Abstract
Natural Killer (NK) cells play important roles in innate defense against infectious agents particularly viruses and also tumors. They mediate their effects through direct cytolysis, release of cytokines and regulation of subsequent adaptive immune responses. NK cells are equipped with sophisticated arrays of inhibitory and activation receptors that regulate their function. In this review we illustrate some of the major evolutionary relationships between NK cell receptors among different animal species and what some of the major mechanisms are that give rise to this diversity in receptor families, including the potential roles of pathogens such as viruses in driving receptor evolution.
1. Introduction
NK cells provide important frontline defenses against infectious agents and tumors. Unlike B and T lymphocytes which require the clonal expansion of very infrequent cells that express appropriate somatically rearranged antigen specific receptors, NK cells use a different strategy to respond to and discriminate infected or tumorigenic cells from normal cells. NK cell subsets that express appropriate receptors to positively recognize and respond to an infectious agent or tumors are relatively abundant compared to antigen-specific T cells or B cells and thus this provides a mechanism for rapid response against potentially life-threatening assaults, in particular to viruses.
NK cells can be activated by a range of soluble factors, including type I interferons, IL-2, IL-12, IL-15 and IL-18, but also by direct cell to cell contact between NK cell receptors and target cell ligands. Molecular cues are sensed and transmitted by an array of inhibitory and activation receptors on individual NK cells. Inhibitory NK receptors can alter NK activation via immunoreceptor tyrosine-based inhibition motifs (ITIMs) in their cytoplasmic tails, which can recruit tyrosine phosphatases (e.g. SHP-1) into receptor signaling complexes. NK activation receptors without ITIM-containing tails, instead carry a basic amino acid within the transmembrane domain for pairing with immunoreceptor tyrosine-based activation motif (ITAM)-containing activation co-receptors (e.g. DAP-12). Thus, a balancing of inhibitory and activation receptor signals are brought together to determine ensuing NK responsiveness.
NK cell inhibitory receptors, the ligands of which include self-MHC class I molecules, are also responsible for self-tolerance and help prevent inappropriate NK cell activation or destruction of normal cells in the body. This feature of NK self-tolerance is acquired when NK cells develop and undergo licensing or arming [1–3]. Once “licensed” or “armed”, inhibitory NK receptors are critical in the mechanism proposed by Kärre described as the ‘missing self’ hypothesis whereby NK cells can attack and kill targets when normal levels of self-MHC class I are reduced or missing [4]. Cellular down-regulation of ligands for inhibitory receptors is therefore a potent stimulus for NK activation, as is frequently the case during viral infection [5].
Despite this, NK cells can also become stimulated when target cells express sufficient specific ligands for NK activation receptors signals to dominate over inhibitory signals. Overall, multiple signals are likely frequently sensed and assimilated by individual NK cells through varied receptors. Whether NK cells respond with cytokine release and/or cytotoxicity is thought to rely on how strong or significant is the sum of all receptor signals. In fact, when inhibitory signals are weak or altogether missing in humans or mice without MHC class I expression, NK cells can become “disarmed” and rendered hyporesponsive through an unknown mechanism [6]. Thus, cells in the bodies of animals without MHC-I expression are still protected from NK attack. A similar mechanism likely normally protects the body from NK cells without any inhibitory receptors for self-MHC class I expression [1, 2].
Vital to early antiviral defenses in the body, many striking features distinguish NK cells in form and function that are well suited for efficiently sensing virus infections. Humans and mice without NK cells because of given genetic deficiencies that restrict their normal development or effector functions, display severe vulnerability to infection and disease caused by herpesviruses [7, 8]. Experimental murine cytomegalovirus (MCMV) infection in mice, including studies with different inbred mouse strains or wild mice, is an important model for investigating early antiviral host defenses and immune responses in the body [9]. Indeed, research in the MCMV model system first revealed that NK cells have and can use Ly49 activation receptors to specifically identify and target virus-infected cells for destruction [9]. More recent work has provided additional examples of NK activation receptors needed to limit virus infection. Not surprisingly therefore, genetic resistance to virus infection has been frequently mapped to the NK gene complex (NKC) on mouse chromosome 6, a genomic region with clustered genes for polymorphic NK cell receptors [10]. The current review serves to highlight some striking NKC features with clustered genes for NK receptors that are specialized for their role in natural killing and antiviral host defenses, which might represent a potential source of natural selection affecting rapid NKC change.
2. NK receptors, polymorphisms and specificity
A battery of structurally diverse cell surface receptors displayed by NK cells belong to superfamilies of C-type lectin or immunoglobulin (Ig) related glycoproteins. Classical MHC class Ia-specific NK cell receptors (NKR) can be broadly grouped into the killer cell Ig-like (KIR) and lectin-like (Ly49, also known as KLRa) receptor families expressed by human or rodent NK cells, respectively. Because KIR-expressing NK cells in humans and other primates display similar functionalities to Ly49-bearing NK cells in rodents and perhaps some other species, similar selective pressures might have led to the acquisition of distinct gene sequences through a process of convergent evolution [11]. Importantly, KIR and Ly49 gene sequences encode polymorphic NK receptors which can either activate or inhibit NK cell effector functions. Separate KIR and Ly49 gene clusters encoding for multiple polymorphic NK receptors hints that tight genetic linkage is potentially advantageous, and further that constant selective pressures are at work to diversify structurally distinct NK receptors for binding their polymorphic ligands. For further discussion of KIR sequence diversity and function and the existence of KIR haplotypes, please see other reviews in this issue.
Membrane-bound Ly49 receptors are displayed by subsets of NK cells in mice. Individual NK cells typically express one to several Ly49 receptor family members and thus, Ly49 receptor expression occurs in a variegated fashion. Expression of multiple polymorphic inhibitory Ly49 receptors is potentially beneficial and may favor effective NK “licensing” or “arming” by maximizing the odds for successful interactions with highly polymorphic self-MHC class I proteins in development. In addition to extending their range of self-awareness, inhibitory Ly49 receptors therefore also improve NK sensing for pathogen intrusion. On the other hand, direct stimulation for NK cells is attained through activation receptors also encoded by related Ly49 genes in the cluster. Less is known about the ligands for Ly49 activation receptors, but at least some also recognize MHC class I or MHC class I-related molecules [12–15].
NK cells in a wide range of mammalian species additionally express CD94/NKG2 (also called KLRD1/KLRC) lectin-like receptor heterodimers which can either activate or inhibit effector cell function. CD94/NKG2 receptors bind non-classical MHC class Ib ligands HLA-E and Qa1b in humans and mice respectively [10]. HLA-E and Qa1b bind and present peptide fragments derived from the leader sequences of classical MHC class I proteins; CD94/NKG2 receptors are thus well adapted to broadly monitor and sense changes in MHC class I expression. MHC class I monitoring may well represent an extremely important functional feature for CD94/NKG2 receptors; KLR-related sequences (cKLR) derived from bony (cichlid) fishes have been recently identified, a finding that underscores their importance in diverse vertebrate species [16]. Further, the cKLR sequences are actually expressed in a minor subset of blood leukocytes and also in the gill and pharyngeal jaw. Though not shown experimentally, one might speculate that cKLR sequences are expressed by killer cells in organs of the fish where innate immune effector function(s) are needed. As with CD94/NKG2 sequences in mammalian species, cKLR genes are also clustered in a narrow genomic NKC-like region [16, 17]. Altogether, there is substantial evidence indicating clustered NKR genes and gene families are an important genomic component retained in a diverse range of vertebrate species.
