Abstract
The Sir2 deacetylase regulates chromatin silencing and lifespan in Saccharomyces cerevisiae1,2. In mice, deficiency for the Sir2 family member SIRT6 leads to a shortened lifespan and a premature ageing-like phenotype3. However, the molecular mechanisms of SIRT6 function are unclear. SIRT6 is a chromatin-associated protein3, but no enzymatic activity of SIRT6 at chromatin has yet been detected, and the identity of physiological SIRT6 substrates is unknown. Here we show that the human SIRT6 protein is an NAD+-dependent, histone H3 lysine 9 (H3K9) deacetylase that modulates telomeric chromatin. SIRT6 associates specifically with telomeres, and SIRT6 depletion leads to telomere dysfunction with end-to-end chromosomal fusions and premature cellular senescence. Moreover, SIRT6-depleted cells exhibit abnormal telomere structures that resemble defects observed in Werner syndrome, a premature ageing disorder4,5. At telomeric chromatin, SIRT6 deacetylates H3K9 and is required for the stable association of WRN, the factor that is mutated in Werner syndrome4,5. We propose that SIRT6 contributes to the propagation of a specialized chromatin state at mammalian telomeres, which in turn is required for proper telomere metabolism and function. Our findings constitute the first identification of a physiological enzymatic activity of SIRT6, and link chromatin regulation by SIRT6 to telomere maintenance and a human premature ageing syndrome.
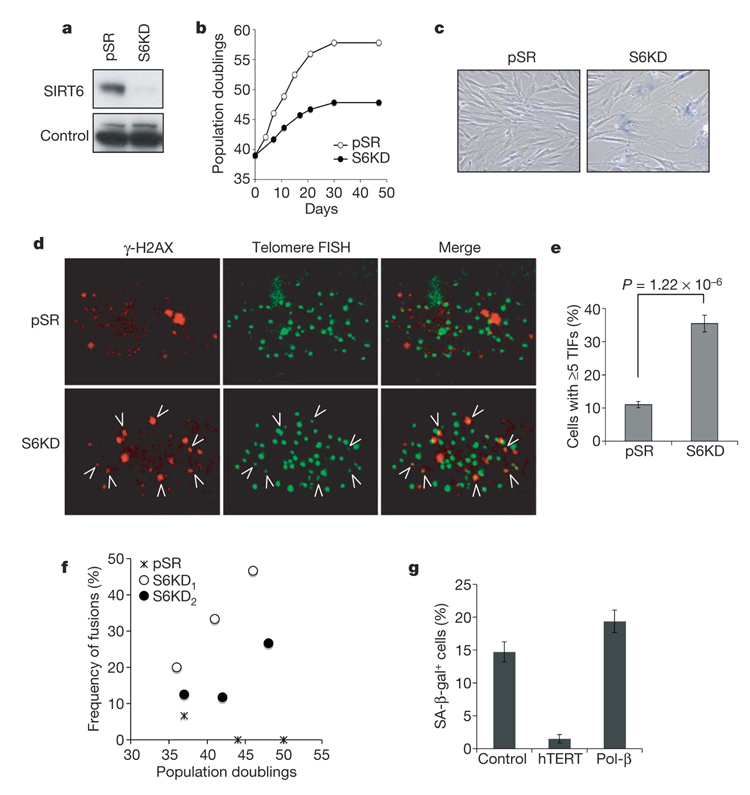
Normal human somatic cells have a finite replicative lifespan, and after prolonged replication they undergo cellular senescence as a result of telomere dysfunction6,7. Despite the ageing-like phenotype of SIRT6-deficient mice, no effect of SIRT6 deficiency on cellular lifespan has been reported. To determine whether SIRT6 influences cellular senescence, retroviral transduction of short hairpin RNAs (shRNAs) was used to stably knock down SIRT6 expression in WI-38 human fibroblasts (Fig. 1a). SIRT6 knockdown (S6KD) cells have a strikingly shortened replicative lifespan, undergoing premature cellular senescence about ten population doublings before control cells, and show increased levels of senescence-associated β-galactosidase (SA-β-gal) staining (Fig. 1b, c, and Supplementary Fig. 2a). These kinetics of premature senescence are similar to those associated with the inactivation of telomere accessory factors (Supplementary Fig. 3a, b)8. Premature cellular senescence was also observed with two independent SIRT6 shRNAs, ruling out off-target shRNA effects, but not with irrelevant shRNAs (Supplementary Fig 2b–d and Supplementary Fig 3c). We conclude that SIRT6 is crucial in maintaining a normal replicative life-span and in preventing the premature senescence of human cells.
