Abstract
Genetic variation in myostatin, a negative regulator of skeletal muscle, in cattle has shown remarkable influence on skeletal muscle, resulting in a double-muscled phenotype in certain breeds; however, DNA sequence variation within this gene in humans has not been consistently associated with skeletal muscle mass or strength. Follistatin and activin-type II receptor B (ACVR2B) are two myostatin-related genes involved in the regulation and signaling of myostatin. We sought to identify associations between genetic variation and haplotype structure in both follistatin and ACVR2B with skeletal muscle-related phenotypes. Three hundred fifteen men and 278 women aged 19–90 yr from the Baltimore Longitudinal Study of Aging were genotyped to determine respective haplotype groupings (Hap Groups) based on HapMap data. Whole body soft tissue composition was measured by dual-energy X-ray absorptiometry. Quadriceps peak torque (strength) was measured using an isokinetic dynamometer. Women carriers of ACVR2B Hap Group 1 exhibited significantly less quadriceps muscle strength (shortening phase) than women homozygous for Hap Group 2 (109.2 ± 1.9 vs. 118.6 ± 4.1 N·m, 30°/s, respectively, P = 0.036). No significant association was observed in men. Male carriers of follistatin Hap Group 3 exhibited significantly less total leg fat-free mass than noncarriers (16.6 ± 0.3 vs. 17.5 ± 0.2 kg, respectively, P = 0.012). No significant associations between these haplotype groups were observed in women. These results indicate that haplotype structure at the ACVR2B and follistatin loci may contribute to interindividual variation in skeletal muscle mass and strength, although these data indicate sex-specific relationships.
Keywords: genetics, skeletal muscle, sex
Differences in muscle mass and strength among individuals are due to both environmental and genetic factors. Muscle mass and strength are highly heritable phenotypes with genetic factors contributing significantly to the variation in lean body mass and muscle strength. For example, Huygens et al. (13) reported that heritable factors accounted for up to 90% of the interindividual variation in muscle mass and ∼60% in strength, while Seeman et al. (27) estimated the genetic component of lean body mass explained 80% of the total variance. Despite the strong evidence for the importance of genetic factors in regulating muscle phenotypes, the identification of specific genes and allelic variants contributing to these phenotypes is in its infancy, and few specific genes have been identified as contributing factors (30).
Despite its relatively short history, myostatin has quickly become a prime candidate to study as a mechanism of muscle development and as a potential therapy for muscle-related disorders. First reported in 1997 by McPherron et al. (23), myostatin (growth and differentiation factor-8) was identified in mice as a transforming growth factor-β (TGF-β) family member that acts as a negative regulator of skeletal muscle growth. Soon after the initial report of myostatin's discovery, several groups identified mutations in the myostatin gene in naturally bred “double-muscled” cattle breeds (10, 24), providing additional evidence for a critical role for myostatin in muscle development.
Following up their initial discovery, Lee and McPherron (17) established putative myostatin receptors (activin-type II receptors A and B; ACVR2A and ACVR2B) and negative regulators (the myostatin propeptide and follistatin) and formalized a basic model of myostatin regulation. With the identification of ACVR2B as the primary myostatin receptor, and the myostatin propeptide (32, 39) and follistatin as negative regulators of myostatin activity, Lee and McPherron (17) put forth a model in which the myostatin COOH-terminal dimer remains in a latent complex with the inhibitory propeptide. This latent complex can be further negatively regulated by binding with follistatin, and on release of the negative regulators, myostatin is free to signal through its receptors, primarily ACVR2B. Lee and McPherron (17) demonstrated that myostatin binding to ACVR2B receptors was specific and saturable. They also demonstrated increased muscle expression of a dominant negative form of ACVR2B increased muscle weights up to 125% more than those of control animals.
Several studies have shown that follistatin can function as a potent myostatin antagonist and plays an important role in vivo. First, follistatin is capable of blocking myostatin activity in both receptor binding and reporter gene assays (17, 40). Second, genetic studies in mice have shown that overexpression of follistatin in muscle can cause dramatic increases in muscle growth. Lee and McPherron (17) generated transgenic mice in which the myosin light chain promoter/enhancer was used to drive the expression of follistatin, and muscle weights were increased by 194–327% relative to control animals. Thus follistatin appears to be a potent myostatin antagonist (17).
Despite the remarkable influence of myostatin on skeletal muscle and the well-established effect of myostatin gene mutations in double-muscled cattle breeds, studies of myostatin genetic variation in humans have shown little association with muscle phenotypes (4, 6, 14, 28). Scheulke and colleagues (26) recently reported a novel loss-of-function mutation in the myostatin gene of a young child who, when born, appeared extraordinarily muscular, with protruding muscles in his thighs and upper arms; however, that mutation is considered rare. Recent linkage analysis work has focused instead on genetic variation in myostatin pathway genes (11, 12), specifically genes involved in the signaling cascade downstream of the myostatin receptor; genes upstream of this signaling cascade have yet to be examined. Therefore, the purpose of this study was to examine the association of genetic variation in the follistatin gene (gene symbol: FST) and the primary myostatin receptor gene, ACVR2B, with skeletal muscle mass and strength in men and women across the adult age span.
