Abstract
Hemoproteins carry out diverse functions utilizing a wide range of chemical reactivity while employing the same heme prosthetic group. It is clear from high-resolution crystal structures and biochemical studies that protein-bound hemes are not planar and adopt diverse conformations. The crystal structure of an H-NOX domain from Thermoanaerobacter tengcongensis (Tt H-NOX) contains the most distorted heme reported to date. In this study, Tt H-NOX was engineered to adopt a flatter heme by mutating proline 115, a conserved residue in the H-NOX family, to alanine. Decreasing heme distortion in Tt H-NOX increases affinity for oxygen and decreases the reduction potential of the heme iron. Additionally, flattening the heme is associated with significant shifts in the N-terminus of the protein. These results show a clear link between the heme conformation and Tt H-NOX structure and demonstrate that heme distortion is an important determinant for maintaining biochemical properties in H-NOX proteins.
The very broad range of chemistry carried out by hemoproteins has attracted a vast amount of attention for many years. Representative examples include oxygen-transporting proteins, like the globins, and potent catalysts involving high-valent iron-oxo complexes, such as the cytochrome P-450s. There are a number of factors that direct and control the type of chemistry carried out by this ubiquitous class of proteins. The coordination environment provided by the protein, for example, plays a significant role in dictating both chemistry and function. The globins have evolved to stabilize the unligated Fe(II) oxidation state and the Fe(II)–O2 complex using a proximal histidine ligand. In contrast, the thiol-ligated cytochrome P450s have a stable Fe(III) oxidation state; however, when reduced to Fe(II), the iron binds O2 and then generates a high-valent iron-oxo complex (formally the Fe(V) oxidation state). This potent oxidant is competent for hydroxylation chemistry of unactivated C-H bonds.
The unique and tunable chemical properties of the heme prosthetic group account for the wide range of functions observed within the hemoprotein family. The tetrapyrrole moiety of the heme is aromatic and as such, hemes in isolation are planar. However, when protein-bound, the heme deviates significantly from planarity (1-5). Although the types of heme distortions found in hemoproteins are energetically unfavorable (6), they are conserved in homologous proteins from different organisms (7), suggesting that heme distortion is important for function. Indeed, out-of-plane heme distortions have been shown to influence the biochemical properties of both non-covalent b-type cytochromes, such as the globins, where the heme is bound to the protein through coordination to the iron atom of an amino acid side chain, as well as the covalent c-type cytochromes, where a porphyrin vinyl group is attached to a protein cysteine residue by a thiol-ether linkage. For example, heme distortion affects the reduction potential (4, 8, 9), as well as ligand binding (3, 10, 11) and spectroscopic characteristics, of cytochrome c and model porphyrins (12, 13). Heme distortion also influences the mechanism of the “on/off” state in heme sensor proteins, such as FixL (14) and the nitrophorins (11), both b-type heme proteins.
Heme distortion and its functional outcomes are complicated. Past work has explored the environment within the pocket and the residues surrounding the bound heme, however, the role of distortion has been experimentally difficult to approach. Fortunately, the highly distorted heme in Tt H-NOX now provides a direct opportunity to address the importance and biochemical properties of heme non-planarity.
The crystal structure of a Heme Nitric oxide/Oxygen Binding (H-NOX) domain from Thermoanaerobacter tengcongensis (Tt H-NOX) contains the most distorted heme observed to date (15). In eukaryotes, H-NOX domains are found as a domain within soluble guanylate cyclase (sGC) as the receptor for nitric oxide (NO) in signaling during vasodilation and neurotransmission (16). In prokaryotes, H-NOX proteins appear to fall into one of two classes. One type is a stand alone protein most often found in a predicted operon with a histidine kinase and less frequently with GGDEF-diguanylate cyclase domain. The other class is fused to methyl-accepting chemotaxis domains in the same open reading frame (17-19). Homology to sGC as well as genomic placement suggest that H-NOX domains in prokaryotes are likely to serve as sensors for gases such as O2 and NO. Recent results with the H-NOX from the facultative aerobe, Shewanella oneidensis, are consistent with this hypothesis (19).
The out-of-plane heme distortions found in Tt H-NOX show large deviations (over 2 Å) from planarity. This distortion appears to be caused by van der Waals interactions in the heme cavity with residue P115 making the largest contribution (Figure 1). P115 is within van der Waals contact with pyrrole-D of the heme, causing the pyrrole to shift out of plane, generating a large kink in the heme propionate group. Multiple sequence alignments show that P115 is invariant among all H-NOX proteins, suggesting that heme distortion is conserved across the entire family. This idea has recently been substantiated as heme distortion is also observed in the crystal structures of Nostoc cyanobacteria H-NOX (Ns H-NOX) (20). Importantly, the same Tt H-NOX proline/heme interaction is maintained in Ns H-NOX, supporting the idea that heme distortion is universal to all H-NOX proteins.
Figure 1. Heme Distortion in wild-type Tt H-NOX.
