Abstract
Although there is compelling evidence that the β amyloid peptide (Aβ) can be centrally involved in Alzheimer’s disease, the natural role (if any) of this peptide remains unclear. Here we use green fluorescent protein (GFP) fusions to demonstrate that the Aβ sequence, like prion domains, can act as a modular aggregation domain when terminally appended to a normally soluble protein. We find that a single amino acid substitution (Leu17 to Pro) in the β peptide sequence can abolish this cis capacity to induce aggregation. Introduction of this substitution into full-length APP (i.e., a Leu613Pro substitution in APP695) alters the processing of APP leading to the accumulation of the C99 C-terminal fragment (CTF). We suggest that in at least some aggregation disease-related proteins the presence of an aggregation domain is not “accidental”, but reflects a selected role of these domains in modulating the trafficking or metabolism of the parental protein.
Keywords: Alzheimer’s disease, C. elegans, transgenic, neurodegeneration, γ-secretase, α-secretase, AICD
Introduction
The β amyloid peptide (Aβ), which is likely to be centrally involved in Alzheimer’s disease (AD) pathology (Hardy and Selkoe, 2002), has an inherent capacity to multimerize and aggregate in vitro and in vivo (Finder and Glockshuber, 2007). It is unknown if the capacity of the β amyloid peptide to multimerize rapidly and aggregate serves a selected function, or if this aggregation ability is an unselected consequence of some other selected capacity. Other larger proteins associated with neurodegenerative diseases, such as prion protein (PrP) and polyglutamine repeat-containing proteins (e.g., huntingtin) show similar aggregation capacities (Koo et al, 1999). The aggregation of polyglutamine repeat proteins depends specifically on the repeat sequence; addition of long polyglutamine repeats to normally soluble proteins, such as Green Fluorescent Protein (GFP), causes aggressive aggregation of the fusion protein (Moulder et al, 1999; Saytal et al, 2000). Similarly, molecular dissection of yeast prion proteins has demonstrated that a glutamine/asparagine-rich domain is responsible for their aggregation, and this domain can convey aggregation when fused to heterologous soluble proteins (DePace et al, 1998; Li and Lindquist, 2000; Osherovich and Weissman, 2001; Osherovich et al, 2004). Si and colleagues identified a neuronal isoform of the Aplysia CPEB protein that contains a glutamine/asparagine-rich domain that can likewise convey prion-like properties when fused to heterologous proteins and expressed in yeast (Si et al, 2003). These authors suggested that the conversion of CPEB to a prion-like state might be important for the function of this protein, as it could allow the establishment of a localized, stable epigenetic switch important for maintaining long-term synaptic changes.
The Aβ peptide is derived by proteolytic cleavage from the Amyloid Precursor Protein (APP), a broadly expressed protein with multiple functions, including a proposed role in synaptic function (Kamenetz et al, 2003). By analogy with the glutamine/asparagine-rich domain of CPEB, the Aβ peptide sequence could function as an aggregation domain and play a role in the function of its “host” protein, APP. We therefore sought to determine 1) if the Aβ sequence could serve as an transferable aggregation domain module, and 2) if interference with the proposed aggregation capacity of the Aβ sequence could modulate APP function. We find that the Aβ sequence can act as an aggregation module, and that APP processing is altered by introduction of a mutation in the Aβ domain that abrogates its aggregation potential.
METHODS
Construction of transgenes and transgenic strains
The human Aβ 3–42 sequence was cloned in frame downstream of GFP in C. elegans expression vectors that use either the myo-3 promoter to drive muscle-specific expression or the snb-1 promoter to drive pan-neuronal expression. An equivalent mammalian expression vector was constructed by inserting the Aβ 3–42 sequence downstream of GFP in the CMV-driven expression vector EGFP-C2 (Clontech). The Leu17Pro mutation was introduced into the C. elegans and mammalian expression constructs (including APP695 and C99-GVP) by in vitro mutagenesis (Gene Tailor kit, Invitrogen). Transgenic C. elegans lines were established by microinjection as previously described (Link, 1995; Fay et al, 1998), using either pRF4 (dominant rol-6 morphological marker) or pCL26 (intestinal GFP expression) as selected markers.
Biochemical measurements of GFP::Aβ aggregation
Sequential extraction of nematode tissue and filter trap assays were performed as previously described (Link et al, 2006). For the sequential extraction experiments, nematode homogenates were prepared in Tris/Triton X-100 immunoprecipitation buffer (Fonte et al, 2002), then initially extracted for 1 hr (4°C) in Tris/glycerol buffer containing protease inhibitors. Insoluble material was pelleted by centrifugation (25,000g × 15 min) and re-extracted in 1% SDS (1 hr, 4°C). The remaining insoluble material was pelleted (25,000g × 15 min centrifugation). Extracts were concentrated, desalted, and fractionated on 4–12% BisTris gels. Immunoblots were performed with anti-GFP antibody (Qbiogene).