3. The NK gene complex (NKC): Clustered KLR genes promote natural killing effector functions and regulation of innate immunity
Yokoyama and Shevach discovered the NKC when they genetically mapped genes for two different NK receptors, NKR-P1 (also called KLRb) and Ly49, on mouse chromosome 6 [18]. NKC regions in rodents and humans (chromosome 12p13) have since been extensively studied with systematic genetic and physical mapping strategies (reviewed in [10, 19]). Early on, separate clusters for Nkrp1 and Ly49 families of genes were mapped in distinct NKC regions [20]. Nkrp1 and Ly49 genes, as with other NKC genes, encode type II membrane-bound surface receptors with one extracellular C-type lectin-like domain (CTLD) [21, 22]. Unlike other C-type lectin superfamily members, most NKC-encoded NKRs lack a functional Ca2+-dependent carbohydrate recognition domain (CRD). The C-type lectin-like domain of NK lectin-like receptors thus typically binds protein ligands instead [23].
Further physical mapping studies revealed additional CD94/NKG2 and C-type lectin domain family 2 (CLEC2) clustered genes and even greater NKC complexity [24–26]. CLEC2 proteins include C-type lectin-related (Clr) ligands Clrg and Clrb recognized by NKR-P1 family members Nkrp1f and Nkrp1d, respectively [23] and other NKC encoded members of the C-type lectin superfamily which display greater tissue distribution profiles than NK lectin-like receptors [27]. The importance of the NKR-P1:Clr recognition process in self-nonself discrimination is emphasized by the recent finding that rat cytomegalovirus encodes a Clr homolog that can specifically target the rat NKR-P1B inhibitory receptor to diminish NK cell activation and anti-viral control [28]. Over time, our understanding of the important role for the NKC gene complex has increased; NKC genes are frequently expressed by NK cells and other immune cells, they are needed in resistance to viral pathogens and malignancy, and NKC genes and regions are conserved among disparate species. More recent comprehensive DNA sequencing for the NKC regions in multiple inbred strains of mice, rats, humans and several additional species has been completed and should allow for the description of the full content of genes as well as more informative NKC comparisons among species.
Thus far, NKC-like regions have been detected in species ranging from bony fishes to humans, an indication of the vital role of the NKC in immunity and an ancient evolutionary origin of this gene cluster. A key conserved feature is clustered/linked genes for Group V (NK lectin-like receptors) and/or Group II (C-type lectin receptors) member sequences of the C-type lectin superfamily. Comparative analyses of the NKCs of humans and other primates, horses, dogs, cattle, mice and rats indicates the NKC region is highly conserved in gene orientation, order, position, structure and sequence homology [10, 27, 29]. NKC conservation and gene orthology supports a widely held view that expansion of related NKC genes and clustered gene families are the result of gene duplications and/or conversions. What is not immediately apparent, however, is the dynamic nature of the NKC and its resident genes. Further analysis of NKC regions of related and unrelated species has revealed considerable complexity and variation.
That CD94/NKG2-related cKLR genes have been identified in teleostean fishes is intriguing; this finding implies that an ancestral KLR gene might have arisen more than 365 million years ago (mya) before the appearance of tetrapods [16]. Direct sequencing of BAC clones containing cKLR sequences further revealed that the CD94/NKG2-related gene sequences are clustered together in the cichlid fish genome [17]. Though related sequences have so far remained elusive in the trout, genetic mapping strategies based on an NK-like trait (i.e. YAC-1 cytotoxicity) revealed the presence of a major quantitative trait locus (QTL) on linkage group 31 in the trout genome [30]. Whether the trait is controlled by one or potentially more sequences is not known, but similar to mammalian species this putative NK effector controlling genetic complex (a trout “NKC“) resides on a linkage group separate from those harboring the MHC or a leukocyte receptor complex (LRC).
As remarkable as a primitive NKC in teleost fishes may appear, subtle variations clearly distinguish fish and mammalian NKC regions. Though the cichlid KLR sequences are most related to mammalian CD94/NKG2 sequences, orthologous matches are not apparent. Instead, fish and mammalian KLR genes are likely to have arisen independently through duplications of distinct ancestral genes in a paralogous fashion [16, 17]. Further, the cichlid NKC might only contain genes for Group V-like activation-type KLR members, and without other CLEC-related gene sequences. Such disparity may divulge NKC complexity and also the extent of selective pressure to maintain clustered genes that can be readily manipulated for the purpose of adapting modified NK receptor sequences toward the unique environs of their hosts.
Another intriguing example is provided by chicken NKC regions, which have been mapped to distinct genomic regions. Genes for two CTLD-containing chicken proteins, B-NK and B-lec were cloned and mapped within the chicken MHC, suggesting that genes for NK lectin-like receptors and their potential ligands might be advantageously physically linked in the chicken genome [31]. For instance, physical linkage in a common haplotype might insure that a given receptor/ligand pair is maintained. Similar to mammalian inhibitory NK lectin-like receptors, the cytoplasmic tail of B-NK includes a canonical ITIM that can recruit SHP-1 and SHP-2 tyrosine phosphatases following ITIM-phosphorylation [31]. That B-NK and B-lec are more similar to mammalian NKC encoded KLR sequences than they are to other chicken C-type lectins further hints that an ancestral KLR sequence existed prior to the divergence of aves and mammals ~330 mya [31]. Curiously, a recent study identified a distinct chicken NKC region by pinpointing a chimeric CD94/NKGA-like sequence that is physically linked with chicken CD69 at an interval spanning a substantial fragment of chromosome 1 (~42-Mb) [32]. As with NKC regions of other species, important disease resistance quantitative trait loci (QTLs) are also linked with chicken NKC and NKC/MHC regions, including resistance to avian coccidiosis [33] and a herpesvirus that causes Marek’s disease [34]. That the chicken genome should encode what appear to be bona fide NK lectin-like receptors at two distinct genomic locations is very interesting and perhaps implicates yet another potential solution for NK receptors to achieve and balance self tolerance with recognition of infection and/or transformation.
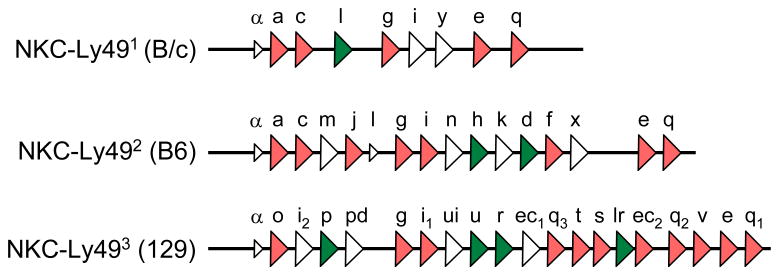
In mice and rat NKC regions, Ly49 genes are separately clustered together [20, 35]. A schematic diagram representing Ly49 genes in the B6 NKC-Ly49 haplotype (NKC-Ly492) corresponding with Yokoyama’s RFLP group 2 (see below) and based on direct sequence analysis [36, 37] is shown in Figure 1. In addition to their role in establishing NK self-tolerance, mouse Ly49 genes also are genetically linked with resistance or susceptibility to viral infection. In fact, MCMV control loci Cmv1 and Cmv3 have been pinpointed to genes for NK activation receptors Ly49H and Ly49P, respectively [13, 38–40]. Ly49Hb6 recognizes the MCMV-encoded m157 class I-like molecule expressed on infected cells to activate NK cells [12, 15], whereas the Ly49Pmamy receptor recognizes virally modified H-2Dk on infected cells [13]. Though refined genetic mapping for Ectromelia virus (ECTV) resistance locus Rmp1 and the MCMV control locus Cmv4 is still ongoing, these traits might also be explained by polymorphic NK activation receptors.