Figure 1. SIRT6 knockdown leads to premature cellular senescence and telomere dysfunction.
a, Western analysis of SIRT6 expression in WI-38 SIRT6 knockdown (S6KD) or control (pSR) cells. b, Serial passaging experiments revealing premature replicative senescence of S6KD WI-38 cells. Passaging was begun at population doubling 39 and cumulative population doublings were calculated after the indicated days. c, Increased SA-β-gal+ staining in S6KD cultures at day 11 of serial passaging. d, e, Increased telomere dysfunction in S6KD WI-38 cells. TIFs were detected (d) by co-localization of γ-H2AX and telomeres, and cells with at least five TIFs were scored (e). Data represent the average of 20 fields. Error bars indicate s.e.m.; n = 158 (pSR); n=110 (S6KD). The P value was calculated with the two-tailed Student’s t-test. f, Increased chromosome end-to-end fusions in S6KD cells observed by spectral karyotype (SKY) analysis. Values represent the numbers of fused chromosomes as a percentage of total metaphases. S6KD1 and S6KD2 are knockdown cells generated with two independent SIRT6-specific shRNAs. g, Stabilizing telomeres by means of hTERT expression reverses the premature senescence of S6KD cells, whereas augmenting BER activity, by expression of the DNA polymerase-β (Pol-β)-dRP lyase domain, does not. hTERT, Pol-β or empty vector were ectopically expressed in S6KD WI-38 cells and passaged in physiological (2%) oxygen conditions. Cells were stained with SA-β-gal at population doubling 36.5. Error bars indicate s.e.m.; n=8 (pSR); n = 16 (S6KD1); n = 20 (S6KD2).
Replicative cellular senescence can result from dysfunctional telomeres, which are recognized by DNA damage response factors and are detected as telomere dysfunction-induced foci (TIFs)9,10. Analysis of TIFs revealed elevated telomere dysfunction in S6KD cells (Fig. 1d, e, and Supplementary Fig. 4a). The telomere signals at TIFs in S6KD cells are weak compared with non-TIF telomere signals, suggesting a subpopulation of telomeres that have undergone significant sequence loss (see below). However, mean telomere length was not significantly reduced in S6KD cells (Supplementary Fig. 5). Together, these observations suggest that S6KD cells undergo accelerated senescence and telomere dysfunction in response to stochastic telomere sequence loss, without increased global telomere erosion.
Loss of proper telomeric protective end structures can lead to dicentric chromosomes as a result of chromosomal end-to-end fusions11. We therefore scored chromosomal end fusions in S6KD and control metaphases in several independent cytogenetic analyses. Non-recurrent chromosomal end-to-end fusions were observed in S6KD cells and were more pronounced at later population doublings, but they were rarely observed in control cells (Fig. 1f and Supplementary Fig. 4b, c). These observations indicate that SIRT6 is critical for maintaining functional telomeres to avert chromosomal instability due to aberrant chromosomal end-to-end fusions.
Several experiments provide further evidence that the premature senescence of S6KD cells is due to telomere dysfunction and not to defective base excision repair (BER), which was previously implicated in the phenotypes of SIRT6 knockout (S6KO) mouse cells3, or to oxidative stress brought on by supraphysiological oxygen conditions of ambient cell culture conditions12. First, telomere stabilization (by the ectopic expression of telomerase (hTERT)) reversed the premature senescence of S6KD, whereas augmenting BER (by the ectopic expression of the DNA polymerase-β dRP lyase domain) did not (Fig. 1g and Supplementary Fig. 6). This DNA polymerase-β domain was previously shown to rescue the hypersensitivity of S6KO mouse cells to alkylating DNA damage agents3. Second, S6KD cells underwent premature senescence even when cultured under low (physiological) oxygen conditions (Supplementary Fig. 7). Together, these findings demonstrate that telomere dysfunction, not BER defects or oxidative stress, underlie the premature senescence phenotype of S6KD cells.