The filter trap assays were performed using Tris/Triton X-100 nematode homogenates cleared of large insoluble material (i.e., cuticle fragments) by low-speed spin (5 min at 325 relative centrifigual force). Replicate homogenate aliquots (5 × 30 μl) were run through a 0.2 μM cellulose acetate membrane pre-soaked in immunoprecipitation buffer. After drying, identical circular punches of the membrane were transferred to flat-bottom wells of a 96-well microtiter plate and total trapped GFP fluorescence was quantified using a microplate reader. Relative trapped GFP fluorescence (Figure 1 H) was calculated by averaging total GFP fluorescence (arbritary units from Tecan GENios microplate reader) from replicate wells and subtracting the background signal from homogenate-only punches.
Figure 1.
Aggregation of GFP::Aβ fusion protein expressed in C. elegans. Panels A–F, fused DIC/epifluorescence images of live transgenic worms. A. C. elegans strain (CL1179, myo-3/GFP transgene) with body wall muscle expression of GFP. Note the even cytoplasmic GFP accumulation in spindle-shaped muscle cells (arrows). B. C. elegans strain (CL1234, snb-1/GFP) with GFP expression in ventral cord neurons. Note GFP accumulation in neuronal cell bodies and axons (arrows). C. C. elegans strain (CL1332, myo-3/GFP::Aβ) with body wall muscle expression of the GFP::Aβ fusion protein. Note perinuclear GFP aggregates (arrows). D. C. elegans strain (CL1358, snb-1/GFP::Aβ) with GFP::Aβ expression in ventral cord neurons. Note GFP fluorescence is restricted to cell body aggregates (arrow). E. C. elegans strain (CL1364, myo-3/GFP::Aβ Leu17Pro) with body wall muscle expression of substituted GFP::Aβ fusion protein. Note restoration of diffuse cytoplasmic GFP fluorescence (arrow). F. C. elegans strain (CL1363, snb-1/GFP::Aβ Leu17Pro) with expression of substituted GFP::Aβ fusion protein in ventral cord neurons. Note restoration of GFP fluorescence throughout neuronal cell bodies and axons (arrows). Size bars = 50 μM. G. Reduced solubility of GFP::Aβ. Disrupted transgenic worms expressing body wall muscle GFP (lanes 1, 3) or GFP::Aβ (lanes 2, 4) were sequentially extracted with Tris pH8.6/10% glycerol buffer and then 1% SDS. Note a significant fraction of GFP is recovered in the Tris/glycerol wash, but measurable recovery of GFP::Aβ requires detergent extraction. H. Preferential recovery of GFP::Aβ by filter trap assay. Cleared homogenates of transgenic worms expressing body wall muscle GFP or GFP::Aβ were passed through a 0.2μM cellulose actetate filter, and retained GFP was assayed by fluorimetry. Bars represent the the average GFP signal (arbritary units) of 5 replicate assays, +/− S.E.M. Note increased retention of GFP::Aβ.
Hippocampal cell culture and transfection
Primary cultures of dissociated neurons from E18 embryonic rat hippocampi were prepared essentially as described (Banker and Goslin, 1998). Cells were maintained in Neurobasal medium (Life-Tech/GIBCO-BRL) supplemented with B-27 and Glutamax. After 7 days in vitro, cells were transfected with 2.0 μg of plasmid DNA using Lipofectamine 2000 (Invitrogen, No. 11668-019), per the manufacturer’s instruction. Cells were maintained in a 37°C/5% CO2 incubator for 12 or 24 hours to allow protein expression.
Immunofluorescence and microscopy of hippocampal cells
Cells were fixed in a solution of 4% paraformaldehyde/4% sucrose in PBS, then blocked in 0.5% fish skin gelatin in PBS for 1 hr at 37°C. After blocking, coverslips were incubated with a polyclonal MAP2 (Chemicon #AB5622, 1:500) antibody overnight at 4°C, which was followed by incubation in Cy3-conjugated goat anti-rabbit (Jackson Labs, 1:500) for 1 hr at 37°C. Coverslips were mounted on glass slides in Elvanol containing 0.5 μg/ml DAPI. Images of GFP expressing cells were acquired using a SPOT RT-SE (Diagnostics Instruments) cooled CCD camera controlled by Metamorph Software (Universal Imaging) with a 60X, 1.4 N.A. Plan Apo objective (Nikon).