Figure 1.
Schematic diagram of representative sequenced NKC-Ly49 haplotypes [36, 37] in BALB/c, C57BL/6 and 129 inbred mouse strains corresponding with RFLP groups identified by Yokoyama and colleagues [18]. Transcriptional orientations for inhibitory (red) and stimulatory (green) Ly49 genes (triangles) are shown. Probable pseudogenes (no fill) are also depicted.
4. Evidence for express NKC alterations through expansions and contractions in KLR gene families
Inasmuch as KLR gene clustering underscores the importance for maintaining NKC linkage groups, selective expansion or contraction in particular NKR gene families increasingly draws our attention towards understanding selective pressure(s) guiding gene duplications and contractions. Plasticity in the KIR/LRC locus is a documented feature, one subject to environmental stimuli that may favor rapid change in primary KIR gene sequences or potential consequent change in function [41, 42]. Recombination between KIR genes through a process of domain shuffling could be a key mechanism for generating novel activating KIR [41, 43], although some KIR genes are the products of gene duplication and subsequent evolution through adaptive point mutations. A recent hypothesis indeed asserts that modern day KIR and KLR receptors, including inhibitory and activation (short tail) types, have descended independently from ancestral genes for inhibitory NK receptors in humans or mice, respectively [41].
KIR/LRC expansion presumably also came at the cost of human Ly49 genes. Ly49 genes have been found in the NKC regions of humans and other primates, cattle, horses and rodents. Only a single residual Ly49 pseudogene remains in the human Ly49 gene cluster and significant alterations in the NKC-Ly49 gene regions of humans and mice are instantly recognizable. On the other hand, Ly49 genes have undergone extensive expansion in some inbred strains of mice and also in the rat. Substantial variation distinguishing mouse and rat Ly49 sequences indicates how rapidly significant changes have occurred and accumulated within NKC-Ly49 regions. Indeed, rodent NKC regions are evolving at a more rapid rate and are less stable than corresponding regions of nonrodent NKCs because of higher birth and death rates of genes within the NKC [27].
Contraction in the cattle NKC-Ly49 genomic region is also evident, even though cattle do express some Ly49 molecules [44]. NKC alterations are not limited to the Ly49 region; instead cattle CD94/NKG2 genes have undergone substantial change relative to their counterparts in other mammalian species [45]. Firstly, four independent cattle CD94 sequences have been identified which display limited polymorphism in contrast with monomorphic CD94 expression in humans and other primates. Birch and Ellis also identified many additional cattle NKGC or NKG2A gene sequences, including one ITIM-containing NKG2A sequence with a basic amino acid in its transmembrane domain and the potential to either inhibit or stimulate effector functions [45]. CD94/NKG2 receptor expansion in the cattle genome implies that their ligands may be more diverse than in other species. A further expectation here is that natural selection has driven expansion of the CD94/NKG2 gene cluster in cattle and thus might also be genetically linked with survival trait variance. Cluster diversity could also be balanced together with KIR/LRC diversity since cattle also express KIR genes.
Interestingly, KIR and Ly49 sequence diversity is a distinguishing feature for NK receptor genes in the horse genome [29]. However, only dysfunctional KIR transcript sequences have so far been identified, suggesting that residual horse KIR genes are pseudogenes. In contrast, multiple functional Ly49 genes encoding horse NK lectin-like receptors have been mapped to chromosome 6q13, a region syntenic with the human NKC on chromosome 12 [29]. Because horse Ly49 genes are all more related to one another than to rodent Ly49 sequences, it suggests that separate Ly49 gene clusters apparently arose independently in rodents and horses, further highlighting the importance and a potential need for clustered NKR genes.
5. Evidence for the existence of NKC haplotypes in mice: (i) NKC haplotypes in inbred mice
Yokoyama and colleagues first appreciated the extent of NKC gene polymorphism in several inbred strains of mice by their distinct restriction fragment length polymorphism (RFLP) groups for Ly49 as well as NKRP1 genes [18, 46]. Genetic mapping strategies with microsatellite markers and other genetic probes extended these earlier findings and established that NKC regions of different inbred mouse strains exist as genetically diverse haplotypes [47, 48]. Indeed, microsatellite-typing revealed common NKC haplotypes in inbred strains corresponding with the prior RFLP analysis. For example, NKC haplotypes in inbred strains A, AKR, BALB/c, C3H, CBA and DBA/2 are highly related and fell into similar microsatellite and RFLP groups for Ly49 (NKC-Ly491) and Nkrp1 (NKC-Nkrp11) gene cluster regions. On the other hand, NKC haplotypes in B6 and 129 are distinct and belong to RFLP groups 2 (NKC-Ly492) or 3 (NKC-Ly493), respectively (Fig. 1). Further studies demonstrated polymorphisms among inbred strains at the level of allelism for specific Ly49 genes such as Ly49A and Ly49C [49–52]. Ly49Cb6 and Ly49Hb6 sequences are only 78% homologous overall, but they exhibit striking homology in the downstream exons coding for the CTLD, thus providing an early indication that evolution may have taken place through a process of recombination between ancestral genes [52]. Analysis of cDNA sequences from the 129/J mouse strain indicated that its NKC contains many full-length Ly49-related coding sequences (Ly49e, g, I, o, p, r, s, t, u, and v). Only Ly49e129, however, proved to be identical to Ly49eb6 [53], with the remaining sequences exhibiting 85.2 to 95.9% sequence homology to B6 Ly49 genes. This provided additional evidence that certain genes present in both the B6 and 129 genomes may represent alleles of the same gene, or at least be ancestrally related based on sequence similarities.
(ii) NKC haplotypes and Ly49 genes in wild mice
An analysis of the degree of natural variation of the NKC region of wild mice revealed that within two geographically distinct populations trapped in Australia there was considerable variability in NKC associated alleles [54]. However, as observed for inbred mouse strains, there was also evidence supporting the concept of the existence of NKC haplotypes shared among individual mice within these populations [54]. Experimental MCMV infection of a specific-pathogen-free population of outbred wild mice with the K181 MCMV laboratory strain revealed that low viral titers in the spleen, which are normally associated with the presence of NK-dependent, Cmv1r-like resistance, were uncommon among this population. Thus, NK cell resistance mechanisms based on Ly49Hb6-like activation receptors binding to an MCMV-encoded m157smith-like ligand are likely to be rare in this population. This was further supported by a study in Adam et al., [55] where they screened six Mus musculus wild-derived inbred strains and found that only one of the six strains, PWK/Pas, exhibited NK cell-dependent resistance to MCMV infection.
In a preliminary analysis of Ly49 gene heterogeneity among a population of wild mice trapped in Australia, we focused on genetic sequences of Ly49 genes belonging to the Ly49C-related and Ly49H-related groups described by Anderson et al. [36]. Of the cDNAs sequenced many were Ly49Cnzb-related (>97% homology) or Ly49I129-related (>95% homology) (Corbett et al, unpublished observations), though a significant proportion of the clones appeared to be pseudogenes. Among the Ly49H-related genes, Ly49Hnzw (>95% homology) and Ly49U129-like (>95% homology) genes were detected, together with related pseudogenes. Only Ly49Hb6-like pseudogenes were detected in this population. Interestingly, Ly49Pmamy-like sequences were also present (Corbett et al, unpublished observations). This preliminary analysis of wild mice has recapitulated findings from inbred mice that significant heterogeneity exists among Ly49 genes [36, 56, 57] and that pseudogenes for both activating and inhibitory receptors are frequent. These data reflect our previous analysis of allelism and haplotypes in wild mice [54] and also indicate that intact Ly49Hb6-like genes are absent or at the very least infrequent.