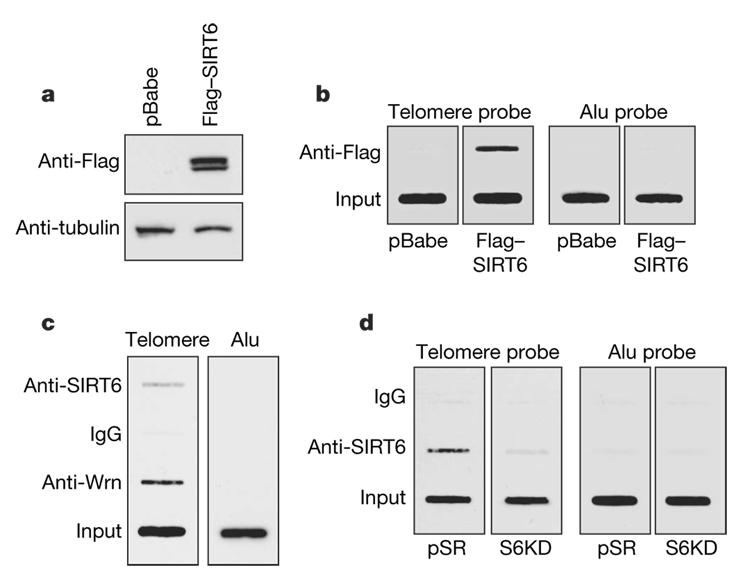
The premature cellular senescence, telomere dysfunction and chromosomal fusions observed in S6KD cells are reminiscent of the cellular phenotype of Werner syndrome (WS)13–16, a hereditary dis-order associated with signs of premature ageing4,5. The WS-defective protein WRN associates with telomeres (in primary human IMR90 cells and U2OS osteosarcoma cancer cells) and regulates telomere processing during S phase13,17. We formed the hypothesis that SIRT6 might function in a similar context. To investigate this possibility, we first examined whether SIRT6 associates with telomeres during S phase by telomere chromatin immunoprecipitation (T-ChIP)18. Cell synchronization, release, and analysis by bromodeoxyuridine/propidium iodide staining were performed to enrich for specific cell-cycle phases (Supplementary Fig. 8). T-ChIP analysis at different time points after release from cell synchronization revealed that SIRT6, like WRN, preferentially associates with telomeric chromatin in S-phase-enriched cultures (Supplementary Fig. 9). SIRT6 occupancy at telomeric chromatin was observed for both recombinant Flag-tagged SIRT6 (Fig. 2a, b) and endogenous SIRT6 (Fig. 2c, d), and in both U2OS osteosarcoma cells (Fig. 2b, c) and primary IMR90 cells (Fig. 2d). As controls, the association of Alu repeat sequences with SIRT6 ChIPs was not above background (Fig. 2b–d), and the SIRT6 T-ChIP signal was abolished in S6KD cells, validating the specificity of the signal (Fig. 2d). Together, these data locate SIRT6 at telomeric chromatin in S phase and suggest a potential role for SIRT6 in regulating replication-associated dynamics in telomere structure.
Figure 2. SIRT6 associates with telomeric chromatin.
a, Western analysis of U2OS cells transduced with pBabe-Flag-SIRT6 or empty virus (pBabe) control. b, T-ChIP analysis with anti-Flag antibodies from the cells shown in a. c, T-ChIP assays in U2OS cells with antibodies specific for SIRT6,WRNor IgG negative control. d, T-ChIP assays with SIRT6 antibodies in S6KD cells or in control pSR IMR90 cells. In b–d, slot-blots are shown of telomere or Alu repeat sequences in ChIP and input DNA. Input loaded represents 5% (b, c) or 2.5% (d) of total.
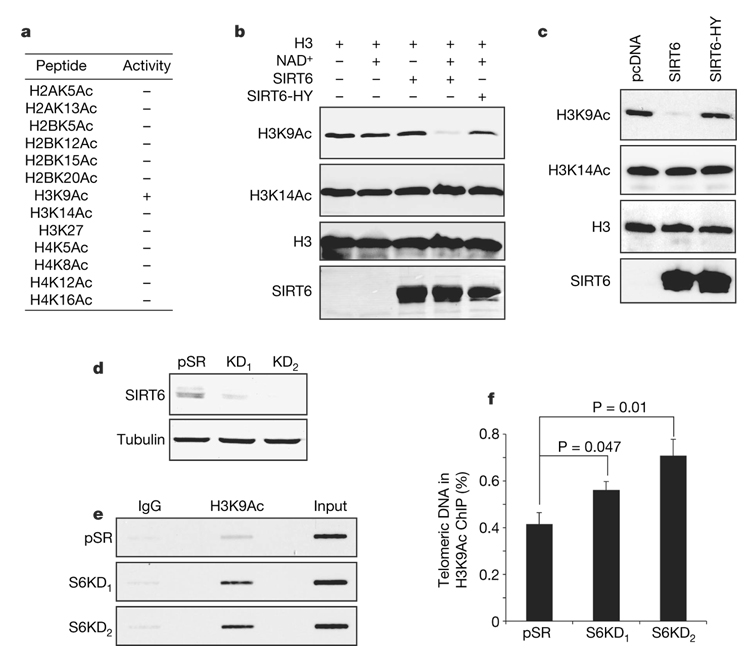
Chromatin at telomeres is enriched for hypoacetylated histone tails19. Although no physiological enzymatic activity for SIRT6 on histones or other trans substrate has yet been observed, we proposed that SIRT6 might regulate chromatin at telomeres by deacetylating a specific histone tail residue. We therefore used mass spectrometry to screen for NAD-dependent SIRT6 deacetylase activity in vitro, on a collection of acetylated histone tail peptides. SIRT6 manifested modest deacetylation activity on peptides containing acetylated H3K9 (H3K9Ac) (Fig. 3a and Supplementary Fig. 10). This activity was highly specific for H3K9Ac, because no deacetylation was detected for 12 other acetylated histone peptides (Fig. 3a and Supplementary Fig. 11). SIRT6 also deacetylated H3K9Ac, but not several other acetylated residues, in the context of purified full-length histone H3, and mutation of a conserved catalytic residue (H133Y) of SIRT6 markedly decreased this activity (Fig. 3b and data not shown). Finally, SIRT6 efficiently and specifically deacetylated H3K9Ac (but not other acetylated histone residues) in 293T cells, whereas the mutant SIRT6 protein did not (Fig. 3c and Supplementary Fig. 12). Together, these observations indicate that SIRT6 is an NAD-dependent deacetylase with specificity for H3K9Ac.