CHO cell transfections and luciferase assay
CHO cells were seeded at 50,000 cells/cm2 in 24-well tissue culture plates (2 cm2/well) in Ham’s F12 supplemented with 10% FBS. One day later, when the cells were approximately 90% confluent, each well was transfected with Lipofectamine 2000 (Invitrogen) together with 100 ng CMV-βgal, 100 ng MH100 (UAS-luciferase) and 100ng of C99/GVP, pCL120 (C99/GVP-L17P) or empty vector pcDNA3.1. All of the plasmids except CL120 were a gift from A. Bergman (Karolinska Institutet), and the luciferase assays were done 18–24 hr post-transfection essentially as described (Bergman et al, 2003). Briefly, cell lysates were prepared, clarified by centrifugation, and kept on ice while aliquots of each lysate were added to tubes containing ATP and D-luciferin, mixed and read luminometrically exactly 20 seconds after mixing, alternating between control and experimental samples. A second aliquot of each lysate was used to measure βgal activity as an indicator of transfection efficiency. The luciferase results were normalized to the βgal acitivity, and statistical significance was determined by Student’s t-test: Two sample assuming equal variances.
Biotinylation of CHO cell surface proteins
CHO cells were transfected with CMV-βgal and either full-length wildtype APP695 or CL146 (APP-Leu613Pro) DNA. At 24 hours after transfection, the cells were biotinylated for 30 min at 4°C using the Pierce Cell Surface Protein Isolation Kit #89881. The cells were harvested by scraping, lysed, and used for immunoblots and determination of transfection efficiency as described above. Transfection efficiencies were approximately equal (APP-Leu613Pro was 92–99% of APP695).
Immunoblots
β galactosidase assay of transfection efficiency was used to normalize samples. Protein was quantitated by a Bradford assay (Pierce), and 20 micrograms of sample were loaded per lane. SDS PAGE sample buffer was used (62.5mM Tris pH 6.8, 1%SDS, 10% glycerol, 2.5% 2-Mercaptoethanol, and 0.02% bromophenol blue), and samples were run at 180 volts on NuPAGE 4–12%Bis-Tris Gel (Invitrogen, NP0321) using MES SDS Running Buffer (Invitrogen NP0002). Gels were transferred to 0.45 micron supported nitrocellulose (GE Osmonics WP4HY00010) using 20% methanol, 39mM glycine, 48mM tris base. Transfer conditions were 21 volts, 108 minutes. Prestained Rainbow size markers (Amersham Biosciences RPN755, RPN800) were used to size bands.
Blots were visualized by Pouceau stain, and then boiled for 3 minutes in PBS. Blots were blocked in TBS-Tween + 5% milk (100 mM Tris7.5, 150 mM NaCl, 0.1 % Tween-20). APP C-terminal antibody CT20 (rabbit polyclonal, gift of C. Eckman) was diluted 1:5000 in blocking solution. Secondary antibody was HRP-conjugated goat anti-rabbit IgG (Sigma) diluted 1:2000. Mouse monoclonal anti-Aβ (Chemicon, 6E10) was diluted 1:1000 in blocking solution. N-terminal APP mouse monoclonal antibody (Chemicon, 22C11) was diluted 1:1000 in PBS-0.1%Tween + 1% BSA. Actin monoclonal antibody JLA20 (Developmental Studies Hybridoma Bank, University of Iowa) was diluted 1:100 in blocking solution. Secondary HRP-conjugated goat anti-rabbit or anti-mouse IgG (Sigma) was diluted 1:2000. Immunoblots were visualized using ECL (Amersham).
ELISA for Aβ40 peptides
CHO cells were transfected with CMV-βgal and either full-length wildtype APP695 or CL146 (APP-Leu613Pro) DNA. The medium was changed at 6 hours, collected at 24 h, and analyzed with a Biosource ELISA kit for human Aβ40 (Invitrogen). The results were normalized to the transfection efficiency of the cells, as determined by βgal activity.
RESULTS
GFP:: Aβ fusion proteins aggregate in invertebrate and mammalian cells
GFP expressed in C. elegans tissues remains cytoplasmic and soluble, even when its expression is driven by high-level promoters (e.g., see Fig 1A, B). Conversely, intracellular expression of Aβ 1–42 in C. elegans results in the formation of immunoreactive deposits and amyloid fibrils (Link et al, 2001). We fused Aβ sequences 3–42 in frame to the C-terminus of GFP, using a convenient EcoRI site present in the Aβ sequence. This GFP::Aβ fusion protein was expressed either in body wall muscle or pan-neuronally, using the promoters from the C. elegans myo-3 and snb-1 genes, respectively. In both muscle and neurons, addition of the Aβ sequence to GFP results in a dramatic redistribution of the GFP into perinuclear aggregates, readily visible in live animals (Fig 1C, D). To determine if the visible aggregation correlated with decreased solubility of the GFP::Aβ fusion, protein lysates from transgenic animals with muscle expression of GFP or GFP::Aβ were sequentially extracted using salt and SDS-containing buffers. As shown in Figure 1G, efficient recovery of GFP::Aβ, but not GFP, requires strong detergent extraction. The insolubility of GFP::Aβ was also demonstrated by filtration of worm lysates through 0.2 μm cellulose acetate filters (Link et al, 2006), where GFP::Aβ is significantly better retained than GFP (Fig 1H). We also attempted to stain GFP::Aβ deposits with the amyloid-specific dye X-34, which can be used to visualize β-amyloid deposits in living C. elegans worms (Link et al, 2001). However, we did not see specific staining of GFP::Aβ deposits, suggesting that addition of the Aβ peptide to GFP cannot drive this normally globular protein into amyloid fibrils (data not shown).