6. Extensive polymorphism and evolution of the rodent Ly49 regions
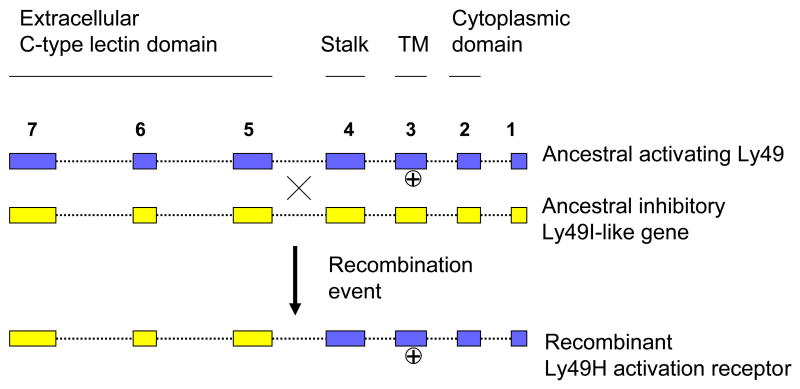
The above studies provided important insights into the notion of NKC polymorphisms, especially for the Ly49 region. Physical maps and full length sequences for the Ly49 region from several inbred mouse strains were needed to fully appreciate evolutionary origins and relationships of individual Ly49 genes and/or alleles. Partial physical maps of the B6 Ly49 region had in part been determined based on YAC, BAC and P1 bacteriophage contigs [20, 58, 59]. Analysis of sequence data available from public databases for the B6 Ly49 region [57] revealed that excluding Ly49b which resides approximately 800 kb telomeric [20, 58], the main centromeric Ly49 cluster comprises 14 genes as well as a number of gene fragments. The protein coding genes, four non-coding pseudogenes and gene fragments including Ly49α and Ly49l are shown in Figure 1. Interestingly, the Ly49l gene in CBA and BALB/c mice codes for an activating receptor [14, 60]. Comparisons of sequence similarities and repetitive elements shared between genes suggest that NKC gene blocks Ly49n-i-g and Ly49m-c-a were probably acquired through duplications from an ancestral gene block. Similarly, the adjacent Ly49n, h, and k genes are highly related to one another and are also thought to have arisen by a gene duplication process. The high degree of similarity of the C-terminal lectin-like domain of Ly49h to that of the inhibitory Ly49i gene suggested that Ly49h may have evolved by a gene conversion or recombination event [52, 57, 61]. In this process, it has been proposed that exons coding for the extracellular domain of a Ly49i-like ancestral gene fused to the exons coding for the transmembrane and intracellular domains of an ancestral activation receptor (Fig. 2).
Figure 2.
Model for gene conversion to give rise to the Ly49H activation receptor. Through a process of recombination between an ancestral activating gene and a Ly49I-like inhibitory gene, the exons encoding the extracellular CTLD of the Ly49I-like gene were fused to the exons encoding the cytoplasmic, stalk and TM domains of the ancestral activating gene.
Direct comparisons between the B6 Ly49 region and that of the 129 mouse strain have been carried out based on BAC contigs [56] and direct sequence comparisons [37]. Excluding the Ly49b gene, the 129 Ly49 cluster spans ~600 kb [56]. Based on comparison of the B6 and 129 NKC-Ly49 gene clusters, it seems probable that Ly49e129, Ly49g129 and Ly49i1129 represent alleles for genes of the same designation in the B6 strain. These and the following Ly49 alleles pairs: Ly49fb6 and Ly49s129; Ly49db6 and Ly49r129; and Ly49hb6 and Ly49u129 together may represent framework genes handed down from an ancestral mouse common to both B6 and 129 [37, 56].
Similar comparative sequence analyses for the BALB/c Ly49 region revealed its 300-kb length and seven intact genes and a pseudogene (Ly49i) [36, 62]. Since Ly49y expression has so far not been detected, it might also be a pseudogene. The BALB/c NKC-Ly49 cluster is much smaller than that of B6 and 129 strains, potentially representing a minimal Ly49 haplotype, which lacks Ly49h- and Ly49d-like genes. Further comparison of the BALB/c, B6 and 129 NKC-Ly49 haplotypes provided further evidence that key conserved genes are likely important framework genes in mice. These are represented by the three gene pairs: Ly49a and c, Ly49g and i, and Ly49e and q [36, 62].
Based on analyses of the distribution and content of repetitive elements in individual Ly49 genes and between genes, models of temporal evolution have been deduced for members of the gene family. These studies further indicate how the particular Ly49 haplotypes present in the inbred strains analyzed may have originated from putative ancestral Ly49 haplotypes [36, 57]. Moreover, the high frequency of pseudogenes together with evidence of homologies among genes (e.g. Ly49C and -I) and of evolution via exon shuffling and recombination between ancestral genes implies that Ly49 genes are likely to be subject to pressures that result in a high rate of birth and death. Curiously however, recombination within the Ly49 gene cluster proper must be exceedingly rare since it was not detected by us in over 2722 meioses, but instead occurred only in flanking recombination hotspots [63]. Likewise, Vidal and colleagues have reported a similar outcome in analyzing 2,657 meioses [13, 64]. Altogether, we still lack a clear understanding of the mechanism responsible for the dynamic nature of the NK gene complex including what underlies its genetic changes and NK cell receptor diversity.
Interestingly, the Ly49q and e genes which reside at the centromeric end of the Ly49 region, together with the more distant telomeric Ly49b gene are conserved among all the inbred strains assessed and share a very high degree of identity between strains [36, 57, 62]. This high degree of conservation likely reflects functional constraints on the protein products encoded by these genes. It is remarkable that they all show expression patterns that are distinct from the core Ly49 proteins. Ly49E, unlike other Ly49 proteins is expressed on fetal NK cells [65]. Ly49B and Ly49Q proteins are not expressed by NK cells, but are preferentially expressed by myeloid lineage cells [66, 67], and indeed in the case of Ly49Q is expressed by IFN-α/β producing plasmacytoid dendritic cells (pDC) [68, 69] and binding of H-2Kb MHC class I molecules by Ly49Q on pDC results suggests a role for Ly49Q in regulating pDC function [70]. Also intriguing, rats have Ly49 genes that are highly similar to Ly49b and Ly49q suggesting that the origin of these genes predates the divergence of rats and mice from an ancestral rodent [35, 71].
Evidence from the rat Ly49 family on rat chromosome 4 has also provided important insights into possible mechanisms of Ly49 evolution. The presence of large numbers of Ly49 genes in rats (34 in the Brown Norway strain, [35]) indicates that the rapid expansion in numbers of Ly49 genes took place by both tandem and genomic block duplication [35, 71]. Rat Ly49-2, -3, -4, -5, and -6 are arranged in a block and show the highest degree of similarity to one another than to other Ly49 genes and it is proposed that they arose by tandem duplication. The block of genes comprising Ly49-23 to -26 is very closely related to the block Ly49-28 to -31 based on the arrangements of repetitive elements within and between genes and these blocks are to have arisen by block duplication [71].