Figure 3. SIRT6 deacetylates lysine 9 of histone H3 at telomeric chromatin.
a, Summary of in vitro deacetylation assays on acetylated histone tail peptides, analysed by mass spectrometry. b, Western analysis showing SIRT6 deacetylation of H3K9 on full-length histone H3 in vitro. Reactions with NAD+, SIRT6 or the catalytic H133Y SIRT6 mutant protein (SIRT6-HY) are indicated. c, SIRT6 deacetylates H3K9 in cells. Western analysis of 293T cells overexpressing SIRT6, SIRT6-HY or empty vector. d, Western analysis of SIRT6 knockdown in U2OS cells used for T-ChIP in e and f. e, T-ChIP analysis showing hyperacetylation of H3K9 at telomeric chromatin in S6KD U2OS cells. f, Quantification of multiple independent T-ChIP experiments as shown in e. Values represent the T-ChIP signal normalized to input signal, and subtracted for background signal in an IgG control. Error bars indicate s.e.m.; n = 4. P values were determined by two-tailed Student’s t-test.
We next sought to identify the physiological context in which SIRT6 deacetylates H3K9Ac. Western analysis of S6KD and control cells did not reveal significant differences in global H3K9Ac levels (data not shown). In contrast, investigation of histone acetylation status at telomeres in S-phase-enriched cultures by T-ChIP revealed H3K9 hyperacetylation in S6KD cells (Fig. 3d–f). SIRT6 is therefore required for the maintenance of the low physiological levels of H3K9 acetylation at telomeric chromatin in S phase, and hyperacetylation of H3K9 in the absence of SIRT6 correlates with telomere dysfunction. H3K9 was also hyperacetylated at telomeric chromatin in S6KO mouse cells, providing in vivo evidence for a physiological role for SIRT6 in deacetylating this histone residue (Supplementary Fig. 13a–c).
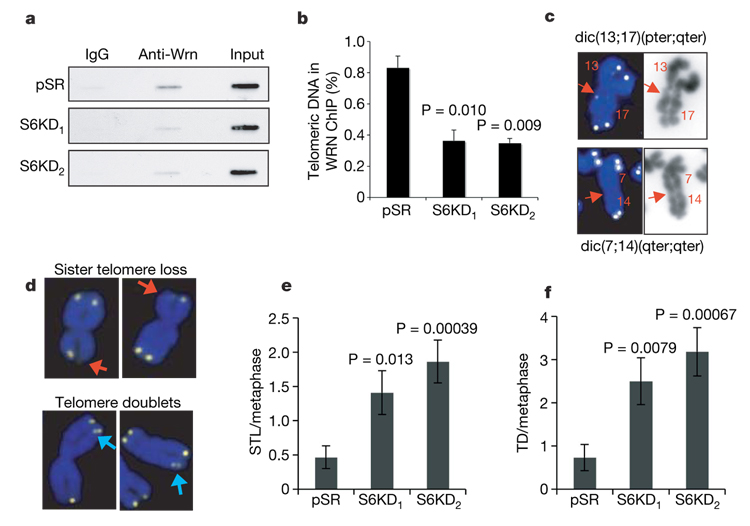
We next proposed that in SIRT6-deficient cells, hyperacetylation of H3K9 at telomeres in S phase might interfere with the association of WRN. WRN occupancy at telomeres was compared in S-phase-enriched S6KD and control cultures by T-ChIP analysis. In both U2OS and IMR90 cells, SIRT6 knockdown significantly inhibited the association of WRN with telomeric chromatin (Fig. 4a, b, and data not shown). We conclude that SIRT6 is required for stabilization of WRN at telomeric chromatin. We note that additional functional or physical interactions between SIRT6 and WRN might exist. However, the association of SIRT6 with telomeres was independent of WRN (Supplementary Fig. 13e), and we have not observed a direct interaction between SIRT6 and WRN at chromatin (data not shown). Thus, our data are consistent with the hypothesis that SIRT6 deacetylation of H3K9Ac at telomeric chromatin leads to an altered chromatin state that is required for efficient WRN association in S phase.