Methods
Subjects
The subjects included in this study were white and came from the Baltimore Longitudinal Study of Aging (BLSA), an ongoing National Institute on Aging-funded investigation of normal aging. Details of the BLSA recruitment methodology are described elsewhere (29). All BLSA subjects received a complete medical history and physical examination. Those with clinical cardiovascular or musculoskeletal disorders that could be adversely affected by exercise testing were excluded. Also, subjects were excluded if they had active neck and back pain, frequent and severe joint pain, any surgery in the past 6 mo, or an abnormal bone scan. Before the study, all subjects received a complete explanation of the purpose and procedures of the investigation and gave their written informed consent. The experimental protocols were approved by the Institutional Review Boards for Medstar Research Institute, Johns Hopkins Bayview Medical Center, and the University of Maryland.
Body composition assessment
Data collected for body composition variables were obtained using methods previously approved by the BLSA (18). Body mass and height were measured to the nearest 0.1 kg and 0.5 cm, respectively, using a Detecto medical beam scale. Total body fat and soft tissue fat-free mass (FFM) and total leg fat and FFM (both legs combined) were assessed by dual-energy X-ray absorptiometry (DEXA) (model DPX-L, Lunar Radiation, Madison, WI) using previously described methods (18). Soft tissue FFM was used as a valid indicator of muscle mass based on previous work (8, 38). Reliability was assessed by performing two total body scans, 6 wk apart, on 12 older men (>65 yr). The difference between the two scans was ∼0.01% for fat and FFM. The scanner was calibrated daily before testing.
Peak torque assessment
Peak torque (PT) (strength) was measured using the Kinetic Communicator isokinetic dynamometer (Kin-Com model 125E, Chattanooga Group, Chattanooga, TN). Shortening-phase PT was measured at angular velocities of 0.52 rad/s (30°/s) and 3.14 rad/s (180°/s) for the dominant knee extensors. The terms “shortening” and “lengthening” are substituted throughout the present study for the more commonly used terms “concentric” and “eccentric,” respectively, on the basis of the recommendations of Faulkner (5). For each test, subjects performed three maximal efforts, separated by 30-s rest intervals, from which the highest value of the three trials was accepted as the PT. PT was assessed by using the Kin-Com computer software (version 3.2). Detailed procedures regarding subject positioning and stabilization, gravity correction, Kin-Com calibration, and test-retest reliability are described elsewhere (18, 21).
Haplotype determination
A graphical genome browser maintained by the International HapMap Project website (http://www.hapmap.org) was used to identify the genomic regions surrounding the candidate genes of interest and retrieve HapMap genotype data for all genotyped markers in the selected regions. A 20-kb region, including the ∼5,500 bases containing the follistatin gene, were downloaded, and a 40-kb region, including the ∼29,000 bases containing the ACVR2B gene, were downloaded from HapMap Public Release no. 19. Genotype data were downloaded from the samples of northern and western European ancestry [Centre d'Etude du Polymorphisme Humain (CEPH) cohort] for each region and imported into Haploview (2). Haploview was used to calculate pairwise measures of linkage disequilibrium (LD) and identify haplotype blocks among the polymorphisms in each region (7). LD plots were constructed using all polymorphisms with a minor allele frequency greater than 5%, and haplotypes were constructed using the method of Gabriel et al. (7) using pairwise r2 values ≥0.8 for establishing block structures. Block structures, haplotypes, and specific single-nucleotide polymorphisms (SNPs) for each gene are described in results, as are the tag polymorphisms used to categorize the major haplotypes for each gene.
Although haplotypes may be more informative than single gene variants, the potentially large number of haplotypes that are present in large haplotype blocks reduces the power of haplotype analysis. The follistatin gene falls within a very large and complex haplotype block. Thus we minimized the haplotype data to include only the four polymorphisms within the follistatin gene, which reduced the number of haplotypes to five. The rationale for inclusion of these four SNPs is that all four fall within the follistatin gene; therefore any true causal variants within the follistatin gene would be linked to one of the five haplotypes analyzed.
Genotyping
DNA was isolated from EDTA-anticoagulated whole blood using standard methods. Genotyping of SNPs was performed using the 5′ nuclease allelic discrimination or TaqMan assay (20) for high-throughput genotyping. Each 12.5 μl PCR contained 1.5 μl (10–20 ng) of genomic DNA, 0.625 μl of 20× diluted SNP mix (SNP rs numbers are shown in results), 4.125 μl DNase-free H2O), and 6.25 5 μl of 2× TaqMan Universal PCR master mix (Perkin-Elmer, Applied Biosystems Division), which is a solution containing buffer, uracil-N-glycosylase, deoxyribonucleotides, uridine, passive reference dye (ROX), and TaqGold DNA polymerase. The PCR cycling protocol consisted of the following: 50° for 2 min, 95° for 10 min, 70 cycles of 92° for 15 s and 60° for 1 min. Fluorescence in each well was measured using an ABI 7300 Real Time PCR System machine (Perkin Elmer, Applied Biosystems Division). Analysis of raw data to determine genotypes was performed by the ABI 7300 Sequence Detection System software. Control samples with sequence-verified genotypes were used for each assay.
Statistical analysis
ANOVA models were used to test for differences in physical characteristics among ACVR2B and follistatin haplotype groups. Physical characteristic data are presented as means ± SE. Analysis of covariance was used to compare means among haplotype groups (Hap Groups) for all outcome variables. Significant covariates, as determined for each analysis, were included in all models and are listed in the legends to Tables 1–6. Analyses were performed within each sex group. Data presented are least squares means ± SE, except where specified. We did not analyze gene × gene interactions or the combined effect of these haplotype groups after determining that statistical power was severely limited for such an analysis. Statistical significance was defined as P < 0.05. For all nonsignificant results, the omnibus P value is shown in Tables 1–6; for all significant results, the specific contrast P values are reported.