To investigate whether the aggregation-induction capacity of Aβ also occurs in mammalian neurons, we constructed an analogous expression vector using CMV promoter-driven EGFP, and transiently transformed primary fetal rat hippocampal neurons. While transformation with an unmodified EGFP expression construct resulted in neurons with EGFP fluorescence throughout the cell body, dendrites, and axons, neurons transformed with EGFP::Aβ had their EGFP fluorescence restricted to the perinuclear region. This restricted distribution was observed at all timepoints and in apparently healthy neurons with intact axons (Fig 2).
Figure 2.

Expression of GFP and GFP fusion proteins in primary rat neurons. Primary cultures of dissociated neurons from E18 embryonic rat hippocampi were transfected with CMV promoter-driven expression vectors expressing GFP (panel A), GFP::Aβ (panel B), or GFP::Aβ Leu17Pro (panel C). Transfected cultures were fixed and stained with anti-MAP2 (red) antibody and DAPI (blue). Note expressed GFP extends from the cell body to the dendrites and axon (arrows, panel A), while GFP::Aβ remains localized to the neuronal cell body (arrow, panel B). In contrast, GFP::Aβ Leu17Pro fluorescence is distributed throughout the neuron as observed for unmodified GFP (arrows, panel C.) MAP2 staining is shown in corner insets (panels A and C) or as fused image (panel B). Size bar = 25μM.
A Leu17Pro substitution in the Aβ sequence interferes with Aβ fusion-induced aggregation
We have previously identified substitutions in the Aβ 1–42 sequence that blocked amyloid formation in the C. elegans model system (Fay et al, 1998). One such substitution, Leu17-to-Pro, targets a central hydrophobic region thought to be critical for Aβ aggregation. This same substitution was subsequently identified in an elegant unbiased study that employed random mutagenesis to reverse the misfolding of an Aβ::GFP fusion protein expressed in E. coli (Wurth et al, 2002). We introduced the Leu17Pro substitution into our C. elegans and mammalian GFP::Aβ expression vectors by in vitro mutagenesis, and found that this substitution countered the Aβ fusion-induced aggregation in all cases (Fig 1 E, F; Fig 2C).
Influence of the Aβ sequence on APP processing and signaling
β-secretase processing of APP results in a 99 residue C-terminal fragment (C99) containing the Aβ sequence at its N-terminus. We hypothesized that this terminal Aβ sequence, via its multimerization capacity, might be able to influence subsequent processing of the APP C99 fragment, specifically its cleavage by γ-secretase. To test this hypothesis, we introduced a Leu613Pro substitution (equivalent to the Leu17Pro Aβ substitution) into an APP695 expression construct and transfected CHO cells with wild type or Leu613Pro APP. Transfected cells were subsequently surface-labeled with biotin hydrazide to allow measurement of both total and cell surface APP and APP processing products. As shown in Figure 3, immunoblot analysis using an antibody specific for the C-terminus of APP revealed that the Leu613Pro substitution leads to a strong increase in both total and cell surface C99, without appreciable effects on the accumulation of full-length APP. Accumulation of the C99 fragment in cells transfected with APP695 Leu613Pro suggests that this substitution leads to reduced γ-secretase processing.
Figure 3.

Effect of Leu-to-Pro substitution on APP processing. CHO cells were transfected with APP695 or APP695 Leu613Pro. The cells were subjected to cell surface biotinylation 24 hr after transfection, then harvested. Total cell extracts or biotinylated material were fractionated by SDS-PAGE and probed with CT-20 antisera, which recognizes the C-terminus of APP. Lanes 1, 3, and 5 contain whole cell extracts; lanes 2, 4, and 6 contain biotinylated cell surface material. Note specific accumulation in both total and cell surface fractions of APP C99 fragment in APP695 Leu613Pro transfected cells, despite levels of full length APP (flAPP, ~105 kd) similar to that expressed in APP695 transfected cells. (The C99 band, but not the C83 band, was recognized by mAb 6E10, confirming that this fragment retains the N-terminal portion of Aβ.)