Figure 4. SIRT6 stabilizes WRN at telomeric chromatin and prevents replication-associated telomere defects.
a, b, T-ChIP experiments showing reduced association of WRN with telomeric chromatin in SIRT6 knockdown cells. a, Representative T-ChIP analysis of WRN occupancy in S6KD and control cells. b, Quantification of three independent experiments as shown in a. c, Representative chromosome end-to-end fusions observed in S6KD cells. Telomere FISH signals are yellow, and DAPI-stained chromosomes are blue. Weak or no telomere signals are detected at the sites of fusion (arrows). dic(13;17)(pter;qter) and dic(7,14)(qter;qter) indicate dicentric chromosomal fusions between the p and q arms of chromosomes 13 and 17, and the q arms of chromosomes 7 and 14, respectively. d, Representative S6KD metaphases showing aberrant telomere signals. Red arrows, sister telomere loss; blue arrows, telomere doublets. e, f, Quantification of sister telomere loss (e) and telomere doublets (f) in S6KD cells and in pSR control cells. pSR, n=15; S6KD1 and S6KD2, n=22. Error bars in b, e and f indicate the s.e.m., and P values were determined by two-tailed Student’s t-test.
WS cells exhibit specific defects that reflect problems with the replication-associated processing and metabolism of telomeres, and analysis of S6KD cells revealed similar abnormalities. First, S6KD metaphases showed elevated levels of missing telomere signals (sister telomere loss) and extra telomere signals (telomere doublets) (Fig. 4d–f), defects observed in WS cells and associated with aberrant telomere processing during replication8,13,20. In addition, chromosome fusions in S6KD cells, as in WRN-defective cells14,15, have weak or no telomere signals at the fusion sites (Fig. 4c). In WS cells this phenotype is proposed to result from stochastic replication-associated telomere loss5,15, and it contrasts with the strong telomere signals observed at sites of chromosome fusions resulting from deficiency for telomere end-capping factors such as telomeric repeat binding factor 2 (TRF2)11. Aberrant replication-associated telomere processing by WRN is proposed to contribute to a delayed completion of S phase observed in WS cells13,14,21,22. Similarly, SIRT6 knockdown in U2OS cells resulted in delayed completion of S phase (Supplementary Fig. 14), which is consistent with a role for SIRT6 in modulating telomeres during replication. Together, these findings suggest that SIRT6 collaborates with WRN at telomeric chromatin to ensure efficient telomere replication and to prevent the accrual of structural abnormalities at telomeres.
Telomeres are specialized structures that function to shield linear chromosome ends from DNA repair, degradation and fusion7. Mammalian telomeres are packaged in an unusual chromatin structure with features of heterochromatin23,24, but relatively little is understood about the role of chromatin modifications on telomere metabolism. In this study we show that human SIRT6 deacetylates H3K9Ac at telomeres to prevent telomere dysfunction. Inactivation of SIRT6 leads to H3K9 hyperacetylation at telomeric chromatin in both human and mouse cells, but the mouse cells do not display the downstream cellular defects observed in human cells, which is consistent with the large functional reserve of mouse telomeres (Supplementary Fig. 13d; data not shown)14,25. Our findings regarding SIRT6 provide a direct link between mammalian telomere dysfunction and a histone modifying enzymatic activity. Our results indicate that deacetylation of H3K9Ac by SIRT6 is important during S phase at replicating telomeres. We propose a model (Supplementary Fig. 1) in which the deacetylation of H3K9Ac by SIRT6 promotes the formation of a specialized telomeric chromatin state that is required for the stable association of S-phase-dependent telomere-processing factors such as WRN, to prevent aberrant sequence loss or metabolism of telomeres. The resulting telomere dysfunction then contributes to premature cellular senescence. Our study has identified a crucial function for SIRT6 in chromatin regulation at mammalian telomeres and provides a new mechanism by which the regulation of chromatin is linked to telomere function, cellular senescence and, potentially, organismal ageing.
