I. The need for antivirals against flaviviruses
In the last decade, the flaviviruses have reemerged as aggressive human pathogens causing an increased number of infections worldwide (Gould and Solomon, 2008; Kuhn, 2006). There are more than 70 related members of the flavivirus genus, a subgroup of the Flaviviridae family. Many of these viruses produce significant human disease and include yellow fever virus (YFV), West Nile virus (WNV), dengue virus (DENV), Japanese encephalitis virus (JEV) and tick-borne encephalitis virus (TBEV). Vaccines are currently available for JEV, YFV, and TBEV. However, disease outbreaks caused by these three viruses continue to be a serious problem in many developing countries. Several promising candidates for WNV vaccines are in progress but may take several years for clinical evaluation (Martin et al., 2007; Monath et al., 2006). Conventional vaccine development for dengue virus (DENV) has been challenging. For instance, prevention of antibody-dependent enhancement (ADE) is of ultimate importance when designing vaccines and requires efforts aimed at eliciting an appropriate immune response against all four serotypes of the virus. Antiviral therapies for these viruses are at a very early stage of development (for a review see (Ray and Shi, 2006). Thus, the flaviviruses have huge disease burdens, and require new approaches to preventing virus replication, pathogenesis, and transmission.
Antivirals previously designed against flaviviruses have primarily focused on inhibition of viral RNA replication. Although these efforts are ongoing, new opportunities for antiviral design have recently emerged based on advances in our knowledge of flavivirus virion structure. These advances include obtaining the pseudo-atomic structures of TBEV subviral particles (Ferlenghi et al., 2001) and of immature and mature DENV (Kuhn et al., 2002; Zhang et al., 2003a; Zhang et al., 2007; Zhang et al., 2004) and WNV (Mukhopadhyay et al., 2003) under different physiological conditions, as well as mature DENV and WNV virus complexed with antibodies and cell-surface attachment molecules (Kaufmann et al., 2006; Lok et al., 2007). X-ray crystallographic analyses and nuclear magnetic resonance (NMR) spectroscopy studies have provided atomic resolution structures of the three flavivirus structural proteins: capsid (C) (Dokland et al., 2004; Ma et al., 2004), pre-membrane (prM) (Li et al., 2008a) and envelope (E) (Bressanelli et al., 2004; Heinz et al., 1991; Huang et al., 2008; Kanai et al., 2006; Modis et al., 2003; Modis et al., 2004; Modis et al., 2005; Mukherjee et al., 2006; Nybakken et al., 2006; Rey et al., 1995; Volk et al., 2007; Yu, Hasson, and Blackburn, 1988; Yu et al., 2004; Zhang et al., 2004). These studies have invited an alternative focus from inhibition of RNA replication towards blocking structural transitions required for efficient virus spread. Similar strategies were previously employed for antiviral design against the rhinoviruses (Badger et al., 1988; Hadfield, Diana, and Rossmann, 1999; Heinz et al., 1989; Rossmann, 1994; Rossmann et al., 2000) and enteroviruses (Padalko et al., 2004; Rossmann et al., 2000), and have recently gained momentum in other fields such as HIV (Copland, 2006; Veiga, Santos, and Castanho, 2006; Wang and Duan, 2007) and influenza (Hsieh and Hsu, 2007).
In this review, we will describe the structural transitions that occur in the flavivirus E protein during the life cycle of the virus, and discuss how these transitions can be targeted for inhibition by antiviral compounds. Specifically, recent advances in the structure determination of the flaviviruses and their component proteins are described with special emphasis on the conformational and translational changes of the E protein as it transitions between the immature, mature and fusion active forms of the virus. Specific surfaces on the E protein are described as potential targets for structure-based antiviral drug design and alternate strategies for viral inhibition are discussed based on the interaction of the E protein with receptor molecules and neutralizing antibodies.
II. Flavivirus replication cycle
Like other positive-strand RNA viruses, flaviviruses replicate in the cytoplasm of susceptible cells (Figure 1). A specific receptor for internalization of these viruses into host cells has not yet been identified. Several cellular molecules capable of mediating virus attachment are known, but none has been conclusively shown to function as virus receptors (Barba-Spaeth et al., 2005; Chu, Buczek-Thomas, and Nugent, 2004; Jindadamrongwech, Thepparit, and Smith, 2004; Krishnan et al., 2007; Lozach et al., 2005; Miller et al., 2008; Navarro-Sanchez et al., 2003; Pokidysheva et al., 2006; Tassaneetrithep et al., 2003).
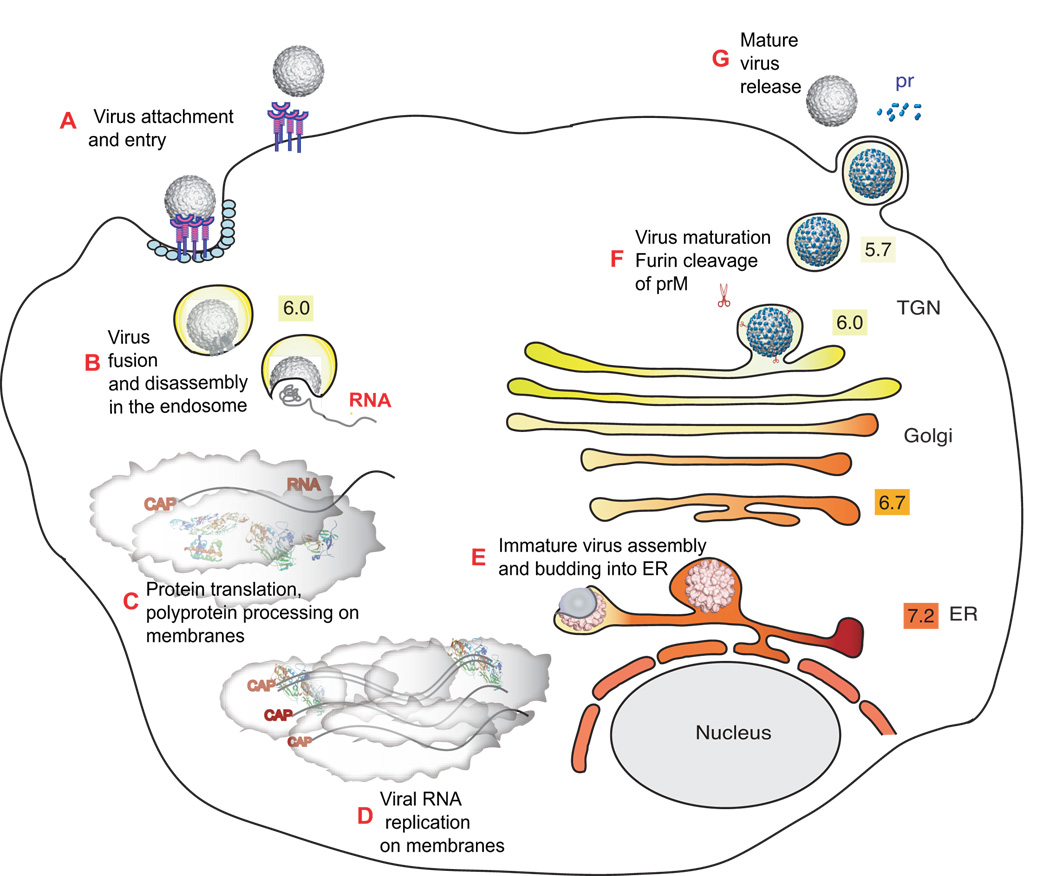
Figure 1. The flavivirus life cycle.