Table 1. Subject characteristics by ACVR2B haplotype for men and women.
| Homozygous Hap Group 1 | Heterozygous Hap Groups 1 and 2 | Homozygous Hap Group 2 | P Value | |
|---|---|---|---|---|
| Men | ||||
| n | 78 (25.1%) | 181 (58.4%) | 51 (16.5%) | |
| Age, yr | 64.6±1.9 | 62.1±1.2 | 59.4±2.4 | 0.23 |
| Height, cm | 176.6±0.8 | 176.2±0.6 | 176.1±1.0 | 0.92 |
| Weight, kg | 86.2±1.5 | 83.6±1.0 | 83.9±1.8 | 0.34 |
| Total body FFM, kg* | 57.8±0.5 | 56.5±0.3 | 57.8±0.7 | 0.09 |
| Total leg FFM, kg*† | 17.7±0.3 | 16.9±0.2 | 17.5±0.4 | 0.15 |
| Women | ||||
| n | 70 (23.7%) | 172 (58.1%) | 54 (18.2%) | |
| Age, yr | 59.4±1.9 | 55.0±1.2 | 59.4±2.1 | 0.06 |
| Height, cm | 163.3±0.8 | 163.3±0.5 | 163.4±0.9 | 0.99 |
| Weight, kg | 68.4±1.5 | 67.5±0.9 | 65.7±1.7 | 0.48 |
| Total body FFM, kg* | 40.1±0.4 | 39.4±0.2 | 39.1±0.5 | 0.19 |
| Total leg FFM, kg*† | 11.3±0.2 | 11.3±0.1 | 11.2±0.3 | 0.89 |
Data are means ± SE. ACVR2B, activin-type II receptor B gene. Hap Group 1 and Hap Group 2 are haplotype groups.
Data are least squares means ± SE.
Sample size for total leg fat-free mass (FFM) data are 76, 157, and 42 for the men, and 70, 172, and 54 for the women, respectively.
Table 6. Total body and leg FFM in carriers and noncarriers of Hap Group 3 in men and women.
| Carriers of Hap Group 3 | Noncarriers of Hap Group 3 | P Value | |
|---|---|---|---|
| Men | |||
| Total body FFM, kg | 56.3±0.5 (111) | 57.4±0.3 (204) | 0.055 |
| Total leg FFM, kg | 16.6±0.3 (93) | 17.5±0.2 (187) | 0.012 |
| Women | |||
| Total body FFM, kg | 39.7±0.3 (107) | 39.3±0.3 (171) | 0.46 |
| Total leg FFM, kg | 11.5±0.2 (97) | 11.2±0.2 (158) | 0.28 |
Data are least square means ± SE; (n), sample size. Age and height were included in the model as significant covariates.
Results
ACVR2B haplotype
Figure 1 shows the haplotype block and haplotype frequency information for the genome sequence containing the ACVR2B gene from the HapMap CEPH database. Haplotypes were grouped to minimize analysis of rare haplotypes and improve statistical power. Genotyping rs2268757, located in intron 1 of the ACVR2B gene, captured the majority of the information of this haplotype block and created two related haplotype groups. Haplotype 1 (frequency, 62%) along with haplotype 4 (frequency, 3%) were considered one group (Hap Group 1) because of their high degree of similarity, sharing eight of nine alleles across the block. Haplotype 2 (frequency, 27%), haplotype 3 (frequency, 5%), and haplotype 5 (frequency, 1%) were grouped as Hap Group 2, sharing a minimum of five of nine alleles to a maximum of eight of nine alleles across the block. Our overall haplotype group frequencies were 54% for Hap Group 1 and 46% for Hap Group 2, which differed somewhat from the frequencies expected as calculated by Haploview using the CEPH data (expected: 65% Hap Group 1; 33% Hap Group 2).
Fig. 1.
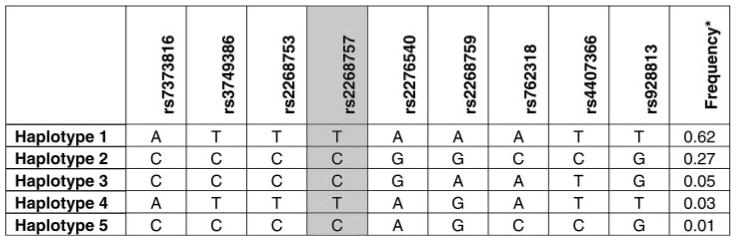
Activin-type II receptor B (ACVR2B) haplotype structure. The tagged polymorphism, rs2268757, is shaded. Haplotypes 1 and 4 were combined to form Haplotype Group 1 (Hap Group 1), while haplotypes 2, 3, and 5 were combined to form Hap Group 2 for the analysis of ACVR2B haplotype structure. *Frequency data from the Centre d'Etude du Polymorphisme Humain (CEPH) cohort within the International HapMap Project database.
Subject characteristics are shown by ACVR2B haplotype group in Table 1. No significant differences existed by ACVR2B haplotype group for any physical characteristic. As shown in Table 2, women heterozygous for Hap Groups 1 and 2 exhibited significantly less quadriceps shortening strength than women homozygous for Hap Group 2 (108.7 ± 2.2 vs. 118.6 ± 4.1 N·m, 30°/s, respectively, P = 0.036). In a combined analysis of carriers of Hap Group 1 vs. homozygous Hap Group 2, the same association was observed (109.2 ± 1.9 vs. 118.6 ± 4.1 N·m, 30°/s, respectively, P = 0.039). These associations were also significant when leg FFM was included as a covariate (P = 0.001). No significant strength differences were observed among ACVR2B haplotype groups in men. Also, no significant differences were observed in total body FFM or total leg FFM in either men or women by ACVR2B haplotype group (Table 1).