7. Dynamics of the host-virus relationship – evolution of host activating receptors and cognate viral ligands
The preceding discussion indicates a complex pattern of polymorphism and dynamic evolution of the Ly49 region. Abi-Rached and Parham [41] provided compelling evolutionary evidence that activating Ly49 receptors have evolved from inhibitory homologs. Wilhelm et al [57] also provided specific models for the evolution of Ly49Hb6 from ancestral genes that have similarity in the extracellular domain to Ly49C, Ly49I and Ly49J. Furthermore, Arase et al, [12] hypothesized that the gene encoding the Ly49H activation receptor may have evolved from genes encoding inhibitory molecules (i.e. Ly49I). While the latter hypothesis is likely correct, several observations suggest that the evolutionary utility for this individual host NK cell recognition process in terms of overall population-wide resistance to MCMV infection may be of limited value. The Ly49U activation receptor from the 129/J strain which is collinear with Ly49Hb6 in the Ly49 region [37, 56] has a highly similar CTLD to that of Ly49Hb6 and is recognized by Ly49C/H/I/U monoclonal antibodies (mAb) such as 1F8 [53]. However, it does not bind to MCMV m157Smith and this results in susceptibility of this mouse strain. Likewise, Ly49Hnzw does not bind m157 even though NZW NK cells are recognized by the Ly49H-specific mAb 3D10 [73]. Nevertheless, NZW NK cells use a Ly49H-independent mechanism to limit MCMV infection. Hence, if Ly49Hb6, Ly49U129 and Ly49Hnzw have a common ancestral origin, the retention of binding capacity to the MCMV m157 molecule in Ly49U129 or Ly49Hnzw has not been conserved.
Passage of MCMV through mouse strains that express the Ly49H activation receptor places sufficient selective pressure on the virus to rapidly mutate which indicates that NK cell surveillance can induce a strong selective pressure for viral evolution [74–76]. As activating Ly49 molecules related to the Ly49H-like grouping of NKC receptors are present in a number of inbred strains [56, 57, 73] and outbred mice (unpublished data, Corbett, Forbes and Scalzo), it may be that individual MCMV strains circulating through mouse populations are subjected to NK cell pressure selecting for sequence variants of m157 that do not directly bind to Ly49H-like activating NK cell receptors. Indeed, sequence analysis of m157 from isolates of MCMV derived from wild mice trapped in Australia indicated this gene was highly variable [76]. Analysis of replication of the m157 variant isolates in Ly49H+ and Ly49H− mouse strains showed that many grew to high levels in Ly49H+ mice. When these m157 sequence variants were tested for their ability to stimulate activation of Ly49-expressing reporter cells, only two isolates G1F and N5, with the highest degree of similarity to the Smith m157 sequence, could activate these reporter cells [76]. These data indicated that the majority of wild-derived MCMV isolates contain m157 sequences that do not bind Ly49Hb6 and hence the benefit of expression of a Ly49Hb6 receptor by an individual mouse might be low. Studies are warranted to determine if in individual wild mice correlations exist between the MCMV m157 sequences present and Ly49 activation receptors that bind those specific sequences.
8. Specificities of activation receptors for classical MHC class I molecules: a role for viruses to modify class I
While the specificity of Ly49H for the MCMV-encoded m157 molecule has provided important insights into NK cell recognition processes, the identification of host ligands recognized by activating Ly49 molecules has not been as exhaustive as for inhibitory Ly49 receptors [14]. In contrast to the well-defined specificity of Ly49H for the MCMV-encoded m157 molecule, conflicting evidence has been presented on the binding specificity of Ly49D. A number of studies have shown that Ly49Db6 interacts with H-2Dd molecules [77–79] and similarly that the related Ly49Pnod binds H-2Dd [80]. Furthermore, studies of the Ly49D-related receptor Ly49R129 also showed moderate binding to H-2Dd, H-2Dk and H-2Ld tetramers [53]. In B6 NK cells that co-express Ly49Db6 and Ly49Gb6 (that also recognizes H-2Dd) inhibition is dominant [81], possibly due to a lower avidity of Ly49Db6 than Ly49Gb6 for H-2Dd. Other studies have not confirmed Ly49Db6 binding of mouse H-2Dd and instead have presented evidence that the receptor binds to hamster MHC class I xenoantigens [82]. It may be that the affinity of the interaction of activating Ly49 molecules with MHC class I is an important consideration; it has been speculated that modification of the class I molecule by infection may be required for optimal recognition by these activation receptors [81]. Recently, evidence has been presented suggesting that Ly49Pmamy activation receptors bind H-2Dk on MCMV-infected cells and thus confer NK-mediated H-2k protection in MCMV-infected mice [13, 83, 84]. This suggests that the H-2Dk class I molecules may be modified in some way perhaps through expression of a viral peptide or by binding of a virally encoded protein to facilitate interaction with the Ly49Pmamy molecule. It will be of interest to determine if this virally modified, class I molecule-dependent mechanism of NK cell recognition of virus-infected cells by Ly49 activation receptors operates for other MCMV NK-mediated resistance mechanisms such as Cmv4-mediated resistance of the PWK/Pas wild-derived mouse strain [55] and for other mouse-specific viruses. This might provide important insights into the evolution of the function and specificity of activating Ly49 receptors. Indeed, our recent studies on genetically determined resistance to Ectromelia virus (ECTV) have shown that in addition to the NKG2D receptor playing a role in NK cell control of ECTV [85] (Scalzo et al, submitted), the Ly49H and Ly49D activation receptors act in concert to effect NK cell-mediated resistance to ECTV (Scalzo et al, submitted).
Lessons learned from the murine model regarding the role of NK cell activation receptors in recognition of viruses might have important parallels in humans. Indeed, recent data has shown that NK cells expressing the KIR3DS1 activation receptor preferentially lyse HIV-1-infected cells expressing the class I HLA-B Bw4-801 molecule [86], so similar mechanisms of NK cell recognition might operate in both humans and rodents and have been selected for by convergent evolution.
9. Concluding Remarks
The NKC region, while being highly heterogeneous among animal species conserves a number of key genes, gene families and gene order and points to the ancient origin in this genetic region among ancestral vertebrates. Some gene lineages in certain species such as the Ly49, Nkrp1 and CLEC families have undergone significant expansion [27]. The data reviewed here pertaining to the interactions of host Ly49 molecules with virally infected cells particularly in the mouse model points to several potential mechanisms by which NK cell activation receptors may have been selected to counter virus infection. However, further work is required to determine if other viral infectious agents may be driving NKR evolution, particularly that of activating Ly49 receptors, and also for human KIR receptors and their interactions with virally infected cells.