METHODS SUMMARY
Telomere-ChIP assay
S-phase T-ChIP analysis was performed essentially as described previously13. In brief, ChIPs were performed by following the Diagenode protocol (http://www.diagenode.com). DNA eluted from the ChIPs was transferred to slot-blots, and telomeric DNA was detected with the TeloTAGGG telomere length assay kit (Roche Applied Science).
Immunofluorescence in situ hybridization TIF assays
Immunofluorescence in situ hybridization (immuno-FISH) analysis of TIFs was performed as described previously26,27. Cells were fixed with paraformaldehyde, washed, and permeabilized in 100% ethanol or 0.2% Triton X-100. After being blocked and immunostained, cells were treated with 1 mg ml−1 dithiobis(succinimidylpropionate), and telomere FISH was performed with the Telomere PNA FISH Kit/fluorescein isothiocyanate (Dako). Laser-scanning confocal microscopy was performed with a Nikon PCM 2000 confocal microscope scanning system.
Cytogenetic chromosome analysis
Metaphase spreads were prepared using standard techniques as previously described28, and stained with 4,6-diamidino-2-phenylindole (DAPI). Detailed procedures of the spectral karyotype (SKY) analysis, including the preparation of SKY probes, slide denaturation, probe hybridization, detection and image acquisition, are described at http://www.riedlab.nci.nih.gov/protocols and in the Supplementary Information. Detection of telomere sequences by telomere FISH was performed with a Telomere PNA FISH Kit/Cy3 (Dako). Acquisition of metaphase spreads with telomere signals and the companion DAPI images for each sample were analysed with Q-Fluoro software (Leica Imaging Systems).
METHODS
Cell culture and retroviral infection
WI-38 and IMR90 cells were obtained from the American Tissue Culture Collection and maintained in DMEM/F12 (1:1) with 10% FBS (Gibco-Invitrogen). U2OS and HeLa cell lines were maintained in DMEM with 10% FBS.
The retrovirus-mediated RNA interference study was performed with the pSuper RNA interference system (Oligoengine). SIRT6 target sequences were as follows: S6KD1, 5′-AAGCTGGAGCCCAAGGAGGAA-3′; S6KD2, 5′-AAGAATGTGCCAAGTGTAAGA- 3′; S6KD3, 5′-CACGGGAACATGTTTGTGGAA-3′. For retroviral packaging, 293T cells were co-transfected with pVPack-VSV-G, pVPack-GP (Stratagene) and the SIRT6 knockdown or pSUPERretro (pSR) control constructs, and viral supernatant was harvested after 48 h. For infections, cells were incubated with viral supernatant in the presence of 1 µgml−1 DEAE-dextran; infected cells were selected after 48 h with puromycin (2 µg ml−1) for 72 h.
Antibodies
The antibodies used were as follows: anti-γ-H2AX (phospho-Ser-139) (Upstate), anti-SIRT6 (ref. 29), anti-Flag (Sigma-Aldrich), anti-WRN (Santa Cruz Biotechnology), anti-H3K9Ac (Sigma-Aldrich, Abcam), and anti-H3K14Ac, anti-H3K23Ac, anti-H4K8Ac, anti-H4K16Ac, anti-H2AK5Ac, anti-H2A, anti-H3 and anti-H4 (Abcam).
Telomere-ChIP assay
Telomere ChIP analysis was performed essentially as described previously13,18. Chromatin immunoprecipitation (ChiP) was performed in accordance with the protocol at the Diagenode website (http://www.diagenode.com). In brief, cultures were synchronized by means of a single thymidine block, then released for various times; cell-cycle distribution was determined by bromodeoxyuridine/propidium iodide analysis. Cell cultures enriched for the indicated cell-cycle phases were crosslinked with 1% formaldehyde for 30 min at 22 °C and the reaction was stopped with 125 mM glycine. Nuclei were isolated by resuspending the cells in swelling buffer (5 mM Tris-HCl pH 8.0, 85 mM KCl, 1% Nonidet P40, 5 mM butyrate and complete protease inhibitor (Roche Molecular Biochemicals)) for 20 min at 4 °C. The isolated nuclei were resuspended in nuclei lysis buffer (50 mM Tris-HCl pH8.0, 10 mM EDTA, 1% SDS) and sonicated with a BioRuptor Sonicator (Diagenode). Samples were immunoprecipitated overnight with anti-H3K9Ac, anti-WRN, anti-Flag or anti-SIRT6 antibody, or the equivalent amount of rabbit IgG (Santa Cruz Biotechnology) at 4 °C. Immunoprecipitates were washed twice with dialysis buffer (50 mM Tris-HCl pH8.0, 2 mM EDTA, 0.2% Sarkosyl) and four times with immunoprecipitation wash buffer (100 mM Tris-HCl pH 8.0, 500 mM LiCl, 1% Nonidet P40, 1% deoxycholic acid sodium salt). After reverse crosslinking, DNA was eluted, purified and transferred to Hybond N+ membrane (Amersham Pharmacia Biotech) using the Minifold II Slot-Blot System (Schleicher & Schuell). Telomeric DNA was detected with the TeloTAGGG telomere length assay kit (Roche Applied Science). The digoxigenin-labelled Alu repeat probe was generated with the DIG DNA labelling kit (Roche Applied Science).