Table 2. Knee extensor PT (shortening phase) values by ACVR2B haplotype in men and women.
| Homozygous Hap Group 1 | Heterozygous Hap Groups 1 and 2 | Homozygous Hap Group 2 | P Value | |
|---|---|---|---|---|
| Men | ||||
| n | 80 | 184 | 49 | |
| PT (30°/s), N·m | 175.7±4.5 | 167.7±3.0 | 172.8±5.8 | 0.31 |
| PT (180°/s), N·m | 117.6±2.9 | 114.2±2.0 | 118.0±3.7 | 0.50 |
| Women | ||||
| n | 61 | 158 | 48 | |
| PT (30°/s), N·m | 110.5±3.6 | 108.7±2.2a | 118.6±4.1b | 0.036 (a vs. b) |
| PT (180°/s), N·m | 72.8±2.4 | 70.4±1.5 | 75.4±2.7 | 0.25 |
Data are least squares means ± SE. Age and height were included in the model as significant covariates. PT, peak torque.
Follistatin haplotype
HapMap database research into the haplotype structure of the genome sequence surrounding the follistatin gene revealed haplotype block and haplotype frequency details as shown in Fig. 2. SNPs rs3797297, rs3756498, rs12152850, and rs12153205 comprise all of the SNPs within the follistatin gene within this block, and these were selected for haplotype determination to minimize the total number of haplotypes under analysis. Within this specific subblock, haplotypes were grouped according to similarity of structure to minimize analysis of the rarest haplotypes. By genotyping rs3797297 and rs12152850 as tag SNPs, the five haplotypes within this region could be identified. Genotyping rs3797297 separated haplotype 3 (Hap Group 3; frequency, 20%) from the rest of the haplotypes. Genotyping rs12152850 separated haplotype 2 (Hap Group 2; frequency, 22%) from the remaining haplotypes. The final group (Hap Group 1; frequency, 54%) consisted of haplotypes 1, 4, and 5, which share a minimum of four of six alleles. Our overall haplotype group frequencies were 62% for Hap Group 1, 17% for Hap Group 2, and 20% for Hap Group 3, which were similar to the frequencies expected as calculated by Haploview using the CEPH data (expected: 54% Hap Group 1, 22% Hap Group 2, 20% Hap Group 3). In some analyses, different combinations of these three haplotype groups were collapsed to improve statistical power and assess unique haplotype associations.
Fig. 2.
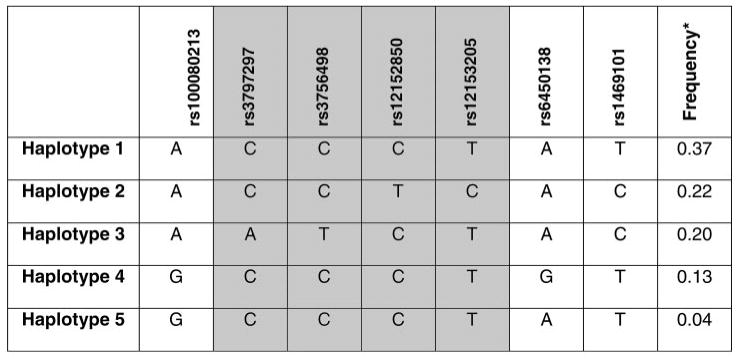
Follistatin (gene symbol: FST) haplotype structure. The shaded region shows the 4 polymorphisms within the follistatin coding region, which were selected for haplotype analysis. The tagged polymorphisms were rs3797297 and rs12152850. Polymorphisms not shaded are outside of the follistatin coding region. Haplotypes 1, 4, and 5 were combined to form Hap Group 1; both haplotype 2 and haplotype 3 were analyzed separately as Hap Group 2 and Hap Group 3, respectively, for the analysis of follistatin haplotype structure. *Frequency data from the CEPH cohort within the International HapMap Project database.
Subject characteristics are shown by follistatin haplotype group in Table 3. Men homozygous for Hap Group 1 were significantly younger than men heterozygous for Hap Groups 1 and 2 (P = 0.015) and men heterozygous for Hap Groups 1 and 3 (P = 0.004). Women homozygous for Hap Group 1 weighed significantly less than women heterozygous for Hap Groups 1 and 2 (P = 0.005), while noncarriers of Hap Group 1 also weighed significantly less than women heterozygous for Hap Groups 1 and 2 (P = 0.045).