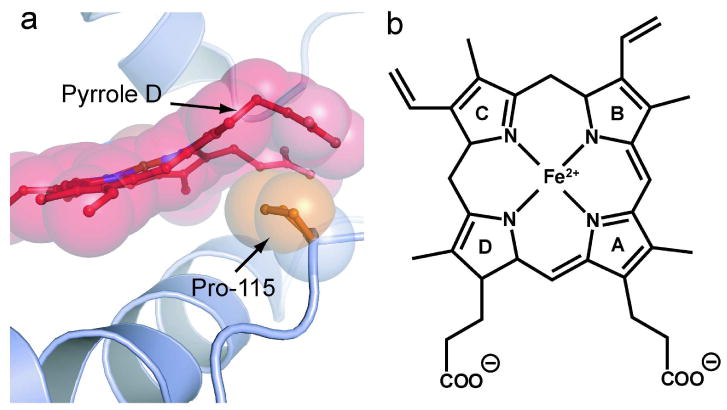
a) Shown is a ball-and-stick and space filling model of P115 and the surrounding heme environment of wild-type Tt H-NOX (15). The invariant P115 (orange) makes the largest contribution to heme (red) distortion in Tt H-NOX. P115 pushes up against pyrrole D, which causes a pronounced kink in the connected propionate group. Out-of-plane distortions of up to 2 Å are observed in Tt H-NOX. b) Heme prosthetic group with pyrrole groups A-D labeled.
The Tt H-NOX crystal structure contains two molecules in the unit cell and one heme (molecule B) is slightly flattened compared to the other (molecule A). The degree of heme distortion is coupled to an N-terminal rotation in the wild-type structure (15). In this work, to directly investigate heme flattening, P115 is mutated to an alanine in Tt H-NOX, a mutation that should allow the heme to become less distorted, and if successful, will allow for the determination of the effects of distortion on both structure and properties. Results obtained show that the heme in P115A is significantly flattened. Additionally, the mutation increases the affinity for oxygen and, in contrast to earlier studies with other hemoproteins (10, 21), decreases the reduction potential of the heme. Finally, a flatter heme in this mutant is correlated to a protein conformational change at the N-terminus where shifts of over 4.9 Å are observed.
Results and Discussion
Design of a planar heme in Tt H-NOX
Guided by visual inspection of the structure of wild-type Tt H-NOX as well as energy minimizations (15) that predict P115 to contribute significantly to heme distortion, the P115A mutant was made. P115A was designed to replace the steric bulk of the cyclic 3-carbon chain with a less bulky methyl group.
Structure determination and analysis of P115A
The crystal structure of the P115A Tt H-NOX domain was solved by molecular replacement and refined to 2.12 Å resolution with a final Rwork value of 20.6% and an Rfree value of 25.3% (Figure 2). A total of four Fe(II)–O2 P115A molecules (Molecules A-D) were built in a monoclinic asymmetric unit cell. Crystallographic data and refinement statistics are summarized in Table 1.
Figure 2. Structural comparison of P115A with wild-type Tt H-NOX.
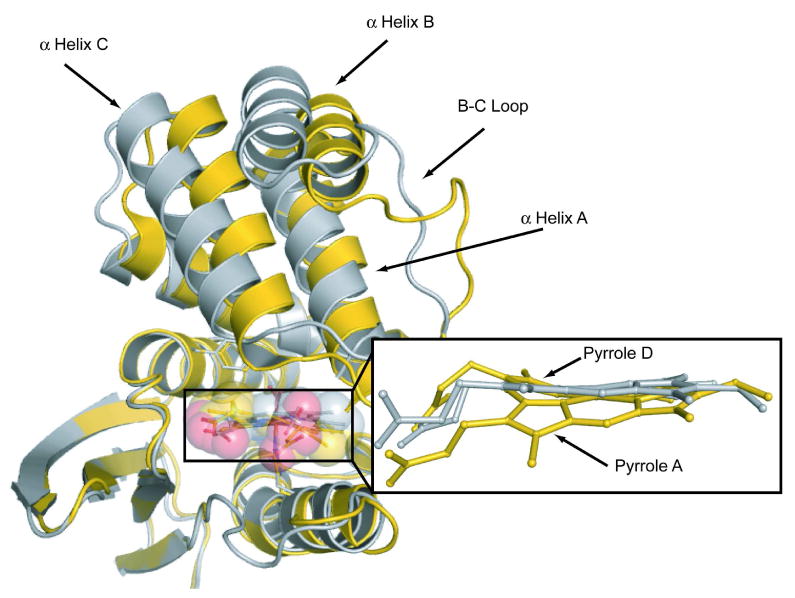
The heme in P115A (silver) is flatter than wild-type (gold). Significant translations are observed in the N-terminal region of the heme-flattened P115A crystal structure. Shown are molecule A from the monoclinic crystal structure of wild-type H-NOX and molecule D from the P115A crystal structure.
TABLE 1. Statistics of crystallographic data collection and refinement statistics.
| Data Collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 116.3, 124.7, 83.6 |
| β (°) | 126.6 |
| Resolutiona (Å) | 50-2.12 (2.20- 2.12) |
| Rmerge (%) | 5.5 (45.5) |
| I / σa | 20.9 (2.7) |
| Completeness (%) | 99.2 (99.0) |
| Redundancy | 4.3 (3.6) |
| Refinement | |
| No. of reflections | 53135 |
| Rwork / Rfreeb (%) | 20.6/25.3 |
| No. atoms | |
| Protein | 3098 |
| Heme | 172 |
| O2 Molecules | 4 |
| Water molecules | 179 |
| Overall B-factors (Å2) | 49 |
| Rms deviation from ideality | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 1.592 |
The values in parentheses relate to highest-resolution shells.
Rfree is calculated for a randomly chosen 5% of reflections.