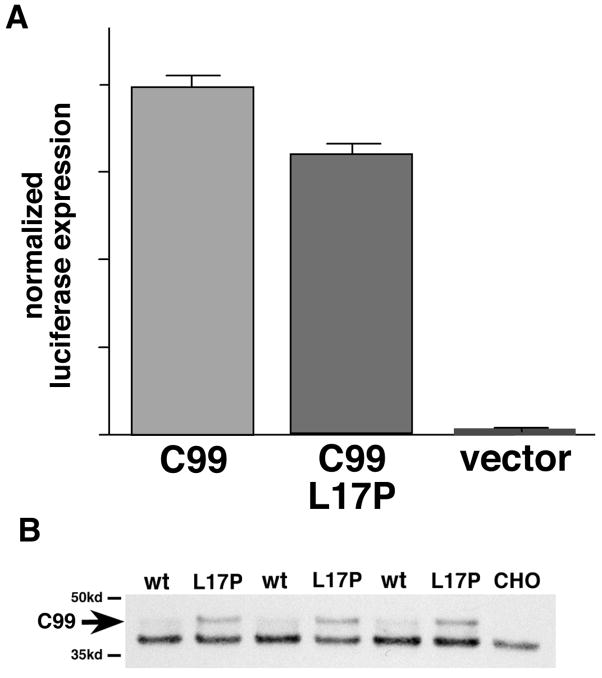
γ-secretase cleavage of APP leads to the release of an intracellular cytoplasmic domain (AICD) that can subsequently translocate to the nucleus and influence gene expression (Kimberly et al, 2001; Leissring et al, 2002). In the experiment described above, we were unable to detect AICD (i.e., C59 or shorter peptides that result from γ/ε cleavage) in any of the transfected cells, likely due to the very short in vivo half-life of this peptide (Cupers et al, 2001). We therefore turned to a sensitive reporter system to determine if the increased accumulation of C99 in cells transfected with APP695 Leu613Pro is mirrored by a decreased production of AICD, as is predicted to result from reduced γ-secretase activity. Karlstrom and colleagues have developed a chimeric reporter construct (C99-GVP) to measure this processing/signaling event by inserting a Gal4VP16 transactivator sequence into the AICD portion of APP C-terminal fragment (Karlstrom et al, 2002; Bergman et al, 2003). Co-transformation of CHO cells with C99-GVP and a UAS-luciferase reporter allows the amount of APP processing to be measured quantitatively by assaying luciferase activity. As shown in Figure 4A, introduction of the Leu17Pro-equivalent substitution into C99-GVP significantly decreased processing as measured by luciferase activity. This reduced release of AICD is associated with an increased accumulation of unprocessed C99-GVP Leu17Pro (Figure 4B), consistent with reduced γ/ε-secretase activity.
Figure 4.
Effect of Leu17-to-Pro substitution on release of APP intracellular domain. A. CHO cells were co-transfected with either the C99-GVP or C99-GVP Leu17Pro reporter constructs (along with UAS/luciferase and a CMV/β-galactosidase plasmid for normalization of transfection efficiencies), harvested 24 hr later, and luciferase activities of cell extracts were determined. (Results are from 6 independent transfection experiments.) Note ~20% decrease in luciferase activity associated with transfection with C99-GVP Leu17Pro (arbitrary luciferase activity units; error bars = S.E.M.; p<0.05). B. Extracts of CHO cells transfected in triplicate as described above were fractionated by SDS-PAGE and probed with mAb 6E10, which recognizes residues 1–16 of Aβ. Note the accumulation of unprocessed C99-GVP specifically in cells transfected with the mutant reporter construct. [The unprocessed C99-GVP runs at ~ 45 kd due to the addition of the Gal4-VP16 transactivator domains (19)].
Although the results described above support an effect of the Leu17Pro substitution on γ/ε cleavage of APP, they do not account for the preferential accumulation of the C99 APP fragment in comparison to accumulation of the APP C83 fragment (Figure 3). One possible explanation for this observation is that the Leu17Pro substitution reduces α-secretase cleavage of APP. Elevated α-secretase levels have been shown to increase AICD production (Kume et al, 2004), and analysis of long Aβ-related peptides has demonstrated that ε-cleavage depends on α-secretase pre-cutting (Kametani, 2004). Thus, the Leu17Pro substitution may secondarily decrease γ/ε cleavage by interfering with α-secretase cleavage. To test this hypothesis, we measured secreted Aβ 1–40 from CHO cells transfected with APP695 or APP695 Leu613Pro using an ELISA assay that only detects full-length Aβ 1–40. Given that the Leu613Pro substitution has a significantly stronger effect on C99 accumulation than on AICD reduction (i.e., γ/ε cleavage), we would predict that reduced α-secretase cleavage of this APP variant would lead to a net overall increase in Aβ secretion. As shown in Figure 5, transfection with APP695 Leu613Pro does lead to an approximately three-fold accumulation of secreted Aβ 1–40.
Figure 5.

Effect of Leu-to-Pro substitution on Aβ40 production. CHO cells were transfected with APP695 or APP695 Leu613Pro, and the amount of Aβ40 released into the medium between 6–24 hr was analyzed by an ELISA for human Aβ40. Note that cells expressing APP695 Leu613Pro secreted approximately 3-fold more Aβ40 than did cells expressing wildtype APP (p<0.001).