A. Virions bind to cell-surface attachment molecules and receptors and are internalized through endocytosis. B. In the low pH of the endosome, viral glycoproteins mediate fusion of viral and cellular membranes, allowing disassembly of the virion and release of RNA into the cytoplasm. C. Viral RNA is translated into a polyprotein that is processed by viral and cellular proteases. D. Viral non-structural proteins replicate the genome RNA. E. Virus assembly occurs at the ER membrane, where capsid protein and viral RNA are enveloped by the ER membrane and glycoproteins to form immature virus particles. F. Immature virus particles are transported through the secretory pathway. In the low pH of the trans-Golgi, network (TGN) furin-mediated cleavage of prM drives maturation of the virus. G. Mature virus is released into the cytoplasm. Numbers shown in colored boxes refer to the pH of the respective compartments.
The flavivirus virion consists of an outer glycoprotein shell and an internal host derived lipid bilayer that surrounds the capsid and viral RNA. During virus entry, envelope (E) proteins forming the glycoprotein shell bind cell to surface receptors that assist in internalizing the virus through clathrin-mediated endocytosis (Gollins and Porterfield, 1984; Ishak, Tovey, and Howard, 1988; van der Schaar et al., 2007). Following internalization, the low pH of the endosome triggers structural rearrangements in the viral glycoproteins that drive fusion of the viral and endocytic membranes to release the viral RNA into the cytoplasm (Bressanelli et al., 2004; Modis et al., 2004). This RNA (~11kb) is directly translated as a single polyprotein that is processed by viral and cellular proteases into 3 structural proteins (C, prM and E) and 7 non-structural proteins (NS1, NS2A/B, NS3, NS4A/B and NS5) (Lindenbach and Rice, 2001). The non-structural proteins actively replicate the viral RNA in replication complexes associated with cellular membranes (Mackenzie, Khromykh, and Westaway, 2001). Newly synthesized RNA and capsid protein are enveloped by glycoproteins prM and E to assemble immature virus particles that bud into the ER. These immature particles are transported through the secretory pathway to the Golgi apparatus. In the low pH environment of the trans-Golgi, furin-mediated cleavage of prM to M drives maturation of the virus. Maturation is also accompanied by significant structural rearrangements of the glycoprotein shell (Elshuber et al., 2003; Kuhn et al., 2002; Li et al, 2008; Li et al., 2008a; Li Long, 2008; Modis et al., 2003; Modis et al., 2005; Mukhopadhyay et al., 2003; Stadler et al., 1997; Yu et al., 2008; Zhang et al., 2003a). Following maturation, virus particles migrate along the surface of naïve cells until they encounter clathrin-coated pits that assist in virus entry (van der Schaar et al., 2007). Because of differences in the cellular environments and the non-lytic nature of the infections, the entry, replication and assembly of these viruses may differ in mosquito cells versus vertebrate cells.
III. The flavivirus E protein as a target for antiviral therapy
Current efforts to develop antivirals against flavivirus entry are focused on the E protein. This protein assumes different conformations in the immature, mature and fusion-activated forms of the virus and plays an important role during virus assembly, maturation and entry (reviewed in (Mukhopadhyay, Kuhn, and Rossmann, 2005; Rey, 2003). E also functions as a receptor-binding ligand, as well as the fusion engine that carries out mixing of viral and cellular membranes. Furthermore, it is a target for most of the antibodies that neutralize the virus.
The E protein forms the glycoprotein shell of the virus (Figure 2). It is a class II fusion protein that shares about ~40% amino acid identity among the flaviviruses. The atomic structure for the ectodomain of E has been solved for DENV-2, DENV-3, WNV and TBEV (Kanai et al., 2006; Modis et al., 2003; Modis et al., 2004; Modis et al., 2005; Nybakken et al., 2006; Rey et al., 1995; Zhang et al., 2004). The proteins consists of three β-barrel domains. Domain I (DI) contains the N-terminus, but is centrally located in the molecule. Domain II (DII) is rather elongated, mediates dimerization of E and also includes the hydrophobic and well conserved fusion peptide at its distal end (Allison et al., 2001). Domain III (DIII) is an immunoglobulin (Ig)-like domain that is predicted to be involved in receptor binding (Bhardwaj et al., 2001; Rey et al., 1995) and antibody neutralization (Beasley and Barrett, 2002; (Crill and Chang, 2004; Crill and Roehrig, 2001; Halstead et al., 2005; Li, Barrett, and Beasley, 2005; Pierson et al., 2007; Stiasny et al., 2006)Sukupolvi-Petty, 2007).