Table 3. Subject characteristics for follistatin haplotype groups in men and women.
| Homozygous Hap Group 1 | Heterozygous Hap Groups 1 and 2 | Heterozygous Hap Groups 1 and 3 | Noncarriers of Hap Group 1 | P Value | |
|---|---|---|---|---|---|
| Men | |||||
| n | 128 (40.7%) | 67 (21.3%) | 83 (26.3%) | 37 (11.7%) | |
| Age, yr | 59.4±1.5a | 65.4±2.0b | 66.1±1.8c | 58.9±2.7d | 0.015 (a vs. b) |
| 0.004 (a vs. c) | |||||
| 0.052 (b vs. d) | |||||
| 0.027 (c vs. d) | |||||
| Height, cm | 176.9±0.7 | 175.3±0.9 | 176.1±0.8 | 175.9±1.2 | 0.53 |
| Weight, kg | 85.4±1.1 | 82.8±1.6 | 83.1±1.4 | 85.6±2.1 | 0.39 |
| Women | |||||
| n | 108 (38.8%) | 53 (19.1%) | 65 (23.4%) | 52 (18.7) | |
| Age, yr | 58.8±1.5 | 55.7±2.2 | 55.8±2.0 | 57.3±2.3 | 0.58 |
| Height, cm | 163.2±0.7 | 164.9±0.9 | 163.1±0.8 | 162.3±0.9 | 0.26 |
| Weight, kg | 65.4±1.2a | 71.3±1.7b | 68.9±1.5c | 66.4±1.7d | 0.005 (a vs. b) |
| 0.066 (a vs. c) | |||||
| 0.045 (b vs. d) | |||||
Data are means ± SE.
As shown in Table 4, men homozygous for Hap Group 1 exhibited significantly more total leg FFM than men heterozygous for Hap Groups 1 and 3 (17.8 ± 0.2 vs. 16.7 ± 0.4 kg, respectively, P = 0.007). They also exhibited significantly greater leg FFM than noncarriers of Hap Group 1 (17.8 ± 0.2 vs. 16.5 ± 0.5 kg, respectively, P = 0.023). Moreover, when men homozygous for Hap Group 1 were compared with men heterozygous for Hap Groups 1 and 3, higher values of total body FFM were indicated (57.4 ± 0.4 vs. 56.1 ± 0.5 kg, respectively, P = 0.06). No significant differences were observed for muscle strength phenotypes in men or for FFM or strength in women for follistatin haplotype groups (data not shown).
Table 4. Total body and leg FFM for follistatin haplotype groups in men and women.
| Homozygous Hap Group 1 | Heterozygous Hap Groups 1 and 2 | Heterozygous Hap Groups 1 and 3 | Noncarriers of Hap Group 1 | P Value | |
|---|---|---|---|---|---|
| Men | |||||
| Total body FFM, kg | 57.4±0.4a (128) | 56.8±0.6 (67) | 56.1±0.5b (83) | 57.9±0.8 (37) | 0.061 (a vs. b) |
| Total leg FFM, kg | 17.8±0.2a (116) | 17.1±0.4 (62) | 16.7±0.3b (72) | 16.5±0.5c (36) | 0.007 (a vs. b) |
| 0.023 (a vs. c) | |||||
| Women | |||||
| Total body FFM, kg | 39.5±0.4 (108) | 39.7±0.4 (53) | 39.5±0.3 (65) | 39.5±0.4 (52) | 0.90 |
| Total leg FFM, kg | 11.1±0.2 (99) | 11.4±0.3 (49) | 11.6±0.3 (61) | 11.4±0.3 (46) | 0.61 |
Data are least square means ± SE; (n), sample size. Age and height were included in the model as significant covariates. Weight was added as an additional significant covariate for the female analysis.
In examining the results from Table 4, we observed that the bulk of noncarriers of Hap Group 1 were carrying haplotype 3 (Hap Group 3). Of the noncarriers of Hap Group 1, only four of these subjects were also not a carrier of Hap Group 3. This led us to perform a subsequent subanalysis focused on Hap Group 3 by collapsing all carriers of Hap Group 3 into one group and all noncarriers of Hap Group 3 into a second group. Subject characteristics for this subanalysis are shown in Table 5. As shown in Table 6, male carriers of Hap Group 3 exhibited significantly less total leg FFM than noncarriers of Hap Group 3 (16.6 ± 0.3 vs. 17.5 ± 0.2 kg, respectively, P = 0.012) and approached a significantly lower total body FFM (56.3 ± 0.5 vs. 57.4 ± 0.3 kg, respectively, P = 0.055). No significant differences were observed in either men or women for muscle strength for the Hap Group 3 subanalysis (data not shown).
Table 5. Subject characteristics for follistatin Hap Group 3 carriers and noncarriers in men and women.
| Carriers of Hap Group 3 | Noncarriers of Hap Group 3 | P Value | |
|---|---|---|---|
| Men | |||
| n | 111 (35.2%) | 204 (64.7%) | |
| Age, yr | 64.3±1.6 | 61.3±1.1 | 0.12 |
| Height, cm | 176.3±0.7 | 176.2±0.5 | 0.88 |
| Weight, kg | 83.8±1.2 | 84.5 ±0.9 | 0.64 |
| Women | |||
| n | 107 (38.5%) | 171 (61.5%) | |
| Age, yr | 56.2±1.5 | 57.9±1.2 | 0.39 |
| Height, cm | 162.7±0.7 | 163.7±0.5 | 0.24 |
| Weight, kg | 67.8±1.2 | 67.3 ±0.9 | 0.72 |
Data are means ± SE.
Discussion
The present study is one of the first to explore haplotype structure of candidate genes and their associations with skeletal muscle phenotypes and shows for the first time that the ACVR2B and follistatin loci may contribute to interindividual variation in muscle mass and strength. The results show that follistatin haplotype 3 is associated with lower skeletal muscle mass in men and that ACVR2B Hap Group 1 is associated with lower skeletal muscle strength in women. Although these associations cannot be verified as causal from the present data, the use of haplotype structure in the present study accounts for a greater fraction of genetic variation than single-polymorphism studies. The results of the present study can be used to generate specific hypotheses regarding the genetic influence of the target genes on muscle phenotypes, and subsequent studies can be aimed at identifying the causal variant(s) associated with each haplotype as well as its mechanism of action (31).