Replicative senescence and senescence associated β-galactosidase assay
Replicative senescence assays were performed with a modified 3T3 protocol as described previously29,30. Senescence-associated b-galactosidase staining31 was detected histochemically at pH 6 with the Senescence Cells Histochemical Staining Kit (Sigma-Aldrich).
Chromosome analysis
Metaphase spreads were prepared as described previously 28. Metaphase spreads screened for gross chromosomal aberrations were stained with DAPI (Roche Diagnostics). Pretreatment of slides containing metaphase spreads for SKY analysis followed protocols described previously32. Detailed procedures of SKY analysis, including the preparation of SKY probes, slide denaturation, probe hybridization, detection and image acquisition, are described at http://www.riedlab.nci.nih.gov/protocols. A total of 15–20 metaphase spreads for each sample were stained with DAPI, imaged and then karyo-typed with BandView (Applied Spectral Imaging); 10–15 metaphase spreads were hybridized with SKY probes for each sample and were analysed with SKYView (Applied Spectral Imaging) with the use of a spectral cube with a custom-designed SKY-3 optical filter (Chroma Technology) and a charge-coupled device camera (Hamamatsu) connected to a DMRXA microscope (Leica). Hybridization of slides for the detection of telomere sequences (telomere FISH) was performed with a Telomere PNA FISH Kit/Cy3 (Dako). The acquisition of 15–20 metaphase spreads with telomere signals and the companion DAPI images for each sample was performed with Q-Fluoro software (Leica Imaging Systems).
Histone deacetylation assays
In vitro deacetylation reactions were performed with 4.5 mg of recombinant SIRT6 and 1 µg of acetylated histone tail peptide or full-length histone substrates, in HDAC buffer (10mM Tris-HCl pH 8.0, 5mM NAD+, 150 mM NaCl) for 1 h at 30 °C. The generation of recombinant SIRT6 proteins was described previously29. Calf thymus histones were from Roche Applied Science. Acetylated histone tail peptides were synthesized at the Yale W. M. Keck peptide synthesis facility. Mass spectrometry was performed at the Stanford University Vincent Coates Foundation Mass Spectrometry Laboratory. For analysis of histone acetylation levels in cells, 293T cells were transiently transfected with pcDNA 3.1 vectors containing Flag-tagged wild-type SIRT6, the catalytically inactive SIRT6-H133Y mutant or an empty virus control, and whole-cell lysates were harvested after 48 h. Western analysis of histone acetylation levels in cells and in vitro deacetylation of full-length histones was performed with modification-specific antibodies.
Immuno-FISH TIF assays
Cells were fixed with paraformaldehyde, washed and then permeabilized in 100% ethanol or 0.2% Triton X-100. After being blocked, cells were immunostained with primary antibody, followed by Alexa Fluor-546-conjugated anti-mouse IgG (H + L) antibody (Molecular Probes). The immuno-stained cells were treated with 1 mg ml−1 dithiobis(succinimidylpropionate) (Pierce Chemical Co.). Telomere FISH was then performed using the Telomere PNA FISH Kit/fluorescein isothiocyanate (Dako). Laser-scanning confocal microscopy was performed with a Nikon PCM 2000 confocal microscope scanning system.
Cell cycle analysis
Cells were fixed with ice-cold 70% ethanol, washed with PBS and incubated in PBS containing 10 µg ml−1 propidium iodide and 0.1 mg ml−1 RNase A. Data were collected on a FACSCalibur flow cytometer with Cell Quest software (BD Biosciences). Percentages of cells in each cell-cycle phase (G1, S and G2/M) were analysed with FlowJo software (TreeStar).