Discussion
Our studies show that fusion of the Aβ sequence (specifically residues 3–42) to a heterologous protein can induce the aggregation of the fusion protein, as has been previously demonstrated for polyglutamine repeats and glutamine/asparagine-rich domains from prion proteins. However, Aβ has neither sequence nor predicted structural similarities to these other aggregation domains, and is thus likely to function by a different molecular mechanism. Nevertheless, an overriding question for all these classes of aggregating proteins is whether the presence of the respective aggregation domains in their natural context is important for the function of the host proteins. In the case of yeast prion [PSI+], it appears that while the prion domain is not necessary for the biochemical function of this protein per se (Ter-Avanesyan et al, 1993), the metastable epigenetic state conferred by the prion domain can confer selective advantages in changing environments (True and Lindquist, 2000). Of perhaps more relevance to APP are the studies of the neuronal isoform of the Aplysia CPEB protein, which have led to the provocative suggestion that the prion-like domain of this protein may function as a self-perpetuating epigenetic switch to maintain activated CPEB in localized synaptic regions (Si et al, 2003). We know of no evidence indicating that the Aβ sequence has true prion-like functions [although small Aβ aggregates can efficiently seed amyloid fibril formation (Jarret and Landsbury, 1993)]. However, the general model that localized aggregation/multimerization is a mechanism for modulating protein function may be applicable to APP.
The relationship between a protein’s conformation in vivo and its subsequent metabolism is difficult to assay directly, in part because of the difficulty of monitoring the in vivo conformation of specific proteins without the addition of components (e.g., FRET probes) that might by themselves alter protein metabolism. We have therefore taken the approach of engineering specific amino acid substitutions that have demonstrable effects on in vivo protein conformation and determining the effects of these substitutions on protein metabolism. Using this approach, we have generated data suggesting that the propensity of the Aβ peptide sequence to drive multimerization may influence α and γ/ε-secretase processing of APP. We have have not, however, directly demonstrated that the APP695 Leu613Pro mutation interferes with APP multimerization (e.g., by cross-linking studies).
There is evidence that APP forms homodimers and homotetramers in vivo, and that this association is mediated at least in part by two conserved regions in the APP ectodomain (Scheuermann et al, 2001). Furthermore, forced dimerization of APP by introduction of a Lys624Cys substitution (resulting in intermolecular disulfide bond formation) was found to increase dramatically the production of Aβ in transfected SY5Y cells, indicating that the multimerization state of APP can profoundly affect its processing. Both the the extracellular loop of of APP (Kaden et al, 2008) and the GxxxG motif in its transmembrane portion (Munter et al, 2007) have also been implicated in the dimerization and γ-secretase processing of APP. Our results suggest that the extracellular portion of the Aβ sequence (including Leu17) may also play a role in the multimerization of APP, with subsequent effects on α- and γ/ε-secretase processing. In this regard, substitutions at Ser26 and Leu28 have also been shown to impair γ-secretase-dependent cleavages (Ren et al, 2007).
The reduction of α-secretase cleavage of APP695 by the Leu613Pro substitution could be the result of this substitution simply altering a consensus protease cleavage site recognized by α-secretase. However, the membrane-associated metalloproteases believed to act as α-secretases (ADAM9,10, and 17; Asai et al, 2003) are not known to have a specific cleavage recognition site. Of particular relevance is the observation that ADAM10 cleaves prion protein in the region of this protein (residues 106–126) known to drive prion multimerization and eventually fibril formation (Checler and Vincent, 2002). The observation that both the Aβ sequence and the prion aggregation domain are cleaved by α-secretase activity suggests that the parental proteins containing these domains may be subject to similar proteolytic regulation. Thus, these domains, despite their lack of sequence similarity, may have analogous functions. The ADAM10 cleavage site in prion protein (..NMKHvMAGA…) has no sequence identity with the ADAM10 cleavage site in APP (..HHQKvLVFF…), consistent with the hypothesis that α-secretase substrate recognition may depend on higher-level protein structure (e.g., multimerization or β-sheet formation) rather than on primary sequence. Studies with the related metalloproteinase ADAMTS-4 also highlight the importance of overall protein structure, rather than specific primary sequence, in cleavage site selection by this class of proteases (Lauer-Fields et al, 2007). These observations support the view that the effects of the Leu613Pro substitution act through influencing APP quarternary structure rather than changing local epitopes.
An alternative mechanism by which the Leu613Pro substitution could alter APP processing would be through modulating the intracellular trafficking of APP. A recent study has shown that the natural “Arctic” FAD mutation of APP (Glu618Gly in APP695, Glu22Gly in Aβ) alters the trafficking of APP, resulting in decreased accumulation of cell surface APP and increased production of intracellular Aβ (Sahlin et al, 2007). However, we have not detected a significant difference in cell surface accumulation between APP695 and APP695 Leu613Pro. Although we cannot exclude the possibility that the Leu613Pro substitution interferes with the binding of some unknown protein(s) that modulate APP processing, the simplest interpretation of our data is that this substitution acts by altering the conformation of the parent protein.