The strong role of myostatin in both muscle development and the maintenance of muscle mass in adults (22) provides a rationale to address whether genetic variation in members of its pathway (e.g., ACVR2B and follistatin) influence muscle phenotypes. The molecular basis for the haplotype associations observed for the ACVR2B and follistatin genes with skeletal muscle phenotypes is uncertain and cannot be addressed by the present study; however, we speculate that these associations may be the result of altered ACVR2B (myostatin receptor) activity, either directly through genetic influence on ACVR2B itself or indirectly through the influence of follistatin. Follistatin has been shown to have a strong affinity for myostatin and can completely prevent myostatin receptor activation and downstream phosphorylation of Smad3 (1). Phosphorylation of Smad3, a key step in the myostatin cascade in negatively regulating skeletal muscle, induces binding of Smad3 to MyoD and represses the activity of the MyoD family of transcription factors, resulting in inhibition of myoblast differentiation (16, 19). On the basis of these relationships, it can be hypothesized that a causal mutation, yet to be identified within the target genes, leads to either increased activity of myostatin (via lower follistatin inhibition) or greater ACVR2B activation, which would result in greater phosphorylation of Smad3. Such influences could be envisioned to affect either total muscle mass or fiber-type composition, the latter of which would have relevance to muscle strength or muscle quality. The soleus muscle of myostatin null mice displays a larger proportion of fast-twitch type II fibers and a reduced proportion of slow-twitch type I fibers compared with wild-type animals (9). Thus higher receptor activation (e.g., due to genetic variation in follistatin haplotype 3, or ACVR2B Hap Group 1) would be envisioned to result in lower muscle mass and/or lower type II fiber proportions. Obviously, further studies will be required to test these hypotheses, but the present results do provide support for the generation of such hypotheses.
There is no obvious explanation for the sex differences observed in the present study (i.e., a relationship between follistatin and muscle mass in men, and a relationship between ACVR2B and muscle strength in women). Previous candidate gene association studies involving skeletal muscle phenotypes by our group (25, 36) and others (3, 33, 34) have also observed sex-specific differences. Although speculative, perhaps the sex differences that have been observed in several studies are partially due to sex-specific gene × hormonal environment interactions.
The genetic analysis performed here may prove important for helping to explain the interindividual variation in muscle mass and strength. Differences in muscle mass and strength may have important implications for sarcopenia, a disorder that affects an estimated 45% of the older population (>60 yr old) with an estimated 18.5 billion dollars in direct healthcare costs in the year 2000 (15). Walston and Fried (37) have argued that higher baseline levels of muscle mass may protect men from reaching a threshold of weakness, while muscle mass loss may put them into a category of frailty. Such a functional threshold has also been suggested by Visser et al. (35), who showed that lower leg strength was independently associated with poorer lower extremity performance. The results from this project and others may eventually provide the data needed for the early identification of individuals genetically susceptible to low levels of muscle mass and strength (i.e., arguably closer to this functional threshold), thus allowing the introduction of more targeted interventions before the onset of associated infirmities.
The present study has several limitations. Because of statistical power limitations regarding the follistatin haplotype analysis, specific haplotype groupings needed to be collapsed. We did not correct for multiple statistical testing, as we view this as an exploratory analysis meant to generate specific hypotheses for future investigations; we recognize the increased chance of false-positive results but argue that false-negative results are more problematic in exploratory investigations. Moreover, this study examines the relationship between only two genes with skeletal muscle phenotypes, and sample size limitations prevented a combined analysis to investigate possible interactions of the two genes. Skeletal muscle mass and strength are complex phenotypes that are likely influenced by multiple genes, environmental factors, and gene × environment interactions. The present study adds the ACVR2B and follistatin genes to a growing list of genes that have been tentatively identified as contributing to interindividual variation in skeletal muscle phenotypes. The importance of all of these genes will need to be confirmed and the interactions among them will also need to be examined in future studies, although the techniques required to perform such a comprehensive analysis are still being established.
In conclusion, this is the first study to explore associations between the haplotype structures of the ACVR2B and follistatin genes with skeletal muscle mass and strength phenotypes. These data indicate that the ACVR2B and follistatin loci may contribute to the interindividual variation in skeletal muscle mass and strength, although these data indicate sex-specific relationships. These results will help generate hypotheses for future studies and, in combination with previous and future studies, will contribute to the growing understanding of the role of genetic variation and its influences on skeletal muscle mass and strength.
Acknowledgments
We thank Jade Clark for technical assistance with follistatin and ACVR2B genotyping and the subjects who made this investigation possible.
GRANTS: The BLSA research was conducted as a component of the Intramural Research Program of the National Institute on Aging (NIA). This work was further sponsored by NIA Grants AG-021500 and AG-022791.
