of DNA mediated by the principal adduct of acrolein. Chem Res Toxicol. 2001;14:1482–1485. doi: 10.1021/tx010127h. [DOI] [PubMed] [Google Scholar]
- 102.Kozekov ID, Nechev LV, Moseley MS, Harris CM, Rizzo CJ, Stone MP, Harris TM. DNA interchain cross-links formed by acrolein and crotonaldehyde. J Am Chem Soc. 2003;125:50–61. doi: 10.1021/ja020778f. [DOI] [PubMed] [Google Scholar]
- 103.Lao Y, Hecht SS. Synthesis and properties of an acetaldehyde-derived oligonucleotide interstrand cross-link. Chem Res Toxicol. 2005;18:711–721. doi: 10.1021/tx0497292. [DOI] [PubMed] [Google Scholar]
- 104.Huang H, Wang H, Qi N, Lloyd RS, Rizzo CJ, Stone MP. Stereochemistry of trans-4-hydroxynonenal-derived exocyclic 1,N2-deoxyguanosine adducts modulates formation of inter-strand cross-links in the 5′-CpG-3′ sequence. Biochemistry. 2008;47 doi: 10.1021/bi8011143. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.VanderVeen LA, Hashim MF, Nechev LV, Harris TM, Harris CM, Marnett LJ. Evaluation of the mutagenic potential of the principal DNA adduct of acrolein. J Biol Chem. 2001;276:9066–9070. doi: 10.1074/jbc.M008900200. [DOI] [PubMed] [Google Scholar]
- 106.Yang IY, Hossain M, Miller H, Khullar S, Johnson F, Grollman A, Moriya M. Responses to the major acrolein-derived deoxyguanosine adduct in Escherichia coli. J Biol Chem. 2001;276:9071–9076. doi: 10.1074/jbc.M008918200. [DOI] [PubMed] [Google Scholar]
- 107.Fernandes PH, Kanuri M, Nechev LV, Harris TM, Lloyd RS. Mammalian cell mutagenesis of the DNA adducts of vinyl chloride and crotonaldehyde. Environ Mol Mutagen. 2005;45:455–459. doi: 10.1002/em.20117. [DOI] [PubMed] [Google Scholar]
- 108.Howell CA, Kondratick CM, Washington MT. Substitution of a residue contacting the triphosphate moiety of the incoming nucleotide increases the fidelity of yeast DNA polymerase zeta. Nucleic Acids Res. 2008;36:1731–1740. doi: 10.1093/nar/gkn023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kouchakdjian M, Eisenberg M, Live D, Marinelli E, Grollman AP, Patel DJ. NMR studies of an exocyclic 1,N2-propanodeoxyguanosine adduct (X) located opposite deoxyadenosine (A) in DNA duplexes at basic pH: Simultaneous partial intercalation of X and A between stacked bases. Biochemistry. 1990;29:4456–4465. doi: 10.1021/bi00470a028. [DOI] [PubMed] [Google Scholar]
- 110.Kouchakdjian M, Eisenberg M, Johnson F, Grollman AP, Patel DJ. Structural features of an exocyclic adduct positioned opposite an abasic site in a DNA duplex. Biochemistry. 1991;30:3262–3270. doi: 10.1021/bi00227a014. [DOI] [PubMed] [Google Scholar]
- 111.Huang P, Eisenberg M. The three-dimensional structure in solution (pH 5.8) of a DNA 9-mer duplex containing 1,N2-propanodeoxyguanosine opposite deoxyadenosine. Restrained molecular dynamics and NOE-based refinement calculations. Biochemistry. 1992;31:6518–6532. doi: 10.1021/bi00143a023. [DOI] [PubMed] [Google Scholar]
- 112.Huang P, Patel DJ, Eisenberg M. Solution structure of the exocyclic 1,N2-propanodeoxyguanosine adduct opposite deoxyadenosine in a DNA nonamer duplex at pH 8.9. Model of pH-dependent conformational transition. Biochemistry. 1993;32:3852–3866. doi: 10.1021/bi00066a004. [DOI] [PubMed] [Google Scholar]