Root mean square (rms) deviation from planarity, normal-coordinate analysis, and interpyrrole angles were calculated for all hemes in P115A (Tables S1 and S2 in Supplemental). The N-terminal and overall rms deviation from wild-type Tt H-NOX were calculated for P115A (Table S3 in Supplemental). Matrix plots based on the differences in α-carbon distances were calculated with the program DDMP (Center for Structural Biology at Yale University, New Haven, CT and Figures S1a-d in Supplemental). These plots show that the C-terminal region overlays well with wild-type, consequently residues 1–83 were used to calculate N-terminal rms deviation. The iron-histidine tilt was calculated using the least squares plane of the 4 pyrrole nitrogens in the heme (the other atoms in the macrocycle were excluded due to the high degree of distortion) and the five imidazole ring atoms of H102 using MOLEMAN2 (22).
Heme distortion in P115A vs. wild-type
All four molecules in the unit cell have flatter hemes than wild-type Tt H-NOX. Molecules C and D (Figure 3) show the most significant change from wild-type (Tables S1 and S2). However, the heme atoms in molecule C have relatively high B-factor values that range from 49 to 79 Å2. Therefore, further analysis is primarily focused on molecule D which can be analyzed with higher certainty with B-factor values that range from 40 to 61 Å2. Molecule A of the wild-type Tt H-NOX is used for comparison since it has the highest degree of heme distortion.
Figure 3. N-terminal movement from wild-type Tt H-NOX (WT) vs. heme distortion in P115A.
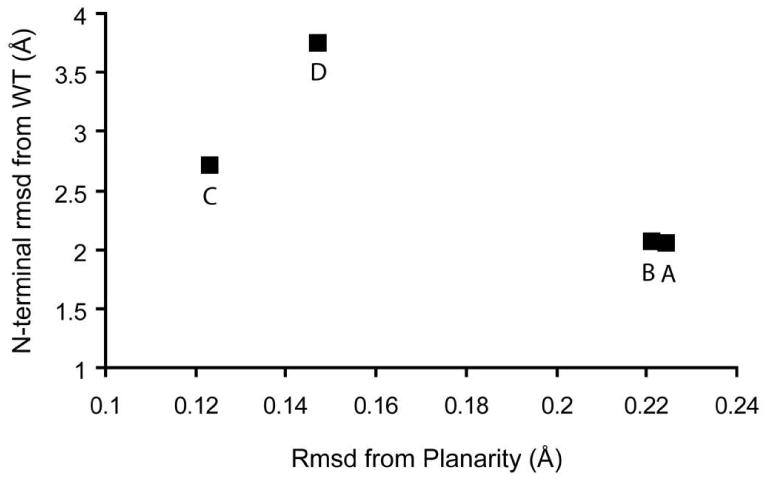
The N-terminal (residues 1–83) rms deviation (Å) was calculated and plotted vs. rms deviation (Å) from planarity for each of the 4 molecules in the asymmetric unit cell (A-D). Wild-type molecule A in the monoclinic space group was used for analysis.
A comparison of P115A molecule D and wild-type Tt H-NOX molecule A is shown in Figure 2. The rms deviation from planarity in P115A has decreased significantly, approximately three-fold compared to wild-type. Specifically, the rms deviation from planarity is 0.147 Å in P115A compared to 0.460 Å in the wild-type Tt H-NOX. The decrease in rms deviation from planarity in P115A from the most distorted heme in wild-type Tt H-NOX is approximately three-fold.
Table S1 shows the major contributions to heme distortion in P115A and wild-type Tt H-NOX as calculated by normal-coordinate structural analysis (7, 23). The major contributors to heme distortion in wild-type Tt H-NOX are saddling and ruffling. In all four P115A molecules (A-D), both the degree of saddling and ruffling decrease with respect to all molecules of wild-type (A and B). For example, saddling and ruffling in P115A molecule D are 0.066 Å and −0.517, respectively, while in wild-type molecule A they are −1.069 Å and −1.105 Å. The angles between the planes of the pyrrole rings within the heme in P115A decrease from that in the wild-type Tt H-NOX (Supplemental Table S2). The pyrrole angles in molecule D range from 0° to 15° whereas in the wild-type Tt H-NOX from 10° to 33°.
Overall, the P115A crystal structure shows that removal of the bulky and conformationally constrained proline, leads to a general flattening of the heme, as demonstrated by the decrease in all inter-pyrrole angles as well as decreased saddling and ruffling displacements, described above. This occurs because the pyrrole D ring and the associated propionate side chain move back into the porphyrin plane, resulting in more space in the heme pocket and allowing the heme to adopt a lower energy conformation. Therefore, the P115A structure illustrates the importance of P115 in maintaining the unusual deviation from planarity of the heme in Tt H-NOX. Because it is invariant across the entire H-NOX family, these results suggest that this residue maintains heme deformation throughout the family.
Heme flattening is associated with a movement of the N-terminus in H-NOX
It was postulated that the degree of heme distortion would be coupled to the conformation of the wild-type structure. Difference distance matrix plots show that the C-terminus in P115A aligns well with wild-type (Figures S1a-d in Supplemental). A plot of heme distortion versus N-terminal movement in P115A (Figure 3) shows a clear trend between heme distortion and N-terminal movement. A significant conformational change is observed in P115A, especially in molecule D, as compared with wild-type Tt H-NOX molecule A (Figure 2 and Table S3). The N-terminal rms deviation between wild-type molecule A and P115A molecule D is 3.76 Å.
Upon heme flattening, the tight network of van der Waals interactions in the heme pocket is lost. In particular, residues I5 and M1, both on α helix A, which are in direct contact with heme pyrrole A and the attached propionate side chain, respectively, undergo significant movement upon heme flattening. Figure 4 shows the P115A heme methyl group connected to pyrrole-A moving into the plane causing I5, which is part of α helix A, to shift away from the heme. Also upon heme flattening, the propionate side chain connected to pyrrole-A pushes M1 away from the heme and the C-terminal region.