References
- 1.Amthor H, Nicholas G, McKinnell I, Kemp CF, Sharma M, Kambadur R, Patel K. Follistatin complexes myostatin and antagonizes myostatin-mediated inhibition of myogenesis. Dev Biol. 2004;270:19–30. doi: 10.1016/j.ydbio.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 2.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 3.Clarkson PM, Devaney JM, Gordish-Dressman H, Thompson PD, Hubal MJ, Urso M, Price TB, Angelopoulos TJ, Gordon PM, Moyna NM, Pescatello LS, Visich PS, Zoeller RF, Seip RL, Hoffman EP. ACTN3 genotype is associated with increases in muscle strength in response to resistance training in women. J Appl Physiol. 2005;99:154–163. doi: 10.1152/japplphysiol.01139.2004. [DOI] [PubMed] [Google Scholar]
- 4.Corsi AM, Ferrucci L, Gozzini A, Tanini A, Brandi ML. Myostatin polymorphisms and age-related sarcopenia in the Italian population. J Am Geriatr Soc. 2002;50:1463. doi: 10.1046/j.1532-5415.2002.50376.x. [DOI] [PubMed] [Google Scholar]
- 5.Faulkner JA. Terminology for contractions of muscles during shortening, while isometric, and during lengthening. J Appl Physiol. 2003;95:455–459. doi: 10.1152/japplphysiol.00280.2003. [DOI] [PubMed] [Google Scholar]
- 6.Ferrell RE, Conte V, Lawrence EC, Roth SM, Hagberg JM, Hurley BF. Frequent sequence variation in the human myostatin (GDF8) gene as a marker for analysis of muscle-related phenotypes. Genomics. 1999;62:203–207. doi: 10.1006/geno.1999.5984. [DOI] [PubMed] [Google Scholar]
- 7.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 8.Gallagher D, Visser M, De Meersman RE, Sepulveda D, Baumgartner RN, Pierson RN, Harris T, Heymsfield SB. Appendicular skeletal muscle mass: effects of age, gender, and ethnicity. J Appl Physiol. 1997;83:229–239. doi: 10.1152/jappl.1997.83.1.229. [DOI] [PubMed] [Google Scholar]
- 9.Girgenrath S, Song K, Whittemore LA. Loss of myostatin expression alters fiber-type distribution and expression of myosin heavy chain iso-forms in slow- and fast-type skeletal muscle. Muscle Nerve. 2005;31:34–40. doi: 10.1002/mus.20175. [DOI] [PubMed] [Google Scholar]
- 10.Grobet L, Martin LJ, Poncelet D, Pirottin D, Brouwers B, Riquet J, Schoeberlein A, Dunner S, Menissier F, Massabanda J, Fries R, Hanset R, Georges M. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet. 1997;17:71–74. doi: 10.1038/ng0997-71. [DOI] [PubMed] [Google Scholar]
- 11.Huygens W, Thomis MA, Peeters MW, Aerssens J, Janssen R, Vlietinck RF, Beunen G. Linkage of myostatin pathway genes with knee strength in humans. Physiol Genomics. 2004;17:264–270. doi: 10.1152/physiolgenomics.00224.2003. [DOI] [PubMed] [Google Scholar]
- 12.Huygens W, Thomis MA, Peeters MW, Aerssens J, Vlietinck R, Beunen GP. Quantitative trait loci for human muscle strength: linkage analysis of myostatin pathway genes. Physiol Genomics. 2005;22:390–397. doi: 10.1152/physiolgenomics.00010.2005. [DOI] [PubMed] [Google Scholar]
- 13.Huygens W, Thomis MA, Peeters MW, Vlietinck RF, Beunen GP. Determinants and upper-limit heritabilities of skeletal muscle mass and strength. Can J Appl Physiol. 2004;29:186–200. doi: 10.1139/h04-014. [DOI] [PubMed] [Google Scholar]
- 14.Ivey FM, Roth SM, Ferrell RE, Tracy BL, Lemmer JT, Hurlbut DE, Martel GF, Siegel EL, Fozard JL, Jeffrey ME, Fleg JL, Hurley BF. Effects of age, gender, and myostatin genotype on the hypertrophic response to heavy resistance strength training. J Gerontol A Biol Sci Med Sci. 2000;55:M641–M648. doi: 10.1093/gerona/55.11.m641. [DOI] [PubMed] [Google Scholar]
- 15.Janssen I, Shepard DS, Katzmarzyk PT, Roubenoff R. The healthcare costs of sarcopenia in the United States. J Am Geriatr Soc. 2004;52:80–85. doi: 10.1111/j.1532-5415.2004.52014.x. [DOI] [PubMed] [Google Scholar]
- 16.Langley B, Thomas M, Bishop A, Sharma M, Gilmour S, Kambadur R. Myostatin inhibits myoblast differentiation by downregulating MyoD expression. J Biol Chem. 2002;277:49831–49840. doi: 10.1074/jbc.M204291200. [DOI] [PubMed] [Google Scholar]
- 17.Lee SJ, McPherron AC. Regulation of myostatin activity and muscle growth. Proc Natl Acad Sci USA. 2001;98:9306–9311. doi: 10.1073/pnas.151270098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindle RS, Metter EJ, Lynch NA, Fleg JL, Fozard JL, Tobin J, Roy TA, Hurley BF. Age and gender comparisons of muscle strength in 654 women and men aged 20–93 yr. J Appl Physiol. 1997;83:1581–1587. doi: 10.1152/jappl.1997.83.5.1581. [DOI] [PubMed] [Google Scholar]