Acknowledgments
The authors acknowledge the support of the National Heath and Medical Research Council of Australia (NH&MRC) and the National Institutes of Health (AI50072 to MGB). The authors also thank Jerome Coudert, Koho Iizuka and Michael Stadnisky for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–42. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Fernandez NC, Treiner E, Vance RE, Jamieson AM, Lemieux S, Raulet DH. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood. 2005;105:4416–23. doi: 10.1182/blood-2004-08-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–13. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 4.Kärre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–8. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 5.Reddehase MJ. Antigens and immunoevasins: opponents in cytomegalovirus immune surveillance. Nature Reviews Immunology. 2002;2:831–44. doi: 10.1038/nri932. [DOI] [PubMed] [Google Scholar]
- 6.Gasser S, Raulet DH. Activation and self-tolerance of natural killer cells. Immunol Rev. 2006;214:130–42. doi: 10.1111/j.1600-065X.2006.00460.x. [DOI] [PubMed] [Google Scholar]
- 7.Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Ann Rev Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 8.Orange JS. Human natural killer cell deficiencies and susceptibility to infection. Microbes Infect. 2002;4:1545–58. doi: 10.1016/s1286-4579(02)00038-2. [DOI] [PubMed] [Google Scholar]
- 9.Scalzo AA, Corbett AJ, Rawlinson WD, Scott GM, Degli-Esposti MA. The interplay between host and viral factors in shaping the outcome of cytomegalovirus infection. Immunol Cell Biol. 2007;85:46–54. doi: 10.1038/sj.icb.7100013. [DOI] [PubMed] [Google Scholar]
- 10.Yokoyama WM, Plougastel BF. Immune functions encoded by the natural killer gene complex. Nature Rev Immunol. 2003;3:304–16. doi: 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]
- 11.Barten R, Torkar M, Haude A, Trowsdale J, Wilson MJ. Divergent and convergent evolution of NK-cell receptors. Trends Immunol. 2001;22:52–7. doi: 10.1016/s1471-4906(00)01802-0. [DOI] [PubMed] [Google Scholar]
- 12.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–6. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 13.Desrosiers MP, Kielczewska A, Loredo-Osti JC, Adam SG, Makrigiannis AP, Lemieux S, et al. Epistasis between mouse Klra and major histocompatibility complex class I loci is associated with a new mechanism of natural killer cell-mediated innate resistance to cytomegalovirus infection. Nature Genet. 2005;37:593–9. doi: 10.1038/ng1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kane KP, Silver ET, Hazes B. Specificity and function of activating Ly-49 receptors. Immunol Rev. 2001;181:104–14. doi: 10.1034/j.1600-065x.2001.1810108.x. [DOI] [PubMed] [Google Scholar]
- 15.Smith HR, Heusel JW, Mehta IK, Kim S, Dorner BG, Naidenko OV, et al. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci U S A. 2002;99:8826–31. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato A, Mayer WE, Overath P, Klein J. Genes encoding putative natural killer cell C-type lectin receptors in teleostean fishes. Proc Natl Acad Sci U S A. 2003;100:7779–84. doi: 10.1073/pnas.1235938100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kikuno R, Sato A, Mayer WE, Shintani S, Aoki T, Klein J. Clustering of C-type lectin natural killer receptor-like loci in the bony fish Oreochromis niloticus. Scand J Immunol. 2004;59:133–42. doi: 10.1111/j.0300-9475.2004.01372.x. [DOI] [PubMed] [Google Scholar]
- 18.Yokoyama WM, Kehn PJ, Cohen DI, Shevach EM. Chromosomal location of the Ly-49 (A1, YE1/48) multigene family. Genetic association with the NK1.1 antigen. J Immunol. 1990;145:2353–8. [PubMed] [Google Scholar]
- 19.Brown MG, Scalzo AA, Matsumoto K, Yokoyama WM. The natural killer gene complex: a genetic basis for understanding natural killer cell function and innate immunity. Immunol Rev. 1997;155:53–65. doi: 10.1111/j.1600-065x.1997.tb00939.x. [DOI] [PubMed] [Google Scholar]
- 20.Brown MG, Fulmek S, Matsumoto K, Cho R, Lyons PA, Levy ER, et al. A 2-Mb YAC contig and physical map of the natural killer gene complex on mouse chromosome 6. Genomics. 1997;42:16–25. doi: 10.1006/geno.1997.4721. [DOI] [PubMed] [Google Scholar]
- 21.Weis WI, Taylor ME, Drickamer K. The C-type lectin superfamily in the immune system. Immunol Rev. 1998;163:19–34. doi: 10.1111/j.1600-065x.1998.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 22.Zelensky AN, Gready JE. The C-type lectin-like domain superfamily. Febs J. 2005;272:6179–217. doi: 10.1111/j.1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]
- 23.Iizuka K, Naidenko OV, Plougastel BF, Fremont DH, Yokoyama WM. Genetically linked C-type lectin-related ligands for the NKRP1 family of natural killer cell receptors. Nat Immunol. 2003;4:801–7. doi: 10.1038/ni954. [DOI] [PubMed] [Google Scholar]
- 24.Ho EL, Heusel JW, Brown MG, Matsumoto K, Scalzo AA, Yokoyama WM. Murine NKG2-D and CD94 are clustered within the natural killer complex and are expressed independently in NK cells. Proceedings of the National Academy of Sciences USA; 1998. pp. 6320–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plougastel B, Dubbelde C, Yokoyama WM. Cloning of Clr, a new family of lectin-like genes localized between mouse NKrp1a and Cd69. Immunogenetics. 2001;53:209–14. doi: 10.1007/s002510100319. [DOI] [PubMed] [Google Scholar]
- 26.Vance RE, Jamieson AM, Raulet DH. Recognition of the class Ib molecule Qa-1(b) by putative activating receptors CD94/NKG2C and CD94/NKG2E on mouse natural killer cells. J Exp Med. 1999;190:1801–12. doi: 10.1084/jem.190.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hao L, Klein J, Nei M. Heterogeneous but conserved natural killer receptor gene complexes in four major orders of mammals. Proc Natl Acad Sci U S A. 2006;103:3192–7. doi: 10.1073/pnas.0511280103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voigt S, Mesci A, Ettinger J, Fine JH, Chen P, Chou W, et al. Cytomegalovirus evasion of innate immunity by subversion of the NKR-P1B:Clr-b missing-self axis. Immunity. 2007;26:617–27. doi: 10.1016/j.immuni.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi T, Yawata M, Raudsepp T, Lear TL, Chowdhary BP, Antczak DF, et al. Natural killer cell receptors in the horse: evidence for the existence of multiple transcribed LY49 genes. Eur J Immunol. 2004;34:773–84. doi: 10.1002/eji.200324695. [DOI] [PubMed] [Google Scholar]
- 30.Zimmerman AM, Evenhuis JP, Thorgaard GH, Ristow SS. A single major chromosomal region controls natural killer cell-like activity in rainbow trout. Immunogenetics. 2004;55:825–35. doi: 10.1007/s00251-004-0645-6. [DOI] [PubMed] [Google Scholar]
- 31.Rogers SL, Gobel TW, Viertlboeck BC, Milne S, Beck S, Kaufman J. Characterization of the chicken C-type lectin-like receptors B-NK and B-lec suggests that the NK complex and the MHC share a common ancestral region. J Immunol. 2005;174:3475–83. doi: 10.4049/jimmunol.174.6.3475. [DOI] [PubMed] [Google Scholar]