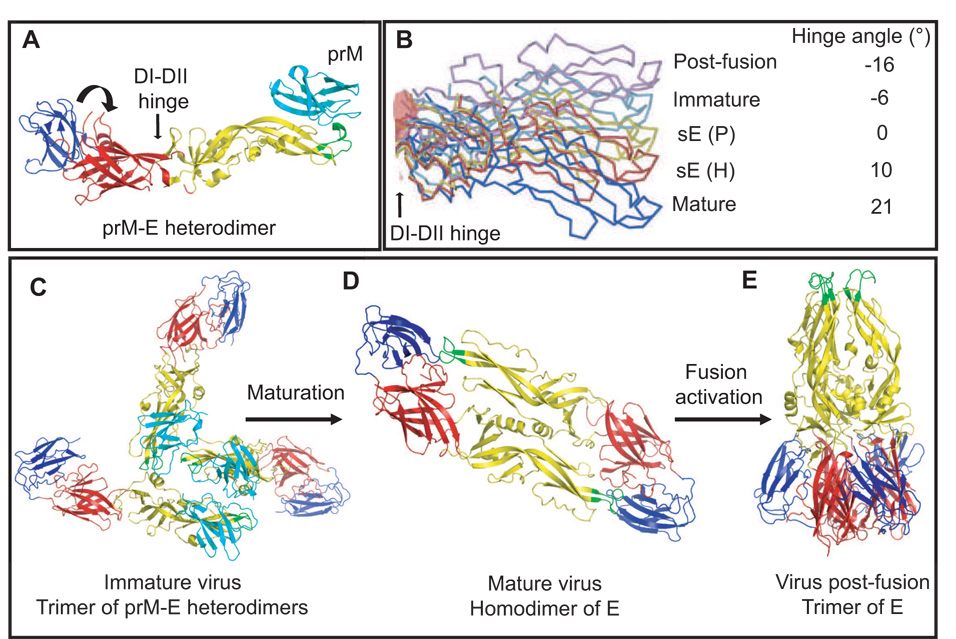
Figure 2. Structure of the flavivirus E protein and its various oligomeric states.
A. A ribbon diagram of the atomic structure of the prM-E heterodimer (Li et al., in press). The E protein is colored as follows: domain I (red), domain II (yellow), domain III (blue) and the fusion peptide (green). The prM protein (cyan) is only found in the immature virus and is shown in its role as a cap structure that protects the fusion peptide. The icosahedral asymmetric unit is shown as a white triangle with the 5-fold and 3-fold axes labeled. The DI-DII hinge is shown with a straight arrow. The movement of DIII with respect to DI during fusion activation is indicated with the circular arrow. B. DII of the E protein is shown as represented by the different structures of E. sE(P), sE(H) and the post-fusion confirmation represent the known crystal structures of E (Modis et al., 2003, 2004; Y. Zhang et al., 2003b; Bressanelli et al., 2004), while the mature and immature conformations represent the fit of the atomic structure of E into the cryo-EM density of the virus (Kuhn et al., 2002; Zhang et al., 2003a; Zhang et al., 2004). The structures were superimposed with respect to DI. Flexibility at the hinge between DI and DII (indicated with an arrow) gives rises to varying hinge angles along the direction of the hinge axis and is suggested to be important in the structural transitions of E during the flavivirus life cycle. C. Oligomeric state of the E protein in the immature particle, D. mature particle, E. fusion active conformation. In the immature particle the E protein forms a trimer of prM-E heterodimers, which must dissociate and form E protein homodimers during maturation. These then undergo further reorganization to form E homotrimers prior to fusion and entry of the virus. Transitions of the E protein are supported by hinge motions that occur between DI-DII and DI-DIII (Modis et al., 2003, 2004; Y. Zhang et al., 2003a,b).
Domains I and II are connected by four polypeptide chains. The hinge angle between these two domains varies by approximately 20° in the various structures of E (Figure 2B). Domain I and III are connected by a single polypeptide linker. Hinge motion at both DI-DII and DI-DIII play an important role during the structural rearrangements of the E protein as it transitions between immature, mature and fusion-active forms of the virus. The DENV-2 crystal showed the presence of a N-octyl-β-d-glucoside (β–OG) molecule that was located in a hydrophobic pocket between DI and DII of selected E protein monomers (Modis et al., 2003). This observation sparked much interest in this pocket as a potential site for binding small molecules that might inhibit conformational changes in the E protein (Li et al., 2008b; Modis et al., 2003; Zhou et al., 2008). The C-terminal region of E, absent in the above structures, is directed towards the viral membrane and consists of a “stem” in the form of two helices connected by a highly conserved sequence, and an “anchor” comprised of two transmembrane antiparallel coiled-coils (Allison et al., 1999; Stiasny et al., 1996; Zhang et al., 2004). The stem-anchor region also undergoes structural transitions during the fusion activation of E.
Structural transitions of the E protein
During the flavivirus life cycle, the virion assumes three main conformational states: immature, mature and fusion-activated. The conformation of the E protein differs in these three states (Figures 2C–E) and therefore, structural rearrangements must occur within the glycoprotein shell to achieve these endpoints. These rearrangements stem from the need to transition between prM-E heterodimers in the immature particle to E homodimers in the mature particle, and finally to E homotrimers in the fusion-activated particle. Understanding these transitions is necessary when considering the inhibition of the E protein and its activity during virus entry. The E protein in each of these conformations is described below.
Conformation of E in the immature virus
Virus particles that bud into the ER are termed “immature” due to the presence of a pre-membrane protein (prM) that must be proteolytically processed during virion maturation. Recent studies by Yu et al have indicated that there are two forms of the immature virus (“spiky” and “smooth”) based on the oligomeric state and arrangement of E proteins on the surface of these particles (Yu et al., 2008). The oligomeric state of the E protein is controlled by the pH of the cellular environment and the presence or absence of prM.