Figure 4. The effect of heme flattening on the N-terminal domain.
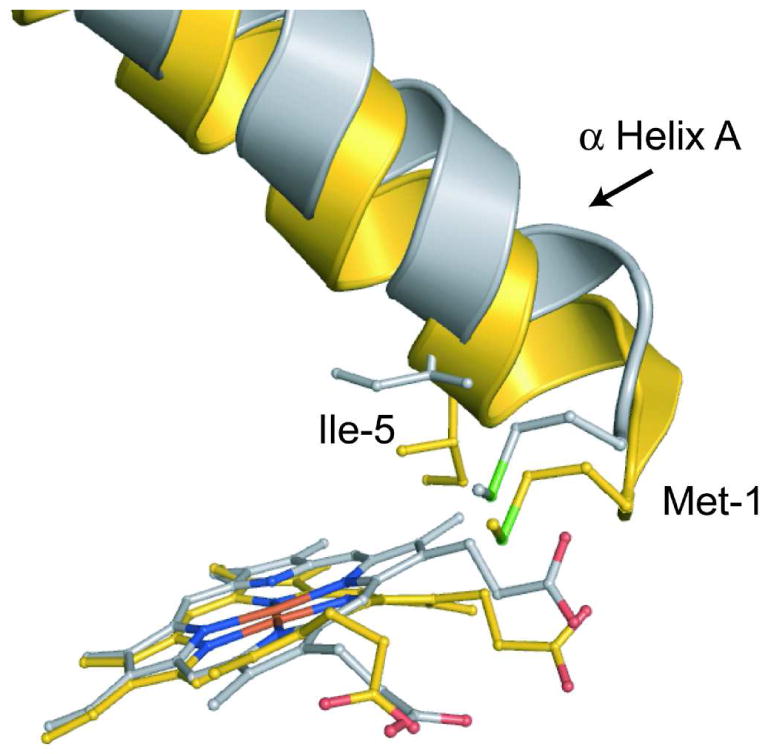
Shown is a comparison of P115A molecule D (silver) and wild-type monoclinic Tt H-NOX molecule A (gold) heme/N-terminal interface. The planar heme makes new contacts with Met1 and Ile5 of α helix A, which shifts the helix away from the C-terminal domain. Shifting of α helix A, along with the rest of the N-terminal region causes shifts over 4.9 Å.
The major conformational shifts observed in P115A molecule D are localized to N-terminal α helices A-D and the loop in between helices B and C (residues 32–45 or loop B-C) (Figure 2). As described in more detail above, α-helix A (residues 1–17) moves in conjunction with pyrrole A shifting back into plane. α-helix C (residues 45–59) and loop B-C move along with α helix A away from the heme and the C-terminal region. The most significant change observed in P115A can be seen in α helix B (residues 19–29), which makes direct contact with helices A and C (Figure 2). Shifts over 4.9 Å in α helix B are observed.
Not surprisingly, the conformation of the protein is intimately tied to the heme conformation. The large deviation in heme conformation from wild-type is caused by the loss of the tight network of van der Waals interactions in the heme pocket. Loss of local contacts with the heme due to heme flattening, especially at the distal pocket, results in N-terminal shifts that significantly alter the conformation of the protein. Clearly, the heme and the local protein environment are working in concert to maintain a particular conformational state in Tt H-NOX.
The molecular surface changes that occur upon heme flattening is a likely mechanism involved in signal transduction in this H-NOX domain. The Tt H-NOX domain is a member of the Tar4 family of receptors and is fused to a predicted methyl-accepting chemotaxis protein (MCP) with two predicted membrane spanning regions between these two domains. A model based on these domains places the H-NOX domain on the same side of the membrane as the MCP, suggesting that intermolecular contacts between the sensor H-NOX and the MCP are involved in the signal transduction mechanism. Thus, ligand-induced H-NOX conformational changes may control methylation of the MCP. Whether heme flattening contributes to the control of methylation has yet to be evaluated.
Higher affinity for O2 in P115A is caused by proximal effects at the heme
It is expected that changes in heme structure will have influence on ligand binding affinity and other heme chemical and physical properties. To evaluate the effects of heme flattening on ligand binding of Tt H-NOX, O2 binding kinetics were measured and compared to the wild-type H-NOX. The association and dissociation rates were measured as previously described for the wild-type Tt H-NOX (24) (Table S4). The dissociation rate of P115A was 0.22 ± 0.01 s-1, compared to 1.22 ± 0.09 s-1 for the wild-type Tt H-NOX. No significant change was observed in the P115A association rate (10.4 ± 1.1 and 13.6 ± 1.0 for P115A and wild-type, respectively), resulting in a protein with a higher affinity for O2 (KD is 21.2 ± 2.1 nM and 89.7 ± 6.2 nM for P115A and wild-type, respectively). Factors such as the distal pocket H-bonding network in Tt H-NOX (24) and the strength of the Fe-His bonds are known to contribute to higher affinity for oxygen in hemoproteins (25, 26). Surprisingly, all Fe-His bond lengths and as well as distal hydrogen bonds to O2 in P115A are within error to the wild-type structure (data not shown) so some other factor must play a role in O2 affinity.
The Fe-histidine tilt, which has also been shown to affect ligand affinity in hemoproteins (27), decreased in P115A compared to wild-type (Supplemental Table S1 and Figure 5). Wild-type Tt H-NOX molecule A has a tilt of 78° whereas the tilt in P115A molecule D is nearly perpendicular with a tilt of 87° (Figure 5). It is likely that flattening the heme in P115A allows for optimized bond overlap between the proximal histidine and iron, thus, creating a stronger proximal bond that stabilizes the Fe(II)–O2 complex and may explain the slower off-rate of in P115A.