Extracellular Aβ peptide has been reported to promote the dimerization of membrane bound APP or the APP C-terminal fragment resulting from β-secretase cleavage, with a subsequent decrease in cell viability (Shaked et al, 2006). This interaction occurs between the Aβ peptide and its cognate sequences in the extracellular portion of APP (i.e., APP 597–624). Thus, the aggregation capacity of the Aβ sequence may also play a role in the modulation of APP function by extracellular Aβ.
Acknowledgments
We would like to thank A. Bergman for providing the C99-GVP and UAS plasmids, and V. Galvan for providing both the APP695 plasmid and extensive experimental advice. The JLA20 monoclonal antibody was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD. We would also like to thank J. Yerg III, D. R. Kipp, and E. Wagner for technical help during the course of this project, and J. Springett for media preparation. This work was supported by NIH grants AG12423 and AG21037 to C.D.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature cited
- Asai M, Hattori C, Szabo B, Sasagawa N, Maruyama K, et al. Putative function of ADAM9, ADAM10, and ADAM17 as APP alpha-secretase. Biochem Biophys Res Commun. 2003;301:231–5. doi: 10.1016/s0006-291x(02)02999-6. [DOI] [PubMed] [Google Scholar]
- Banker G, Goslin K. Culturing Nerve Cells. 2. MIT Press; 1998. [Google Scholar]
- Bergman A, Religa D, Karlstrom H, Laudon H, Winblad B, et al. APP intracellular domain formation and unaltered signaling in the presence of familial Alzheimer’s disease mutations. Exp Cell Res. 2003;287:1–9. doi: 10.1016/s0014-4827(03)00117-4. [DOI] [PubMed] [Google Scholar]
- Checler F, Vincent B. Alzheimer’s and prion diseases: distinct pathologies, common proteolytic denominators. Trends Neurosci. 2002;25:616–20. doi: 10.1016/s0166-2236(02)02263-4. [DOI] [PubMed] [Google Scholar]
- DePace AH, Santoso A, Hillner P, Weissman JS. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell. 1998;93:1241–52. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- Fay DS, Fluet A, Johnson CJ, Link CD. In vivo aggregation of beta-amyloid peptide variants. J Neurochem. 1998;71:1616–25. doi: 10.1046/j.1471-4159.1998.71041616.x. [DOI] [PubMed] [Google Scholar]
- Jarrett JT, Lansbury PT., Jr Seeding “one-dimensional crystallization” of amyloid: a pathogenic mechanism in Alzheimer’s disease and scrapie? Cell. 1993;73:1055–8. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- Kaden D, Munter LM, Joshi M, Treiber C, Weise C, et al. Homophilic interactions of the amyloid precursor protein (APP) ectodomain are regulated by the loop region and affect beta-secretase cleavage of APP. J Biol Chem. 2008;283:7271–9. doi: 10.1074/jbc.M708046200. [DOI] [PubMed] [Google Scholar]
- Kamenetz F, Tomita T, Hsieh H, Seabrook G, Borchelt D, et al. APP processing and synaptic function. Neuron. 2003;37:925–37. doi: 10.1016/s0896-6273(03)00124-7. [DOI] [PubMed] [Google Scholar]
- Kametani F. Secretion of long Abeta-related peptides processed at epsilon-cleavage site is dependent on the alpha-secretase pre-cutting. FEBS Lett. 2004;570:73–6. doi: 10.1016/j.febslet.2004.06.034. [DOI] [PubMed] [Google Scholar]
- Karlstrom H, Bergman A, Lendahl U, Naslund J, Lundkvist J. A sensitive and quantitative assay for measuring cleavage of presenilin substrates. J Biol Chem. 2002;277:6763–6. doi: 10.1074/jbc.C100649200. [DOI] [PubMed] [Google Scholar]
- Kimberly WT, Zheng JB, Guenette SY, Selkoe DJ. The intracellular domain of the beta-amyloid precursor protein is stabilized by Fe65 and translocates to the nucleus in a notch-like manner. J Biol Chem. 2001;276:40288–92. doi: 10.1074/jbc.C100447200. [DOI] [PubMed] [Google Scholar]
- Koo EH, Lansbury PT, Jr, Kelly JW. Amyloid diseases: abnormal protein aggregation in neurodegeneration. Proc Natl Acad Sci U S A. 1999;96:9989–90. doi: 10.1073/pnas.96.18.9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krobitsch S, Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc Natl Acad Sci U S A. 2000;97:1589–94. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume H, Maruyama K, Kametani F. Intracellular domain generation of amyloid precursor protein by epsilon-cleavage depends on C-terminal fragment by alpha-secretase cleavage. Int J Mol Med. 2004;13:121–5. [PubMed] [Google Scholar]