- 113.Moe JG, Reddy GR, Marnett LJ, Stone MP. 1H NMR characterization of a duplex oligodeoxynucleotide containing propanodeoxyguanosine opposite a two-base deletion in the (CpG)3 frameshift hotspot of Salmonella typhimurium hisD3052. Chem Res Toxicol. 1994;7:319–328. doi: 10.1021/tx00039a008. [DOI] [PubMed] [Google Scholar]
- 114.Weisenseel JP, Moe JG, Reddy GR, Marnett LJ, Stone MP. Structure of a duplex oligodeoxynucleotide containing propanodeoxyguanosine opposite a two-base deletion in the (CpG)3 frameshift hotspot of Salmonella typhimurium hisD3052 determined by 1H NMR and restrained molecular dynamics. Biochemistry. 1995;34:50–64. doi: 10.1021/bi00001a007. [DOI] [PubMed] [Google Scholar]
- 115.Weisenseel JP, Reddy GR, Marnett LJ, Stone MP. Structure of the 1,N2-propanodeoxyguanosine adduct in a three-base DNA hairpin loop derived from a palindrome in the Salmonella typhimurium hisD3052 gene. Chem Res Toxicol. 2002;15:140–152. doi: 10.1021/tx010107f. [DOI] [PubMed] [Google Scholar]
- 116.Plum GE, Grollman AP, Johnson F, Breslauer KJ. Influence of an exocyclic guanine adduct on the thermal stability, conformation, and melting thermodynamics of a DNA duplex. Biochemistry. 1992;31:12096–12102. doi: 10.1021/bi00163a019. [DOI] [PubMed] [Google Scholar]
- 117.Shanmugam G, Goodenough AK, Kozekov ID, Guengerich FP, Rizzo CJ, Stone MP. Structure of the 1,N2-etheno-2′-deoxyguanosine adduct in duplex DNA at pH 8.6. Chem Res Toxicol. 2007;20:1601–1611. doi: 10.1021/tx7001788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zaliznyak T, Lukin M, Johnson F, de los Santos C. Solution structure of duplex DNA containing the mutagenic lesion 1,N2-etheno-2′-deoxyguanine. Biochemistry. 2008;47:4606–4613. doi: 10.1021/bi7022514. [DOI] [PubMed] [Google Scholar]
- 119.Shanmugam G, Kozekov ID, Guengerich FP, Rizzo CJ, Stone MP. Structure of the 1,N2-ethenodeoxyguanosine adduct opposite cytosine in duplex DNA: Hoogsteen base pairing at pH 5.2. Chem Res Toxicol. 2008;21:1795–1805. doi: 10.1021/tx8001466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, showing chemical shifts of the mismatched duplex at pH 5.5; Table S2, showing chemical shifts of the mismatched duplex at pH 8.9; Table S3, showing NOE restraints utilized in the rMD calculation at pH 5.5; Table S4, showing NOE restraints utilized in the rMD calculation at pH 8.9, Table S5, showing torsion angles of the rMD structure at pH 5.5; Table S6, showing torsion angles of the rMD structure at pH 8.9; Figure S1, showing force field parameters for the HNE-dG adduct; Figure S2, showing 1H NMRs of the mismatched duplex at pH 5.5, 7.3, and 8.9; Figure S3, showing the exchange NOE correlations of the two sets of resonances at pH 7.3; Figure S4, showing NOE crosspeaks of the imino protons at pH 5.5 and 8.9; Figure S5, showing NOE crosspeaks of the protonated A18 and C6 amino protons at pH 5.5; Figure S6, showing NOE crosspeaks associated with HNE protons at pH 5.5 and 8.9; Figure S7, showing chemical shift perturbations of the mismatched duplex at pH 8.9; Figure S8, showing chemical shift comparisons of the mismatched duplex at pH 5.5 and 8.9; Figure S9, showing residue-by-residue sixth root residuals (R1x) at pH 5.5 and 8.9; and Figure S10, showing CPK models of average structures of the mismatched duplex at pH 5.5 and 8.9. This material is available free of charge via the internet at http://pubs.acs.org.