Figure 5. Comparison of the iron-histidine bond geometry.
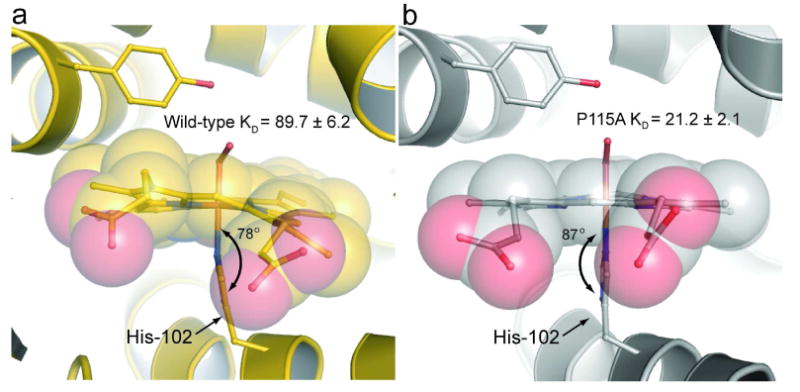
Shown are the iron-histidine tilts of a) wild-type Tt H-NOX (gold) and b) P115A (silver). The least squares plane of the heme was calculated using the 4 pyrrole nitrogens of the heme (other atoms were excluded due to the high degree of distortion) and the five imidazole ring atoms of H102 using MOLEMAN2 (22). Wild-type has a tilt of 78° whereas P115A has a tilt of 87°.
We have speculated that Tt H-NOX serves as an oxygen sensor for the obligate anaerobe T. tengcongensis based on the fact that O2 binding to this H-NOX is very tight (KD= 90 nM). The fold of Tt H-NOX may tune the affinity for oxygen by distorting the heme so that the KD is set for the appropriate physiological response. The situation may not be this simple, however. Taylor and colleagues showed that Desulfovibrio vulgaris Hildenborough, a sulfate-reducing bacterium thought to be an obligate anaerobe, actually preferred an oxygen concentration of 0.02 to 0.04% (0.24 to 0.48 mM). This O2 concentration was also shown to support growth (28). Hence, sensors for O2 are likely tuned to respond to changing environmental conditions and complex physiological responses.
Reduction potential decreases in P115A
To determine whether chemical properties of the heme are sensitive to heme conformation the reduction potential was measured for P115A and Tt H-NOX. P115A has a significantly lower reduction potential than wild-type. The reduction potentials of P115A and wild-type Tt H-NOX are −3.8 ± 10.2 and 167.0 ± 6.7 mV versus the standard hydrogen electrode (SHE) (Figure 6), respectively.
Figure 6. Reduction potential of wild-type Tt H-NOX and P115A.
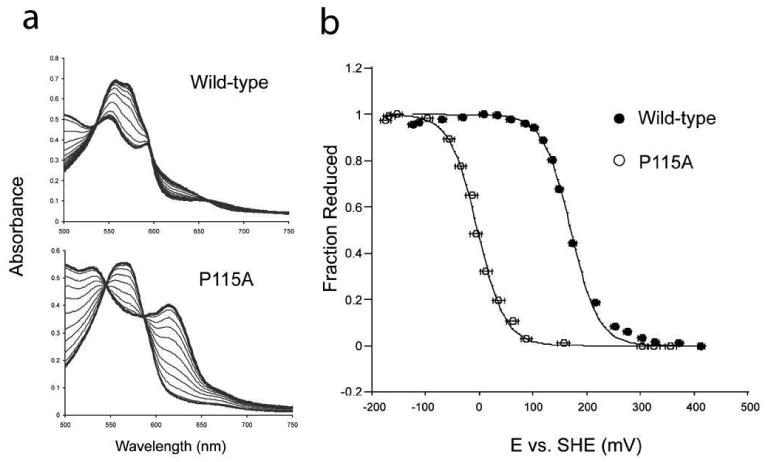
a) Shown are the titration spectra for wild-type Tt H-NOX and P115A. b) Titration curves for wild-type (•) and P115A (o). The reduction potentials of P115A and wild-type Tt H-NOX were determined against the standard hydrogen electrode (SHE). The ratio of reduced Fe2+ to oxidized Fe3+ heme was measured based on their α/β maximum at approximately 557 (reduced) nm for wild-type. The difference absorbance of the α/β maximum for reduced and the α/β minimum for oxidized was used to calculate the fraction reduced for P115A. The voltage against the SHE was measured for both oxidative and reductive titrations of wild-type and P115A. Error bars represent the standard error.
Previous studies have suggested that factors such as electrostatic interactions near the heme pocket control hemoprotein reduction potentials (29-32). In particular, it has been demonstrated that the reduction potential increases as a function of decreasing dielectric constant (33, 34), however, we cannot definitively comment on a change in the dielectric environment in P115A vs. the wild-type protein. While the P115A mutant was designed to specifically address the role heme distortion, the mutation could introduce other changes such as an alteration in the dielectric of the heme environment.
Implications for the role of heme distortion in H-NOX proteins
Our results show that (i) P115 is important for maintaining heme distortion in Tt H-NOX, (ii) that heme distortion maintains a particular molecular oxygen KD and a particular heme iron reduction potential, presumably at their physiologically relevant values, and (iii) that heme distortion is correlated with movement in the N-terminal region of the Tt H-NOX. Thus, the heme and the surrounding environment work in conjunction to maintain conformation and function in Tt H-NOX.