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgements
We thank S. Artandi, J. Karlseder, B. North, E. Verdin, J. Lipsick, H. Wen, L. Christensen and Chua and Gozani laboratory members for reagents, technical assistance and/or advice; and Regeneron Pharmaceuticals for SIRT6 knockout mice. This work was supported by grants from the National Institutes of Health (NIH) (to K.F.C., O.G., H.Y.C., R.A.M. and T.L.A.K.), the American Federation for Aging Research/Paul Beeson Scholar Award and the Department of Veterans Affairs Merit Review (to K.F.C.) and the Burroughs Wellcome Fund and Searle Scholar Award (to O.G.), and by funds from the Intramural Research Program of the NIH, the National Cancer Institute, the Center for Cancer Research and the National Institutes on Aging.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.Landry J, et al. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl Acad. Sci. USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 3.Mostoslavsky R, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 4.Cheng WH, Muftuoglu M, Bohr VA. Werner syndrome protein: Functions in the response to DNA damage and replication stress in S-phase. Exp. Gerontol. 2007;42:871–878. doi: 10.1016/j.exger.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 5.Multani AS, Chang S. WRN at telomeres: implications for aging and cancer. J. Cell Sci. 2007;120:713–721. doi: 10.1242/jcs.03397. [DOI] [PubMed] [Google Scholar]
- 6.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 8.van Overbeek M, de Lange T. Apollo, an Artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr. Biol. 2006;16:1295–1302. doi: 10.1016/j.cub.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 9.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 10.d’Adda di Fagagna F, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 11.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 12.Sherr CJ, DePinho RA. Cellular senescence: mitotic clock or culture shock? Cell. 2000;102:407–410. doi: 10.1016/s0092-8674(00)00046-5. [DOI] [PubMed] [Google Scholar]
- 13.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 14.Laud PR, et al. Elevated telomere-telomere recombination in WRN-deficient, telomere dysfunctional cells promotes escape from senescence and engagement of the ALT pathway. Genes Dev. 2005;19:2560–2570. doi: 10.1101/gad.1321305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crabbe L, Jauch A, Naeger CM, Holtgreve-Grez H, Karlseder J. Telomere dysfunction as a cause of genomic instability in Werner syndrome. Proc. Natl Acad. Sci. USA. 2007;104:2205–2210. doi: 10.1073/pnas.0609410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wyllie FS, et al. Telomerase prevents the accelerated cell ageing of Werner syndrome fibroblasts. Nature Genet. 2000;24:16–17. doi: 10.1038/71630. [DOI] [PubMed] [Google Scholar]
- 17.Opresko PL, et al. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 18.Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 19.Benetti R, Garcia-Cao M, Blasco MA. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nature Genet. 2007;39:243–250. doi: 10.1038/ng1952. [DOI] [PubMed] [Google Scholar]
- 20.Ariyoshi K, Suzuki K, Goto M, Watanabe M, Kodama S. Increased chromosome instability and accumulation of DNA double-strand breaks in Werner syndrome cells. J. Radiat. Res. (Tokyo) 2007;48:219–231. doi: 10.1269/jrr.07017. [DOI] [PubMed] [Google Scholar]
- 21.Takeuchi F, Hanaoka F, Goto M, Yamada M, Miyamoto T. Prolongation of S phase and whole cell cycle in Werner’s syndrome fibroblasts. Exp. Gerontol. 1982;17:473–480. doi: 10.1016/s0531-5565(82)80009-0. [DOI] [PubMed] [Google Scholar]
- 22.Poot M, Hoehn H, Runger TM, Martin GM. Impaired S-phase transit of Werner syndrome cells expressed in lymphoblastoid cell lines. Exp. Cell Res. 1992;202:267–273. doi: 10.1016/0014-4827(92)90074-i. [DOI] [PubMed] [Google Scholar]
- 23.Tommerup H, Dousmanis A, de Lange T. Unusual chromatin in human telomeres. Mol. Cell. Biol. 1994;14:5777–5785. doi: 10.1128/mcb.14.9.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blasco MA. The epigenetic regulation of mammalian telomeres. Nature Rev. Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- 25.Chang S, et al. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nature Genet. 2004;36:877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- 26.Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21CIP1, but not p16INK4a. Mol. Cell. 2004;14:501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- 27.Dimitrova N, de Lange T. MDC1 accelerates nonhomologous end-joining of dysfunctional telomeres. Genes Dev. 2006;20:3238–3243. doi: 10.1101/gad.1496606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Padilla-Nash HM, et al. Jumping translocations are common in solid tumor cell lines and result in recurrent fusions of whole chromosome arms. Genes Chromosom. Cancer. 2001;30:349–363. doi: 10.1002/gcc.1101. [DOI] [PubMed] [Google Scholar]
- 29.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chua KF, et al. Mammalian SIRT1 limits replicative life span in response to chronic genotoxic stress. Cell Metab. 2005;2:67–76. doi: 10.1016/j.cmet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 31.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schrock E, et al. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.