- Lauer-Fields JL, Minond D, Sritharan T, Kashiwagi M, Nagase H, Fields GB. Substrate conformation modulates aggrecanase (ADAMTS-4) affinity and sequence specificity. Suggestion of a common topological specificity for functionally diverse proteases. J Biol Chem. 2007;282:142–50. doi: 10.1074/jbc.M605236200. [DOI] [PubMed] [Google Scholar]
- Leissring MA, Murphy MP, Mead TR, Akbari Y, Sugarman MC, et al. A physiologic signaling role for the gamma-secretase-derived intracellular fragment of APP. Proc Natl Acad Sci U S A. 2002;99:4697–702. doi: 10.1073/pnas.072033799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lindquist S. Creating a protein-based element of inheritance. Science. 2000;287:661–4. doi: 10.1126/science.287.5453.661. [DOI] [PubMed] [Google Scholar]
- Link CD. Expression of human beta-amyloid peptide in transgenic Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1995;92:9368–72. doi: 10.1073/pnas.92.20.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link CD, Fonte V, Hiester B, Yerg J, Ferguson J, et al. Conversion of green fluorescent protein into a toxic, aggregation-prone protein by C-terminal addition of a short peptide. J Biol Chem. 2006;281:1808–16. doi: 10.1074/jbc.M505581200. [DOI] [PubMed] [Google Scholar]
- Link CD, Johnson CJ, Fonte V, Paupard M, Hall DH, et al. Visualization of fibrillar amyloid deposits in living, transgenic Caenorhabditis elegans animals using the sensitive amyloid dye, X-34. Neurobiol Aging. 2001;22:217–26. doi: 10.1016/s0197-4580(00)00237-2. [DOI] [PubMed] [Google Scholar]
- Moulder KL, Onodera O, Burke JR, Strittmatter WJ, Johnson EM., Jr Generation of neuronal intranuclear inclusions by polyglutamine-GFP: analysis of inclusion clearance and toxicity as a function of polyglutamine length. J Neurosci. 1999;19:705–15. doi: 10.1523/JNEUROSCI.19-02-00705.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munter LM, Voigt P, Harmeier A, Kaden D, Gottschalk KE, et al. GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Abeta42. Embo J. 2007;26:1702–12. doi: 10.1038/sj.emboj.7601616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherovich LZ, Cox BS, Tuite MF, Weissman JS. Dissection and design of yeast prions. PLoS Biol. 2004;2:E86. doi: 10.1371/journal.pbio.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106:183–94. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- Ren Z, Schenk D, Basi GS, Shapiro IP. Amyloid beta-protein precursor juxtamembrane domain regulates specificity of gamma-secretase-dependent cleavages. J Biol Chem. 2007;282:35350–60. doi: 10.1074/jbc.M702739200. [DOI] [PubMed] [Google Scholar]
- Sahlin C, Lord A, Magnusson K, Englund H, Almeida CG, et al. The Arctic Alzheimer mutation favors intracellular amyloid-beta production by making amyloid precursor protein less available to alpha-secretase. J Neurochem. 2007;101:854–62. doi: 10.1111/j.1471-4159.2006.04443.x. [DOI] [PubMed] [Google Scholar]
- Satyal SH, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, et al. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2000;97:5750–5. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann S, Hambsch B, Hesse L, Stumm J, Schmidt C, et al. Homodimerization of amyloid precursor protein and its implication in the amyloidogenic pathway of Alzheimer’s disease. J Biol Chem. 2001;276:33923–9. doi: 10.1074/jbc.M105410200. [DOI] [PubMed] [Google Scholar]
- Senut MC, Suhr ST, Kaspar B, Gage FH. Intraneuronal aggregate formation and cell death after viral expression of expanded polyglutamine tracts in the adult rat brain. J Neurosci. 2000;20:219–29. doi: 10.1523/JNEUROSCI.20-01-00219.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked GM, Kummer MP, Lu DC, Galvan V, Bredesen DE, Koo EH. Abeta induces cell death by direct interaction with its cognate extracellular domain on APP (APP 597–624) Faseb J. 2006;20:1254–6. doi: 10.1096/fj.05-5032fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115:879–91. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Kushnirov VV, Dagkesamanskaya AR, Didichenko SA, Chernoff YO, et al. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol. 1993;7:683–92. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–83. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- Wurth C, Guimard NK, Hecht MH. Mutations that reduce aggregation of the Alzheimer’s Abeta42 peptide: an unbiased search for the sequence determinants of Abeta amyloidogenesis. J Mol Biol. 2002;319:1279–90. doi: 10.1016/S0022-2836(02)00399-6. [DOI] [PubMed] [Google Scholar]