As noted by Shelnutt and colleagues, porphyrin deformations are often conserved within functional classes of proteins, strongly supporting the idea that these deformations are important for function (1). P115 is likely to be important for the function of the H-NOX family since it is conserved in the entire class of proteins. Heme distortion in Tt H-NOX is essential for maintaining its structural conformation and biochemical properties. The data show that heme distortion contributes to maintain a specific KD for oxygen and reduction potential of the heme iron. Additionally, heme distortion is related to the conformation of Tt H-NOX. The heme and the surrounding environment work in conjunction to maintain a particular conformation and fold.
Conclusions
The influence of heme distortion on function has been a long-standing question and certainly related to the diverse chemistry exhibited by this large class of proteins. While many structural and biochemical studies over the last several decades have made note of heme distortion, a systematic approach has not appeared. The challenge is to trap the heme in different conformations under the similar conditions (pH, salinity, temperature, etc.) and ligation state. The H-NOX family, for the reasons outlined above, provides the opportunity to carry out such an investigation, and the first step in this regard is reported here. As discussed, heme distortion in the nitrophorins is significant (9, 11). Walker and colleagues attempted to trap a flattened heme in nitrophorin 2 (10). They speculated that distal pocket mutations in the protein would flatten the heme and speculated further about a change in redox potential. However, very little change in heme conformation was observed between the hemes of both the wild-type protein and the mutant crystal structures (PDB codes: 2A3F and 2ALL). The H-NOX results presented in this paper show a heme trapped in different conformational states generated by a single point mutation. The differences in ligand binding and redox potential correlate with the changes in heme distortion; however, our results do not agree with a previous report that shows opposite trend in heme ruffling vs. redox potential in cytochromes c3 (21). Clearly, the redox potential and ligand binding will depend on multiple factors including the heme environment, ligands to the iron and covalent vs. non-covalent heme as well as the degree of heme distortion. The large movement of the N-terminal region of the protein was unexpected and provides a clear example of protein conformation linked to the heme cofactor. The movement was presaged by what we previously observed in molecule A and B in our first structure (15), however, it was uncertain whether the heme conformation and N-terminal shift was an outcome of the crystallization. Our results here unambiguously show the importance of heme conformation in protein structure and chemistry.
Methods
Expression of P115A mutant of Tt H-NOX in E. coli
Mutagenesis was carried out using the QuikChange® protocol from Stratagene. Cell culture procedures and purification of P115A for kinetic were carried out as previously described (18). Cell culture and expression procedures for P115A crystallization and redox potentiometry were carried out as described above except the growth media was Terrific Broth.
Purification of P115A for crystallization
Cell lysis and thermal treatment were carried out as described (18). The supernatant after thermal treatment was concentrated to 10 mL using a Vivaspin concentrator (Sartorius, 10 kDa) and loaded onto a Superdex 200 HiLoad 26/60 gel filtration column (Pharmacia) that was equilibrated with buffer A (50 mM TEA, pH 7.5, 50 mM NaCl, and 5% glycerol) at a flow rate of 0.5 mL/min. Fractions containing P115A were pooled on the basis of the intensity of the red/brown color and applied to a POROS HQ 7.9 mL (1 × 10 cm, 10 μm) anion-exchange column (Applied Biosystems) that had been equilibrated with buffer A. The flow rate was 10 mL/min and the flow-through was collected. Aliquots (2 mL) of P115A were then loaded onto a Superdex 75 HiLoad 26/60 gel filtration column (Pharmacia) that was equilibrated with buffer A at a flow rate of 0.5 mL/min. P115A was isolated as the Fe(II)–O2 complex (18), and stored at −80 °C.
Crystallization of Tt P115A
Samples of P115A were equilibrated with 20 mM TEA (pH 7.5) and concentrated to 30 mg/mL. Crystals were grown by hanging drop vapor diffusion by mixing 1 μL of the protein solution with 1 μL of the reservoir solution and equilibrating against a 750 μL reservoir of 0.1 M NaSCN, 0.1 M Tris (pH 9.1), 0.2 M (NH4)2(SO4), and 18% (w/v) PEG 8000 at 16 °C. Crystals began to appear within 24 hrs. Cryoprotection was achieved by transferring the crystals stepwise into mother liquor solutions containing 10%, 15% glycerol, and ending with 20% glycerol and 5% xylitol. Crystals of P115A were obtained in the C2 space group, flash frozen in liquid propane, and stored in liquid nitrogen.
X-ray data collection, phasing and refinement
X-ray data were collected by using synchrotron radiation at beamline 8.2.1 at the Advanced Light Source, Lawrence Berkeley National Laboratory. Diffraction images were collected at 100K with 10 s exposure time and 1° oscillations per frame. Data were processed with the HKL2000 suite (35). Molecular replacement was carried out with Phaser (36) using wild-type H-NOX (PDB ID 1U55) as a search model. Model building was carried out by using the programs O (37) and Coot (38). Refinement was carried out by using CNS (39) and Phenix (40) with TLS refinement parameters incorporated. The final model includes 4 P115A molecules in the asymmetric unit. The structure of P115A was refined to a final Rwork of 20.6% (Rfree = 25.3%) at 2.12Å.
Kinetics
The on-rate of O2 binding to heme and dissociation of O2 from heme were measured as previously described (24).
Redox potentiometry
Potentiometric titrations were performed as described previously (41). The change in the Fe oxidation state in Tt H-NOX was monitored by the absorbance change at α/β region maximum (∼557 nm). For P115A the oxidation state change was measured by the difference of absorbance of the α/β maximum of the oxidized and the α/β minimum of the reduced oxidation state spectra.