- 32.Chiang HI, Zhou H, Raudsepp T, Jesudhasan PR, Zhu JJ. Chicken CD69 and CD94/NKG2-like genes in a chromosomal region syntenic to mammalian natural killer gene complex. Immunogenetics. 2007;59:603–11. doi: 10.1007/s00251-007-0220-z. [DOI] [PubMed] [Google Scholar]
- 33.Zhu JJ, Lillehoj HS, Allen PC, Van Tassell CP, Sonstegard TS, Cheng HH, et al. Mapping quantitative trait loci associated with resistance to coccidiosis and growth. Poult Sci. 2003;82:9–16. doi: 10.1093/ps/82.1.9. [DOI] [PubMed] [Google Scholar]
- 34.Kaufman J. The simple chicken major histocompatibility complex: life and death in the face of pathogens and vaccines. Philos Trans R Soc Lond B Biol Sci. 2000;355:1077–84. doi: 10.1098/rstb.2000.0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nylenna O, Naper C, Vaage JT, Woon PY, Gauguier D, Dissen E, et al. The genes and gene organization of the Ly49 region of the rat natural killer cell gene complex. Eur J Immunol. 2005;35:261–72. doi: 10.1002/eji.200425429. [DOI] [PubMed] [Google Scholar]
- 36.Anderson SK, Dewar K, Goulet ML, Leveque G, Makrigiannis AP. Complete elucidation of a minimal class I MHC natural killer cell receptor haplotype. Genes Immun. 2005;6:481–92. doi: 10.1038/sj.gene.6364232. [DOI] [PubMed] [Google Scholar]
- 37.Makrigiannis AP, Patel D, Goulet ML, Dewar K, Anderson SK. Direct sequence comparison of two divergent class I MHC natural killer cell receptor haplotypes. Genes Immun. 2005;6:71–83. doi: 10.1038/sj.gene.6364154. [DOI] [PubMed] [Google Scholar]
- 38.Brown MG, Dokun AO, Heusel JW, Smith HRC, Beckman DL, Blattenberger EA, et al. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science. 2001;292:934–7. doi: 10.1126/science.1060042. [DOI] [PubMed] [Google Scholar]
- 39.Daniels KA, Devora G, Lai WC, O'Donnell CL, Bennett M, Welsh RM. Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J Exp Med. 2001;194:29–44. doi: 10.1084/jem.194.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SH, Girard S, Macina D, Busa M, Zafer A, Belouchi A, et al. Susceptibility to mouse cytomegalovirus is associated with deletion of an activating natural killer cell receptor of the C-type lectin superfamily. Nature Genet. 2001;28:42–5. doi: 10.1038/ng0501-42. [DOI] [PubMed] [Google Scholar]
- 41.Abi-Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J Exp Med. 2005;201:1319–32. doi: 10.1084/jem.20042558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A. 2000;97:4778–83. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajalingam R, Parham P, Abi-Rached L. Domain shuffling has been the main mechanism forming new hominoid killer cell Ig-like receptors. J Immunol. 2004;172:356–69. doi: 10.4049/jimmunol.172.1.356. [DOI] [PubMed] [Google Scholar]
- 44.McQueen KL, Wilhelm BT, Harden KD, Mager DL. Evolution of NK receptors: a single Ly49 and multiple KIR genes in the cow. Eur J Immunol. 2002;32:810–7. doi: 10.1002/1521-4141(200203)32:3<810::AID-IMMU810>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 45.Birch J, Ellis SA. Complexity in the cattle CD94/NKG2 gene families. Immunogenetics. 2007;59:273–80. doi: 10.1007/s00251-006-0189-z. [DOI] [PubMed] [Google Scholar]
- 46.Yokoyama WM, Ryan JC, Hunter JJ, Smith HRC, Stark M, Seaman WE. cDNA cloning of mouse NKR-P1 and genetic linkage with Ly-49: identification of a natural killer cell gene complex on mouse chromosome 6. J Immunol. 1991;147:3229–36. [PubMed] [Google Scholar]
- 47.Brown MG, Scalzo AA, Stone LR, Clark PY, Du Y, Palanca B, et al. Natural killer gene complex (Nkc) allelic variability in inbred mice: evidence for Nkc haplotypes. Immunogenetics. 2001;53:584–91. doi: 10.1007/s002510100365. [DOI] [PubMed] [Google Scholar]
- 48.Lee SH, Gitas J, Zafer A, Lepage P, Hudson TJ, Belouchi A, et al. Haplotype mapping indicates two independent origins for the Cmv1(s) susceptibility allele to cytomegalovirus infection and refines its localization within the Ly49 cluster. Immunogenetics. 2001;53:501–5. doi: 10.1007/s002510100359. [DOI] [PubMed] [Google Scholar]
- 49.Brennan J, Mahon G, Mager DL, Jefferies WA, Takei F. Recognition of class I major histocompatibility complex molecules by Ly-49: specificities and domain interactions. J Exp Med. 1996;183:1553–9. doi: 10.1084/jem.183.4.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Held W, Roland J, Raulet DH. Allelic exclusion of Ly49-family genes encoding class I MHC-specific receptors on NK cells. Nature. 1995;376:355–8. doi: 10.1038/376355a0. [DOI] [PubMed] [Google Scholar]
- 51.Sundbäck J, Karre K, Sentman CL. Cloning of minimally divergent allelic forms of the natural killer (NK) receptor Ly-49C, differentially controlled by host genes in the MHC and NK gene complexes. J Immunol. 1996;157:3936–42. [PubMed] [Google Scholar]
- 52.Takei F, Brennan J, Mager DL. The Ly-49 family: genes, proteins and recognition of class I MHC. Immunol Rev. 1997;155:67–77. doi: 10.1111/j.1600-065x.1997.tb00940.x. [DOI] [PubMed] [Google Scholar]
- 53.Makrigiannis AP, Pau AT, Saleh A, Winkler-Pickett R, Ortaldo JR, Anderson SK. Class I MHC-binding characteristics of the 129/J Ly49 repertoire. J Immunol. 2001;166:5034–43. doi: 10.4049/jimmunol.166.8.5034. [DOI] [PubMed] [Google Scholar]
- 54.Scalzo AA, Manzur M, Forbes CA, Brown MG, Shellam GR. NK gene complex haplotype variability and host resistance alleles to murine cytomegalovirus in wild mouse populations. Immunol Cell Biol. 2005;83:144–9. doi: 10.1111/j.1440-1711.2005.01311.x. [DOI] [PubMed] [Google Scholar]
- 55.Adam SG, Caraux A, Fodil-Cornu N, Loredo-Osti JC, Lesjean-Pottier S, Jaubert J, et al. Cmv4, a new locus linked to the NK cell gene complex, controls innate resistance to cytomegalovirus in wild-derived mice. J Immunol. 2006;176:5478–85. doi: 10.4049/jimmunol.176.9.5478. [DOI] [PubMed] [Google Scholar]
- 56.Makrigiannis AP, Pau AT, Schwartzberg PL, McVicar DW, Beck TW, Anderson SK. A BAC contig map of the Ly49 gene cluster in 129 mice reveals extensive differences in gene content relative to C57BL/6 mice. Genomics. 2002;79:437–44. doi: 10.1006/geno.2002.6724. [DOI] [PubMed] [Google Scholar]
- 57.Wilhelm BT, Gagnier L, Mager DL. Sequence analysis of the Ly49 cluster in C57BL/6 mice: a rapidly evolving multigene family in the immune system. Genomics. 2002;80:646–61. doi: 10.1006/geno.2002.7004. [DOI] [PubMed] [Google Scholar]
- 58.Depatie C, Lee SH, Stafford A, Avner P, Belouchi A, Gros P, et al. Sequence-ready BAC contig, physical, and transcriptional map of a 2-Mb region overlapping the mouse chromosome 6 host-resistance locus Cmv1. Genomics. 2000;66:161–74. doi: 10.1006/geno.2000.6186. [DOI] [PubMed] [Google Scholar]