Immature virus particles have a diameter of ~600Å (Figures 3A and C). They have a spiky surface protein shell consisting of prM and E proteins, which form 180 heterodimers that are arranged as 60 trimeric spikes. The three E proteins within each spike are tilted such that the long axis of the protein forms a ~25Å angle with the surface of the particle (Zhang et al., 2003b; Zhang et al., 2004). This arrangement places the fusion peptide on DII at the furthest point from the viral surface and DIII in close proximity to the viral membrane. Recently, the structure of the prM-E heterodimer was solved by X-ray crystallography (Li et al., 2008a). Fitting of this prM-E structure into the 12.5Å cryo-electron microscopy (cryo-EM) map of immature (spiky) DENV-2 (Zhang et al., 2004) indicates that the pr peptide portion of prM extends linearly along the E protein surface remaining on the inside edge of the spike. This places the M protein along the dimerization interface on DII, a location where it would prevent homodimerization of E (observed in the mature particle). The prM protein forms a cap-like structure that protects the fusion peptide of E and prevents premature fusion of the virus with host cell membranes (Guirakhoo, Bolin, and Roehrig, 1992; Zhang et al., 2003b). This orientation also allows the carbohydrate moieties on prM proteins to form a hydrophilic surface on the immature virus particle. Conserved histidine residues within the prM and E proteins are appropriately located within the interface of the heterodimer and suggest a pH-mediated interaction between the two proteins (Li et al., 2008).
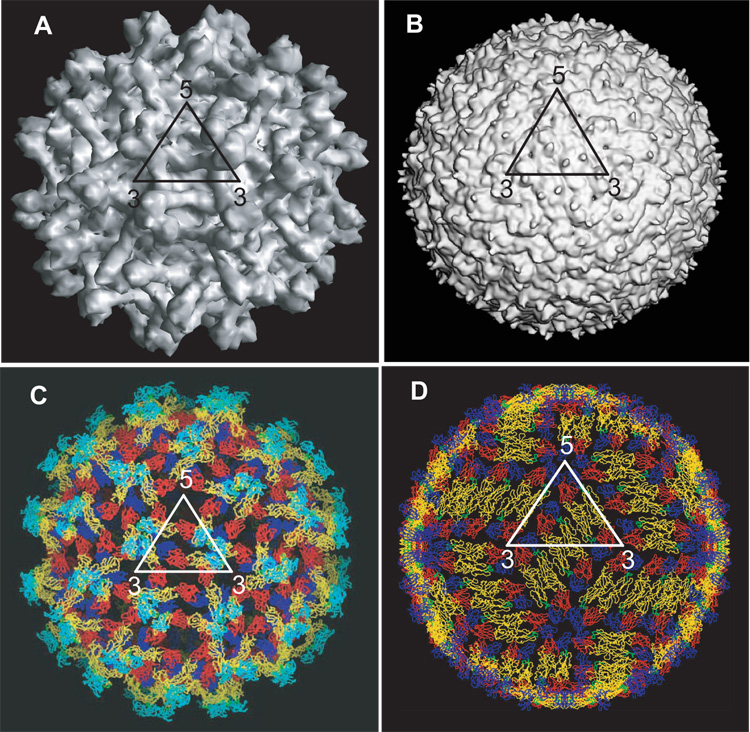
Figure 3. Structure of flaviviruses.
A. A surface shaded view of the cryo-EM reconstruction of immature DENV-2 showing the spiky surface features (Y. Zhang et al., 2004). The map is calculated to 12.5Å. B. A surface shaded view of the cryo-EM reconstruction of mature DENV-2 showing the relatively smooth surface features (map is calculated to 14Å resolution) (W. Zhang et al., 2003a). C. Fit of the atomic coordinates of the E protein Cα residues into the immature virus (Li et al., in press) and D. mature virus (W. Zhang et al., 2003a), showing the arrangement of the E proteins on the surface of the virion. The E proteins are colored as in figure 2. Note the difference in orientation of the E proteins as well as the surface packing of the E proteins in the immature and mature viruses.
When these immature “spiky” virus particles transit through the Golgi apparatus, they encounter a low pH environment that triggers significant rotational and translational movements within the glycoprotein lattice. These movements result in the transition of the E proteins from prM-E heterodimers (Figure 3C) to antiparallel E homodimers (Figure 3D) that lie flat against the viral membrane. The resulting particle has a “smooth” morphology, but is still considered to be immature, due to the presence of prM protecting the fusion peptide on E. The transition from spiky to smooth morphology is reversible, becoming irreversible only when furin cleavage of prM (maturation) has occurred. Furin cleavage occurs in the trans-Golgi and results in the processing of prM to pr and M. The cleaved pr portion remains associated with E and is only released from the virus particle following exit into a neutral pH environment (Yu et al., 2008). Therefore, following maturation, release of the particle into a neutral pH environment does not cause any further re-arrangement of the E proteins, but instead drives the release of the pr peptide from the virion (Yu et al., 2008). The ‘mature’ particles are competent for fusion and entry into new host cells.
Conformation of E in the mature virus
Mature flavivirus particles (diameter ~500Å) have a relatively smooth surface with the lipid bilayer membrane completely covered by the envelope (E) and membrane (M) protein shell (Figure 3B and D). This shell consists of 180 copies of the E protein arranged as 90 homodimers forming a herringbone pattern or so-called protein rafts that lie flat on the viral surface. The E protein homodimers within these rafts consist of two monomers associated in an antiparallel orientation with DII forming the primary dimerization interface (Zhang et al., 2004; Modis et al., 2003; Modis et al., 2005; Kanai et al., 2006; Nybakken et al., 2006). The fusion loop at the distal end of DII (now missing its pr cap) is buried in a pocket between DI and DIII (see Figure 2D, where the fusion loop is shown in green). Comparison of the E protein in the mature and immature viruses indicates that flexibility at the hinge between DI and DII (angular difference of 27°) primarily underlies the ability of E to adopt distinct conformations in these two particles (Figure 3).