- 19.Liu D, Black BL, Derynck R. TGF-beta inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes Dev. 2001;15:2950–2966. doi: 10.1101/gad.925901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Flood SJ, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 21.Lynch NA, Metter EJ, Lindle RS, Fozard JL, Tobin JD, Roy TA, Fleg JL, Hurley BF. Muscle quality. I. Age-associated differences between arm and leg muscle groups. J Appl Physiol. 1999;86:188–194. doi: 10.1152/jappl.1999.86.1.188. [DOI] [PubMed] [Google Scholar]
- 22.McCroskery S, Thomas M, Maxwell L, Sharma M, Kambadur R. Myostatin negatively regulates satellite cell activation and self-renewal. J Cell Biol. 2003;162:1135–1147. doi: 10.1083/jcb.200207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 24.McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA. 1997;94:12457–12461. doi: 10.1073/pnas.94.23.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roth SM, Schrager MA, Lee MR, Metter EJ, Hurley BF, Ferrell RE. Interleukin-6 (IL6) genotype is associated with fat-free mass in men but not women. J Gerontol A Biol Sci Med Sci. 2003;58:B1085–B1088. doi: 10.1093/gerona/58.12.b1085. [DOI] [PubMed] [Google Scholar]
- 26.Schuelke M, Wagner KR, Stolz LE, Hubner C, Riebel T, Komen W, Braun T, Tobin JF, Lee SJ. Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med. 2004;350:2682–2688. doi: 10.1056/NEJMoa040933. [DOI] [PubMed] [Google Scholar]
- 27.Seeman E, Hopper JL, Young NR, Formica C, Goss P, Tsalamandris C. Do genetic factors explain associations between muscle strength, lean mass, and bone density? A twin study. Am J Physiol Endocrinol Metab. 1996;270:E320–E327. doi: 10.1152/ajpendo.1996.270.2.E320. [DOI] [PubMed] [Google Scholar]
- 28.Seibert MJ, Xue QL, Fried LP, Walston JD. Polymorphic variation in the human myostatin (GDF-8) gene and association with strength measures in the Women's Health and Aging Study II cohort. J Am Geriatr Soc. 2001;49:1093–1096. doi: 10.1046/j.1532-5415.2001.49214.x. [DOI] [PubMed] [Google Scholar]
- 29.Shock NW, Gruelich RC, Andres RA, Arenberg D, Costa PT, Lakatta EG, Tobin JD. Normal Human Aging. The Baltimore Longitudinal Study of Aging. Washington, DC: U.S. Government Printing Office; 1984. [Google Scholar]
- 30.Stewart CE, Rittweger J. Adaptive processes in skeletal muscle: molecular regulators and genetic influences. J Musculoskelet Neuronal Interact. 2006;6:73–86. [PubMed] [Google Scholar]
- 31.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thies RS, Chen T, Davies MV, Tomkinson KN, Pearson AA, Shakey QA, Wolfman NM. GDF-8 propeptide binds to GDF-8 and antagonizes biological activity by inhibiting GDF-8 receptor binding. Growth Factors. 2001;18:251–259. doi: 10.3109/08977190109029114. [DOI] [PubMed] [Google Scholar]
- 33.van Rossum EF, Voorhoeve PG, te Velde SJ, Koper JW, Delemarre-van de Waal HA, Kemper HC, Lamberts SW. The ER22/23EK polymorphism in the glucocorticoid receptor gene is associated with a beneficial body composition and muscle strength in young adults. J Clin Endocrinol Metab. 2004;89:4004–4009. doi: 10.1210/jc.2003-031422. [DOI] [PubMed] [Google Scholar]
- 34.Van Pottelbergh I, Goemaere S, Nuytinck L, De PA, Kaufman JM. Association of the type I collagen alpha1 Sp1 polymorphism, bone density and upper limb muscle strength in community-dwelling elderly men. Osteoporos Int. 2001;12:895–901. doi: 10.1007/s001980170043. [DOI] [PubMed] [Google Scholar]
- 35.Visser M, Newman AB, Nevitt MC, Kritchevsky SB, Stamm EB, Goodpaster BH, Harris TB. Reexamining the sarcopenia hypothesis. Muscle mass versus muscle strength. Health, Aging, and Body Composition Study Research Group. Ann NY Acad Sci. 2000;904:456–461. [PubMed] [Google Scholar]
- 36.Walsh S, Zmuda JM, Cauley JA, Shea PR, Metter EJ, Hurley BF, Ferrell RE, Roth SM. Androgen receptor CAG repeat polymorphism is associated with fat-free mass in men. J Appl Physiol. 2005;98:132–137. doi: 10.1152/japplphysiol.00537.2004. [DOI] [PubMed] [Google Scholar]
- 37.Walston J, Fried LP. Frailty and the older man. Med Clin North Am. 1999;83:1173–1194. doi: 10.1016/s0025-7125(05)70157-7. [DOI] [PubMed] [Google Scholar]
- 38.Wang ZM, Visser M, Ma R, Baumgartner RN, Kotler D, Gallagher D, Heymsfield SB. Skeletal muscle mass: evaluation of neutron activation and dual-energy X-ray absorptiometry methods. J Appl Physiol. 1996;80:824–831. doi: 10.1152/jappl.1996.80.3.824. [DOI] [PubMed] [Google Scholar]
- 39.Yang J, Ratovitski T, Brady JP, Solomon MB, Wells KD, Wall RJ. Expression of myostatin pro domain results in muscular transgenic mice. Mol Reprod Dev. 2001;60:351–361. doi: 10.1002/mrd.1097. [DOI] [PubMed] [Google Scholar]
- 40.Zimmers TA, Davies MV, Koniaris LG, Haynes P, Esquela AF, Tomkinson KN, McPherron AC, Wolfman NM, Lee SJ. Induction of cachexia in mice by systemically administered myostatin. Science. 2002;296:1486–1488. doi: 10.1126/science.1069525. [DOI] [PubMed] [Google Scholar]