- 59.McQueen KL, Freeman JD, Takei F, Mager DL. Localization of five new ly49 genes, including three closely related to ly49c. Immunogenetics. 1998;48:174–83. doi: 10.1007/s002510050421. [DOI] [PubMed] [Google Scholar]
- 60.Makrigiannis AP, Etzler J, Winkler-Pickett R, Mason A, Ortaldo JR, Anderson SK. Identification of the Ly49L protein: evidence for activating counterparts to inhibitory Ly49 proteins. J Leukoc Biol. 2000;68:765–71. [PubMed] [Google Scholar]
- 61.Arase H, Lanier LL. Specific recognition of virus-infected cells by paired NK receptors. Rev Med Virol. 2004;14:83–93. doi: 10.1002/rmv.422. [DOI] [PubMed] [Google Scholar]
- 62.Proteau MF, Rousselle E, Makrigiannis AP. Mapping of the BALB/c Ly49 cluster defines a minimal natural killer cell receptor gene repertoire. Genomics. 2004;84:669–77. doi: 10.1016/j.ygeno.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 63.Scalzo AA, Wheat R, Dubbelde C, Stone L, Clark P, Du Y, et al. Molecular genetic characterization of the distal NKC recombination hotspot and putative murine CMV resistance control locus. Immunogenetics. 2003;55:370–8. doi: 10.1007/s00251-003-0591-8. [DOI] [PubMed] [Google Scholar]
- 64.Depatie C, Muise E, Lepage P, Gros P, Vidal SM. High-resolution linkage map in the proximity of the host resistance locus Cmv1. Genomics. 1997;39:154–63. doi: 10.1006/geno.1996.4498. [DOI] [PubMed] [Google Scholar]
- 65.Van Beneden K, Stevenaert F, De Creus A, Debacker V, De Boever J, Plum J, et al. Expression of Ly49E and CD94/NKG2 on fetal and adult NK cells. J Immunol. 2001;166:4302–11. doi: 10.4049/jimmunol.166.7.4302. [DOI] [PubMed] [Google Scholar]
- 66.Gays F, Aust JG, Reid DM, Falconer J, Toyama-Sorimachi N, Taylor PR, et al. Ly49B is expressed on multiple subpopulations of myeloid cells. J Immunol. 2006;177:5840–51. doi: 10.4049/jimmunol.177.9.5840. [DOI] [PubMed] [Google Scholar]
- 67.Toyama-Sorimachi N, Tsujimura Y, Maruya M, Onoda A, Kubota T, Koyasu S, et al. Ly49Q, a member of the Ly49 family that is selectively expressed on myeloid lineage cells and involved in regulation of cytoskeletal architecture. Proc Natl Acad Sci U S A. 2004;101:1016–21. doi: 10.1073/pnas.0305400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kamogawa-Schifter Y, Ohkawa J, Namiki S, Arai N, Arai K, Liu Y. Ly49Q defines 2 pDC subsets in mice. Blood. 2005;105:2787–92. doi: 10.1182/blood-2004-09-3388. [DOI] [PubMed] [Google Scholar]
- 69.Toyama-Sorimachi N, Omatsu Y, Onoda A, Tsujimura Y, Iyoda T, Kikuchi-Maki A, et al. Inhibitory NK receptor Ly49Q is expressed on subsets of dendritic cells in a cellular maturation- and cytokine stimulation-dependent manner. J Immunol. 2005;174:4621–9. doi: 10.4049/jimmunol.174.8.4621. [DOI] [PubMed] [Google Scholar]
- 70.Tai LH, Goulet ML, Belanger S, Troke AD, St-Laurent AG, Mesci A, et al. Recognition of H-2K(b) by Ly49Q suggests a role for class Ia MHC regulation of plasmacytoid dendritic cell function. Mol Immunol. 2007;44:2638–46. doi: 10.1016/j.molimm.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 71.Hao L, Nei M. Genomic organization and evolutionary analysis of Ly49 genes encoding the rodent natural killer cell receptors: rapid evolution by repeated gene duplication. Immunogenetics. 2004;56:343–54. doi: 10.1007/s00251-004-0703-0. [DOI] [PubMed] [Google Scholar]
- 72.Scalzo AA, Lyons PA, Fitzgerald NA, Forbes CA, Yokoyama WM, Shellam GR. Genetic mapping of Cmv1 in the region of mouse chromosome 6 encoding the NK gene complex-associated loci Ly49 and musNKR-P1. Genomics. 1995;27:435–41. doi: 10.1006/geno.1995.1074. [DOI] [PubMed] [Google Scholar]
- 73.Rodriguez M, Sabastian P, Clark P, Brown MG. Cmv1-independent antiviral role of NK cells revealed in murine cytomegalovirus-infected New Zealand white mice. J Immunol. 2004;173:6312–8. doi: 10.4049/jimmunol.173.10.6312. [DOI] [PubMed] [Google Scholar]
- 74.French AR, Pingel JT, Kim S, Yang L, Yokoyama WM. Rapid emergence of escape mutants following infection with murine cytomegalovirus in immunodeficient mice. Clin Immunol. 2005;115:61–9. doi: 10.1016/j.clim.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 75.French AR, Pingel JT, Wagner M, Bubic I, Yang L, Kim S, et al. Escape of mutant double-stranded DNA virus from innate immune control. Immunity. 2004;20:747–56. doi: 10.1016/j.immuni.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 76.Voigt V, Forbes CA, Tonkin JN, Degli-Esposti MA, Smith HR, Yokoyama WM, et al. Murine cytomegalovirus m157 mutation and variation leads to immune evasion of natural killer cells. Proc Natl Acad Sci USA. 2003;100:13483–8. doi: 10.1073/pnas.2233572100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.George TC, Mason LH, Ortaldo JR, Kumar V, Bennett M. Positive recognition of MHC class I molecules by the Ly49D receptor of murine NK cells. J Immunol. 1999;162:2035–43. [PubMed] [Google Scholar]
- 78.George TC, Ortaldo JR, Lemieux S, Kumar V, Bennett M. Tolerance and alloreactivity of the Ly49D subset of murine NK cells. J Immunol. 1999;163:1859–67. [PubMed] [Google Scholar]
- 79.Nakamura MC, Linnemeyer PA, Niemi EC, Mason LH, Ortaldo JR, Ryan JC, et al. Mouse Ly-49D recognizes H-2Dd and activates natural killer cell cytotoxicity. J Exp Med. 1999;189:493–500. doi: 10.1084/jem.189.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Silver ET, Gong DE, Chang CS, Amrani A, Santamaria P, Kane KP. Ly-49P activates NK-mediated lysis by recognizing H-2Dd. J Immunol. 2000;165:1771–81. doi: 10.4049/jimmunol.165.4.1771. [DOI] [PubMed] [Google Scholar]
- 81.Mason LH, Willette-Brown J, Mason AT, McVicar D, Ortaldo JR. Interaction of Ly-49D+ NK cells with H-2Dd target cells leads to Dap-12 phosphorylation and IFN-γ secretion. J Immunol. 2000;164:603–11. doi: 10.4049/jimmunol.164.2.603. [DOI] [PubMed] [Google Scholar]
- 82.Furukawa H, Iizuka K, Poursine-Laurent J, Shastri N, Yokoyama WM. A ligand for the murine NK activation receptor Ly-49D: activation of tolerized NK cells from beta 2-microglobulin-deficient mice. J Immunol. 2002;169:126–36. doi: 10.4049/jimmunol.169.1.126. [DOI] [PubMed] [Google Scholar]
- 83.Dighe A, Rodriguez M, Sabastian P, Xie X, McVoy M, Brown MG. Requisite H2k role in NK cell-mediated resistance in acute murine cytomegalovirus-infected MA/My mice. J Immunol. 2005;175:6820–8. doi: 10.4049/jimmunol.175.10.6820. [DOI] [PubMed] [Google Scholar]
- 84.Xie X, Dighe A, Clark P, Sabastian P, Buss S, Brown MG. Deficient major histocompatibility complex-linked innate murine cytomegalovirus immunity in MA/My. L-H2b mice and viral downregulation of H-2k class I proteins. J Virol. 2007;81:229–36. doi: 10.1128/JVI.00997-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fang M, Lanier LL, Sigal LJ. A Role for NKG2D in NK Cell-Mediated Resistance to Poxvirus Disease. PLoS Pathog. 2008;4:e30. doi: 10.1371/journal.ppat.0040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204:3027–36. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]