Flavivirus entry and the post-fusion conformation of E
Several distinct events contribute to flavivirus entry into target cells. Initially, viral glycoproteins interact with molecules on the surface of host cells as a point of attachment. Following attachment, specific cell surface receptors mediate endocytosis of the virus. In the endosome, the fusion peptide of the virus becomes exposed at the distal end of the E protein and is inserted into the host membrane. Low pH-induced structural rearrangements within E then bring the transmembrane domains anchored in the viral membrane closer to the fusion peptide, forming a hairpin structure that promotes fusion of the viral and host membranes. The flavivirus fusion mechanism is similar to that of other other viruses, such as the alphaviruses, with class II fusion mechanisms and is attributed to their very similar secondary and tertiary fusion protein structure (reviewed in Kielian et al., 2000; Heinz and Allison, 2001; Staisny and Heinz, 2006; Mukhupadhyay et al., 2004; Earp et al., 2005; Harrison et al., 2005; Schibli and Weissenhorn 2004; Cohen and Melikyan 2004; Jardetzky and Lamb, 2004).
Flavivirus fusion has a maximum pH threshold of 6.6–6.8 (Ueba and Kimura, 1977; Gollins and Porterfield, 1986; Summers et al., 1989; Randolph and Stollar, 1990; Guirakhoo et al., 1991, 1993; Despres et al., 1993; McMinn et al., 1996; Corver et al., 2000; Stiasny et al., 2003). In the late endosome, the E homodimers within the mature virion dissociate and re-arrange into fusion-active homotrimers (Allison et al., 1995; Stiasny et al., 2002, 2007). In this conformation, the E proteins are in a parallel orientation to one another within the trimer, extending vertically away from the virion surface. As a result, the fusion loop on DII that was previously buried in the DI/DIII pocket in the homodimer becomes exposed and available to insert into a target host cell membrane.
The post-fusion structures of the DENV and TBEV E protein have been solved by X-ray crystallography (Modis et al., 2004; Bressanelli et al., 2004). The DENV post-fusion E structure is shown in Figure 2E. The structures suggest that the E protein trimers differ markedly from the mature homodimers due to the rotation and translation of the three domains relative to one another. In the E protein of DENV-2, domain II rotates approximately 30° relative to domain I through the movement of a hairpin that resides between the two domains. This hairpin induces a switch between an “open” (β-OG bound) and “closed” conformation of a hydrophobic pocket that was observed in the selected crystal structures of E. Residues within this pocket have been shown to influence the pH threshold of fusion (Modis et al., 2003).
The largest displacement is found in DIII, which folds over DI, rotating approximately 70° to bring its C-terminus closer to the fusion loop in DII. This displacement is mediated by a 10-residue linker (residues 290–299 in DENV-2) that was previously found to be disordered in the E homodimer, but assumes a short β-strand configuration in the trimer. Furthermore, the aromatic residues (W101 and F108, TBEV) that are buried in the pocket between DI/DIII of the dimer become exposed in the trimer, suggesting an interaction of this region with the aliphatic groups in the lipid bilayer during fusion. The fusion peptide loops exposed at one end of the trimer still maintain the conformation observed in the dimer, but several polar groups in the loop are exposed, suggesting that the fusion loop only interacts with the polar head groups of the lipid outer leaflet and does not penetrate much deeper (only ~6Å) (Modis et al., 2004; Stiasny et al., 2004; Bresanelli et al., 2004). These post-fusion conformations of E suggest that in the final fusogenic form of the E trimer, the fusion peptide loops are juxtaposed with the transmembrane domains of E, forming a hairpin-like structure. These structural transitions of E present ideal targets that can be explored as surfaces for structure-based antiviral design.
Inhibition of protein-protein interactions
Several groups have identified molecules that interfere with flavivirus entry. Liao and Kielian (2005) demonstrated that a recombinant form of DENV-2 DIII that included helix I of the E protein blocked flavivirus entry by specifically inhibiting virus fusion. However, DIII from Semliki Forest virus, an alphavirus, did not inhibit DENV-2 entry. The alphaviruses were also blocked at the fusion step by their own DIII proteins. Therefore, the authors suggest that exogenous DIII proteins could function as inhibitors of class II fusion mechanisms. Their study also revealed that DIII functioned by binding to fusion intermediates following low pH-induced trimerization and prevented hairpin formation (Liao and Kielian, 2005).
Chu et al. have also shown that a recombinant form of WNV DIII blocked entry of WNV into Vero and mosquito cells while it only effectively blocked DENV-2 entry into mosquito cells (Chu et al., 2005). The corresponding DIIIs of DENV-1 and DENV-2 could inhibit the entry of the respective viruses into HepG2 and mosquito cells. Murine polyclonal antibodies generated against these soluble DIII proteins were capable of neutralizing these viruses in plaque reduction assays (Chin et al., 2006). Several peptides derived from a murine brain cDNA library were shown to inhibit WNV in vitro at a concentration of 2.6–67 µM. These peptides reduced viremia and fatality in mice during a challenge with WNV, and some were also found to cross the blood-brain barrier (Bai et al., 2007).
Other molecules that have been explored as entry inhibitors include sulfated polysaccharides, polyoxotungstates, and sulfated galactomannans (Talarico et al., 2005, 2007; Pujol et al., 2002; Ono et al., 2003), sulfated glycosaminoglycans, heparin and suramin (Chen et al., 1997; Marks et al., 2001; Lee et al., 2006; Lee and Lobigs, 2000, 2002; Goto et al., 2003; Mandle et al., 2001). The biological effects of these molecules that block flavivirus entry may be explained and potentially improved by analysis of the vast array of structural data currently available on these viruses and their component proteins. These structural studies provide insight into the role of the E protein in the flaivivirus life cycle and present several promising targets for the design of entry inhibitors. Three specific regions on the E protein have emerged from structural studies as targets for the rational design of antivirals against these viruses. They include the β-OG pocket, the E-protein rafts in the mature virus and the E-protein homotrimer.
The β-OG binding pocket
The discovery by Modis and colleages of a ligand binding pocket buried at the hinge between DI and DII and its movement in the fusion activation of E made it a prime target for the design of compounds that might inhibit required structural transitions of the virus (Modis et al., 2003). Several groups including ours have undertaken a targeted drug discovery search for biologically active compounds that bind in this pocket and inhibit virus assembly and entry (Li et al., submitted; Zhou et al., submitted).