Supplementary Material
This material is available free of charge via the internet.
Acknowledgments
This work was supported by the National Institutes of Health grant GM070671 and a Eugene Cota-Robles Fellowship to C.O. We are grateful to Debora Makino, Meindert Lamers, Xeuwu Zhang, Nick Levinson, and members of the Kuriyan lab for assistance and advice during structure refinement. We thank Wendy Belliston-Bittner, Jay Winkler, and Harry Gray at the Beckman Institute Laser Resource Center at the California Institute of Technology for their essential help in measuring oxygen association rates. We thank Katelyn Connell and Matthew Volgraf for initial studies with the P115A mutant. We thank members of the M.A.M. and J.K. laboratories for helpful discussions and review of the manuscript.
Footnotes
Data Deposition: The atomic coordinates have been deposited in the Protein Data Bank, ww.pdb.org (PDB ID 3EEE; RCSB ID code rcsb049218).
References
- 1.Shelnutt JA, Song XZ, Ma JG, Jia SL, Jentzen W, Medforth CJ. Nonplanar porphyrins and their significance in proteins. Chem Soc Rev. 1998;27:31–41. [Google Scholar]
- 2.Zbylut SD, Kincaid JR. Resonance Raman evidence for protein-induced out-of-plane distortion of the heme prosthetic group of mammalian lactoperoxidase. J Amer Chem Soc. 2002;124:6751–6758. doi: 10.1021/ja012578u. [DOI] [PubMed] [Google Scholar]
- 3.Li D, Stuehr DJ, Yeh SR, Rousseau DL. Heme distortion modulated by ligand-protein interactions in inducible nitric oxide synthase. J Biol Chem. 2004;279:26489–26499. doi: 10.1074/jbc.M400968200. [DOI] [PubMed] [Google Scholar]
- 4.Ravikanth M, Chandrashekar TK. Nonplanar porphyrins and their biological relevance - ground and excited-state dynamics. Coordination Chemistry. 1995;82:105–188. [Google Scholar]
- 5.Weichsel A, Andersen JF, Roberts SA, Montfort WR. Nitric oxide binding to nitrophorin 4 induces complete distal pocket burial. Nature Structural Biology. 2000;7:551–554. doi: 10.1038/76769. [DOI] [PubMed] [Google Scholar]
- 6.Anderson KK, Hobbs JD, Luo LA, Stanley KD, Quirke JME, Shelnutt JA. Planar Nonplanar Conformational Equilibrium in Metal Derivatives of Octaethylporphyrin and Meso-Nitrooctaethylporphyrin. J Amer Chem Soc. 1993;115:12346–12352. [Google Scholar]
- 7.Jentzen W, Ma JG, Shelnutt JA. Conservation of the conformation of the porphyrin macrocycle in hemoproteins. Biophys J. 1998;74:753–763. doi: 10.1016/S0006-3495(98)74000-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barkigia KM, Chantranupong L, Smith KM, Fajer J. Structural and theoretical-models of photosynthetic chromophores - implications for redox, light-absorption properties and vectorial electron flow. J Amer Chem Soc. 1988;110:7566–7567. [Google Scholar]
- 9.Maes EM, Roberts SA, Weichsel A, Montfort WR. Ultrahigh resolution structures of nitrophorin 4: heme distortion in ferrous CO and NO complexes. Biochem. 2005;44:12690–12699. doi: 10.1021/bi0506573. [DOI] [PubMed] [Google Scholar]
- 10.Shokhireva T, Berry RE, Uno E, Balfour CA, Zhang H, Walker FA. Electrochemical and NMR spectroscopic studies of distal pocket mutants of nitrophorin 2: stability, structure, and dynamics of axial ligand complexes. Proc Natl Acad Sci U S A. 2003;100:3778–3783. doi: 10.1073/pnas.0536641100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts SA, Weichsel A, Qiu Y, Shelnutt JA, Walker FA, Montfort WR. Ligand-induced heme ruffling and bent no geometry in ultra-high-resolution structures of nitrophorin 4. Biochem. 2001;40:11327–11337. doi: 10.1021/bi0109257. [DOI] [PubMed] [Google Scholar]
- 12.Ma JG, Laberge M, Song XZ, Jentzen W, Jia SL, Zhang J, Vanderkooi JM, Shelnutt JA. Protein-induced changes in nonplanarity of the porphyrin in nickel cytochrome c probed by resonance Raman spectroscopy. Biochem. 1998;37:5118–5128. doi: 10.1021/bi972375b. [DOI] [PubMed] [Google Scholar]
- 13.Ryeng H, Ghosh A. Do nonplanar distortions of porphyrins bring about strongly red-shifted electronic spectra? Controversy, consensus, new developments, and relevance to chelatases. J Amer Chem Soc. 2002;124:8099–8103. doi: 10.1021/ja0202659. [DOI] [PubMed] [Google Scholar]
- 14.Gong W, Hao B, Mansy SS, Gonzalez G, Gilles-Gonzalez MA, Chan MK. Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc Natl Acad Sci U S A. 1998;95:15177–15182. doi: 10.1073/pnas.95.26.15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pellicena P, Karow DS, Boon EM, Marletta MA, Kuriyan J. Crystal structure of an oxygen-binding heme domain related to soluble guanylate cyclases. Proc Natl Acad Sci U S A. 2004;101:12854–12859. doi: 10.1073/pnas.0405188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denninger JW, Marletta MA. Guanylate cyclase and the •NO/cGMP signaling pathway. Biochim Biophys Acta. 1999;1411:334–350. doi: 10.1016/s0005-2728(99)00024-9. [DOI] [PubMed] [Google Scholar]