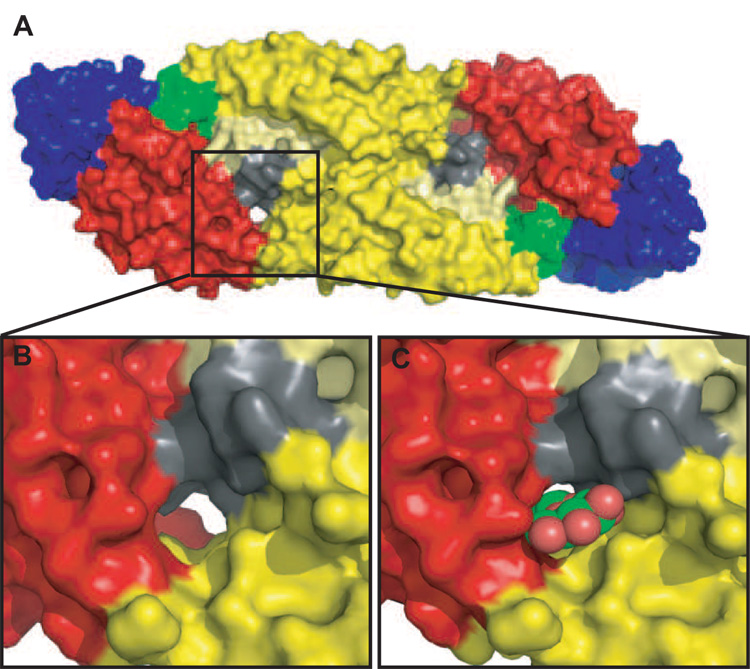
The β–OG pocket forms a channel with open access at both ends (Figure 4), allowing linear molecules of varying lengths to be accommodated. The channel is lined by hydrophobic residues that have been shown through mutagenesis studies to influence the pH threshold of fusion (Modis et al., 2003). The movement of a loop (kl), that resides between DI and DII (shown in grey in Figure 4) controls access to the channel for small hydrophobic molecules. In the β–OG-bound structure of DENV-2 E, the kl loop formed a salt bridge and hydrogen bond with the ij loop (shown in pale yellow) in the dimer partner (within the homodimer of E) forming an “open” conformation of the pocket. This conformation exposes the hydrophobic core and accommodates binding of a single β–OG molecule (Figure 4C). In the absence of β–OG, a “closed” conformation of the pocket was observed, with the kl loop burying the underlying hydrophobic residues (as seen in the crystal structures of TBEV, DENV-3, WNV and selected structures of DENV-2). It has been proposed that the varying conformations of this pocket induced by the movement of the kl loop play an important role during fusion. Specifically, the loop assists in the movement of DII and allows the fusion peptide at the distal end of DII to be directed towards the host cell membrane. The β-OG pocket therefore may prove to be an ideal target for the inhibition of viral fusion and entry. Binding of small molecules that “pry open” the pocket may trigger conformational changes similar to those induced by low pH and induce premature fusion. Alternatively, inhibitors binding in the pocket may also prevent the structural transitions necessary for maturation and fusion activation of the virus (Modis et al., 2003).
Figure 4. β-OG binding pocket in the E protein.
A. A space-filled representation of the atomic structure of the E protein homodimer (Modis et al., 2003; Y. Zhang et al., 2003). The E proteins are colored as in Figure 2, with the exception of the kl loop (residues 268–280 in DENV-2) (shaded in grey) that is suggested to control the ‘open’ and ‘closed’ confirmation of the β-OG pocket (boxed). This kl loop interacts with the ij loop in DII (residues 237–253 of DENV-2 and shaded in pale yellow) during these conformational changes. B. An enlarged view of the β-OG pocket pocket in its ‘apo’ conformation. Note that the pocket forms a channel with open access at both ends. C. An enlarged view of the pocket with β-OG occupying the pocket.
Observations from the picornaviruses provide precedence for these hypotheses (Pavear et al.,1989; Rossmann 1994; Rossmann et al., 2000; Badger et al., 1988; Heinz et al., 1989; Smith et al., 1986; Fox et al., 1986; Kim et al., 1993; Reisdorph, 2003). The picornavirus virion consists of four structural proteins (VP1-4), with VP1-3 forming the external protein shell and VP4 remaining inside the capsid (Rossmann, 1985). VP4 is released from the capsid upon uncoating. In these viruses, a deep canyon is formed at the junction between VP1 and VP2/3 that functions as a receptor binding site as well as an antigenic site for neutralizing antibodies. This canyon is found around the five-fold vertices in the capsid. A pocket within VP1 in the canyon floor was found to bind antiviral compounds (WIN drugs) (Smith et al., 1986; Badger et al., 1988; Kim et al., 1993; Reisdorph, 2003). Binding of these compounds inhibited uncoating and entry of the virus by preventing “breathing” of the capsid. It was also observed that conformational changes occurred in residues forming the canyon floor upon binding the compounds (up to 4Å movement in Cα positions) that could potentially prevent receptor binding and attachment to host cells.
To identify small molecules that directly bind the β–OG pocket and functionally inhibit the virus, we have used a hierarchical computational approach to screen three compound libraries (a total of 143,000 compounds) from the National Cancer Institute (Zhou et al., submitted; Li et al., submitted). The 45 top-scoring compounds selected from the computational docking approach were further visually screened for drug-like properties and ideal structural characteristics for interacting with the β-OG pocket. A group of 23 compounds were tested for cytotoxicity (CC50) in baby hamster kidney cells (BHK) and inhibitory activity against YFV (Zhou et al., submitted; Li et al., submitted).
Three approaches were used to assess virus spread. Initially, the effect of the compounds was monitored using a YFV expressing the fire-fly luciferase gene. BHK cells were infected at low multiplicity and levels of luciferase activity in infected cell lysates was measured at various compound concentrations. Nine compounds inhibited virus spread at inhibitory concentrations (IC50) between 20–500 µM. In a second approach, pseudo-infectious particles (PIPS) obtained using a replicon system were produced in cells either treated with compounds or untreated. These PIPS are generated through sequential transfection of BHK cells with two RNA transcripts independently expressing the viral non-structural proteins and the viral structural proteins. The particles produced by this method are only capable of a single round of infection. Therefore, PIPs provide a direct measure of virus assembly and release, as well as virus entry, requiring all of the structural transitions of E to be completed. As originally designed, 5 of the 9 compounds inhibited virus maturation/entry and/or fusion.
In a third approach, a replicon expressing the viral non-structural proteins and the Renilla luciferase gene was used to monitor the effect of these compounds on viral RNA replication. The replicon is capable of autonomous replication but cannot spread from cell to cell due to the lack of the structural proteins (Jones et al., 2005). Luciferase activity of cells transfected with this construct provides a direct measure of the replication of the viral RNA. Therefore, it should identify compounds that may influence only viral RNA replication. Of the 9 compounds tested, 4 affected RNA replication. Promisingly, there was no overlap between the compounds that affected the structural transitions of E and those affecting viral RNA replication (Zhou et al., submitted; Li et al., submitted).
NMR studies show that one of the compounds directly binds the DENV E protein and competes with β–OG (Zhou et al., submitted). In the DENV-2 E structure, the β–OG molecule is oriented with the glucosyl head group in the channel’s mouth and the hydrocarbon chain projecting deep into the channel’s cavity. Hydrogen bonds between the pocket residues and the β–OG molecule fix its orientation in the cavity (Modis et al., 2003). Similarly, several of the compounds mentioned above are predicted to form hydrogen bonds with pocket residues that lie on the top of the channel (Zhou et al., submitted). These interactions potentially contribute to the binding free energy, as well as specificity, as observed with HIV-1 reverse transcriptase inhibitors (Zhou et al., 2004, 2005).
Second- and third-generation compounds were designed from these initial lead compound hits. Promising reactive groups such as thiazole rings and aromatic rings were maintained while eliminating cytotoxic groups such as α,β-unsaturated ketones. Tighter binding was explored by increasing the number of potential hydrogen bonds between the pocket residues and the compounds. These efforts were rewarded by the design and synthesis of two compounds that showed IC50 values ranging from 1–5 µM. Computational docking indicated that they bound into a channel in the pocket that increased in hydrophobicity with increasing depth from the entrance to the bottom. Three electrostatic/steric cavities within this channel accommodated various parts of the two compounds. The most potent compound was smaller and bound very deeply into the channel and interacted with a cavity surrounded by hydrophylic residues (Zhou et al., submitted; Li et al., submitted).
E-protein rafts
The E-protein rafts (Figure 3D) that densely pack against the viral membrane in the mature virus present another promising target for the design of flaviviral entry inhibitors. These rafts provide ideal protein surfaces for docking small molecules that might interfere with protein-protein interactions. Molecules that stabilize the dimers in the mature virus and prevent downstream structural rearrangements could also be effective. Such inhibitors could prevent the transformation of E into the low pH-induced trimer conformation required for fusion and entry into naïve cells.
A similar computational docking protocol as mentioned above has been used to identify potential binding surfaces that would accommodate such inhibitors. Two pockets within the rafts have currently been identified and used for high throughput screening of compounds from the same NCI library. Based on initial screens, 14 out of 42 potential compounds have been classified as inhibitors of virion morphogenesis (i.e. they do not affect viral RNA replication). Further analyses of these compounds as potential entry inhibitors are ongoing (La Bauve, Zhou, Kuhn and Post, unpublished data).
Fusion-active trimer of E
The fusion-active state of viruses has long been a target for inhibition as observed in viruses with class I fusion mechanisms (Copeland, 2006; Eckert and Kim, 2001; Veiga et al., 2006; Rusconi et al., 2007; Esté and Telenti, 2007). The active fusion core of viruses such as influenza and the human immunodeficiency virus constitutes a 6-helix bundle consisting of two helices from each fusion protein in the trimer (Skehel and Wiley, 2000; Melikyan et al., 2000; Russel et al., 2001; Chang et al., 2008). Its formation can be prevented by antiviral peptides that mimic the helices and compete with the protein-protein interactions that give rise to the fusion core (Hsieh and Hsu JT., 2007; Stevens and Donis, 2007).
Similarly, the fusion core of the flaviviruses could be targeted to prevent virus entry. As discussed above, preliminary studies have been carried out using exogenous DIII proteins to trap fusion intermediates (Liao and Kielian, 2005; Chu et al., 2005; Chin et al., 2007). The DIII proteins bound a fusion intermediate following trimerization indicating that a trimer intermediate with a relatively prolonged lifetime existed and could be blocked prior to fusion (Liao and Kielian, 2005). These studies could be further extended to prevent the formation of the trimer, by sterically inhibiting the movement between DI and DIII and preventing the folding over of DIII (Figure 2). The existence of intermediates prior to trimer formation has been previously observed (Stiasny et al., 2002, 2007). These studies have indicated that a monomeric E intermediate capable of interacting with membranes in a pre-hairpin conformation does exist prior to the formation of E homotrimers. In addition, Stiasny et al., also showed that DIII relocation and trimer formation were concomitant and occurred after the membrane binding of the monomeric E pre-hairpin. These intermediates are targets that could be considered during the design of entry inhibitors.
Although class I and II fusion proteins are structurally distinct, the end result remains the same. The former undergoes a refolding step prior to forming its fusion active conformation, and the latter re-orients domains with limited refolding. Ultimately, a hairpin structure is formed with the fusion loop and transmembrane regions juxtaposed in the same membrane. Therefore, lessons learned from class I fusion proteins could be adapted to inhibit flavivirus fusion and entry.
IV. Other potential sites for the design of flaviviral entry inhibitors
Apart from the binding pockets within the E protein that are obvious targets for the design of antivirals, several other interactions involving the E protein provide promising targets for the design of entry inhibitors. Studies have shown that attachment molecules are required for virus entry. The interaction of E with these molecules have been studied in detail using structural and biochemical techniques and can be pursued as an option for interrupting virus entry. In addition, the interaction of the E protein with neutralizing antibodies has been studied extensively. Structural information gleaned from these interactions can be utilized to design novel entry inhibitors with increased efficacy.