- 17.Iyer LM, Anantharaman V, Aravind L. Ancient conserved domains shared by animal soluble guanylyl cyclases and bacterial signaling proteins. BMC Genomics. 2003;4:5. doi: 10.1186/1471-2164-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karow DS, Pan D, Tran R, Pellicena P, Presley A, Mathies RA, Marletta MA. Spectroscopic characterization of the soluble guanylate cyclase-like heme domains from Vibrio cholerae and Thermoanaerobacter tengcongensis. Biochem. 2004;43:10203–10211. doi: 10.1021/bi049374l. [DOI] [PubMed] [Google Scholar]
- 19.Price MS, Chao LY, Marletta MA. Shewanella oneidensis MR-1 H-NOX regulation of a histidine kinase by nitric oxide. Biochem. 2007;46:13677–13683. doi: 10.1021/bi7019035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma X, Sayed N, Beuve A, van den Akker F. NO and CO differentially activate soluble guanylyl cyclase via a heme pivot-bend mechanism. EMBO J. 2007;26:578–588. doi: 10.1038/sj.emboj.7601521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma JG, Zhang J, Franco R, Jia SL, Moura I, Moura JJ, Kroneck PM, Shelnutt JA. The structural origin of nonplanar heme distortions in tetraheme ferricytochromes c3. Biochem. 1998;37:12431–12442. doi: 10.1021/bi981189i. [DOI] [PubMed] [Google Scholar]
- 22.Kleywegt GJ, Jones TA. Software for handling macromolecular envelopes. Acta Crystallogr D Biol Crystallogr. 1999;55:941–944. doi: 10.1107/s0907444999001031. [DOI] [PubMed] [Google Scholar]
- 23.Shelnutt JA. Normal-coordinate structural decomposition and the vibronic spectra of porphyrins. J Prophyr Phthal. 2001;5:300–311. [Google Scholar]
- 24.Boon EM, Huang SH, Marletta MA. A molecular basis for NO selectivity in soluble guanylate cyclase. Nature Chem Biol. 2005;1:53–59. doi: 10.1038/nchembio704. [DOI] [PubMed] [Google Scholar]
- 25.Giangiacomo L, Ilari A, Boffi A, Morea V, Chiancone E. The truncated oxygen-avid hemoglobin from Bacillus subtilis: X-ray structure and ligand binding properties. J Biol Chem. 2005;280:9192–9202. doi: 10.1074/jbc.M407267200. [DOI] [PubMed] [Google Scholar]
- 26.Capece L, Marti MA, Crespo A, Doctorovich F, Estrin DA. Heme protein oxygen affinity regulation exerted by proximal effects. J Amer Chem Soc. 2006;128:12455–12461. doi: 10.1021/ja0620033. [DOI] [PubMed] [Google Scholar]
- 27.Friedman JM, Rousseau DL, Ondrias MR, Stepnoski RA. Transient Raman study of hemoglobin: structural dependence of the iron-histidine linkage. Science. 1982;218:1244–1246. doi: 10.1126/science.7146910. [DOI] [PubMed] [Google Scholar]
- 28.Johnson MS, Zhulin IB, Gapuzan ME, Taylor BL. Oxygen-dependent growth of the obligate anaerobe Desulfovibrio vulgaris Hildenborough. J Bacteriol. 1997;179:5598–5601. doi: 10.1128/jb.179.17.5598-5601.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mauk AG, Moore GR. Control of metalloprotein redox potentials: What does site-directed mutagenesis of hemoproteins tell us? J Biol Inorg Chem. 1997;2:119–125. [Google Scholar]
- 30.Mao JJ, Hauser K, Gunner MR. How cytochromes with different folds control heme redox potentials. Biochem. 2003;42:9829–9840. doi: 10.1021/bi027288k. [DOI] [PubMed] [Google Scholar]
- 31.Kennedy ML, Silchenko S, Houndonougbo N, Gibney BR, Dutton PL, Rodgers KR, Benson DR. Model hemoprotein reduction potentials: The effects of histidine-to-iron coordination equilibrium. J Amer Chem Soc. 2001;123:4635–4636. doi: 10.1021/ja0156441. [DOI] [PubMed] [Google Scholar]
- 32.Kassner RJ. Effects of nonpolar environments on the redox potentials of heme complexes. Proc Natl Acad Sci U S A. 1972;69:2263–2267. doi: 10.1073/pnas.69.8.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tezcan FA, Winkler JR, Gray HB. Effects of ligation and folding on reduction potentials of heme proteins. J Amer Chem Soc. 1998;120:13383–13388. [Google Scholar]
- 34.Cowley AB, Kennedy ML, Silchenko S, Lukat-Rodgers GS, Rodgers KR, Benson DR. Insight into heme protein redox potential control and functional aspects of six-coordinate ligand-sensing heme proteins from studies of synthetic heme peptides. Inorg Chem. 2006;45:9985–10001. doi: 10.1021/ic052205k. [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 37.Kleywegt GJ, Jones TA. Efficient rebuilding of protein structures. Acta Crystallogr D Biol Crystallogr. 1996;52:829–832. doi: 10.1107/S0907444996001783. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 40.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 41.Dutton PL. Redox potentiometry: determination of midpoint potentials of oxidation-reduction components of biological electron-transfer systems. Methods Enzymol. 1978;54:411–435. doi: 10.1016/s0076-6879(78)54026-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This material is available free of charge via the internet.