Attachment molecules
Several attachment molecules important for flavivirus entry have been identified. The C-type lectin, dendritic cell-specific ICAM3 grabbing nonintegrin (DC-SIGN) has been shown to be essential for DENV infections through its interaction with carbohydrate moieties on the E protein (Navarro-Sanchez et al., 2003; Tassaneetrihep et al., 2003). Depending on the virus from which it is derived, one or two N-linked glycosylation sites are found on the E protein. Asn153 is conserved among all flaviviruses while Asn67 is unique to DENV (Rey, 2003). N-linked glycosylation of Asn67 is required for DENV growth in mammalian cells (Bryant et al., 2007; Mondotte et al., 2007). The above-cited studies by Navarro-sanchez et al. and Tassaneetrihep et al. have demonstrated that both soluble DC-SIGN and antibodies against DC-SIGN inhibit DENV infection. However, Lozach et al. (2005) showed that internalization of DC-SIGN was not necessary for DENV infectivity. It therefore probably does not function as a specific receptor, but allows for virus attachment and concentration on the cell surface.
Structural insight into the interaction of DENV E with DC-SIGN has been obtained through a cryoEM reconstruction of DENV-2 in complex with the carbohydrate recognition domain (CRD) of human DC-SIGN (Podishevskaya et al., 2006). The CRD bound the Asn67 residue in the DENV E protein. Interestingly, binding did not induce conformational changes in the E protein on the mature virus, although such changes may occur when full-length DC-SIGN binds to E. The stoichiometry of binding between the CRD and the E proteins on the virion surface left one E molecule in the asymmetric unit unoccupied and the putative DIII receptor binding domain of each E molecule free to bind the receptor (on the icosahedral 5 fold and 3 fold axes). Based on the number of DC-SIGN molecules that interact with the virus, the oligomeric state of DC-SIGN on the surface of a cell is a tetramer, so binding of DC-SIGN could therefore promote clustering of the virus on the cell surface and assist in receptor binding. The authors suggested that the binding of the carbohydrate moieties to DC-SIGN mimics normal cellular processes and therefore functions to protect the receptor-binding domain from immune surveillance and neutralization. By contrast, WNV binds the related C-type lectin, DC-SIGNR, during dendritic cell infections (Davis et al., 2006), while YFV does not have any glycan modification on E and therefore attaches to cells in a lectin-independent manner (Barba-Spaeth et al., 2005).
Another C-type lectin receptor, the mannose receptor (MR), has recently been shown to bind DENV, JEV and TBEV through a mechanism similar to that of DC-SIGN. However, the ligand specificity of MR (terminal mannose, fucose and N-acetyl glucosamine) differs from that of DC-SIGN (high-mannose oligosaccharides and fucosylated glycans). The authors propose that MR has the potential for being a DENV receptor (rather than just an attachment molecule), because it is constitutively internalized and found mainly in the endocytic pathway, in contrast to DC-SIGN, which is mainly localized in the plasma membrane (Miller et al., 2008).
Other molecules that have been implicated in assisting flavivirus entry include heparan sulfate (Hung et al.,1999; Kroschewski et al., 2003; Liu etal., 2004), αvB3 integrin (Chu and Ng, 2004), Rab5 (Krishnan et al., 2007), HSC70 (Ren et al., 2007) and BiP (Jindadamrongwech et al., 2004). αvB3 integrin is an endothelial cell receptor that is implicated in WNV and JEV entry of vertebrate cells. Integrin binding has been predicted for the flaviviruses, because domain III of most flavivirus envelope proteins has an RGD-type motif important for integrin-ligand interactions (van der Most et al., 1999). Other viruses, including foot-and-mouth disease virus, coxsackie virus and adenovirus, also interact with integrins in an RGD-dependent manner (Roivainen et al., 1991; Reider et al., 1994; Bai et al., 1993). However, the binding of WNV to αvB3 integrin was independent of the RGD motif. Antibodies against this integrin, as well as soluble forms of the protein, inhibited WNV and JEV entry into permissive cells. RNAi studies also supported the observation that this particular integrin might serve as a receptor for WNV (Chu and Ng, 2004). Rab5 GTPase is a key regulator of traffic to the early endosome and has been implicated in DENV and WNV entry. Dominant negative inhibition or RNAi-based inhibition of Rab5 (but not Rab7, a late endosomal GTPase) significantly reduced DENV and WNV entry and replication in Hela cells (Krishnan et al., 2007).
Neutralizing epitopes on E
The humoral immune response plays an important role during flavivirus infections and several antibodies that are effective in neutralizing these viruses have been identified (Halstead, 1989; Halstead, Porterfield, and O'Rourke, 1980)Kaufman et al., 1987, 1989; Phillpotts et al., 1987; Johnson and Roehrig, 1999; Mathews and Roehrig, 1984; Roehrig et al., 1998, 2001; Roehrig, 2003; Diamond et al., 2003; Sukupolvi-Petty et al., 2007). The interaction of the E protein with these antibodies provides insight into epitopes that might be accessible during structural transitions of the virus and presents novel avenues of antibody-mediated therapeutic intervention (Crill and Chang, 2004; Crill and Roehrig, 2001; Halstead et al., 2005; Li, Barrett, and Beasley, 2005; Pierson et al., 2007; Stiasny et al., 2006)Sukupolvi-Petty etal., 2007; Kaufmann etal., 2007; Lok etal., 2008; Beasley and Barrett, 2002; Lin et al., 1994; Lin and Wu 2003; Oliphant etal., 2005, 2006; Roehrig et al., 1998; Sanchez et al., 2005). Specifically, antibodies or Fabs can interfere with virus attachment, membrane fusion, and internalization mediated by the E protein, and they can also trap intermediates during the structural transition between mature and fusion-active forms of the virus. Although the primary neutralizing epitopes lie on DIII, cross-reactive epitopes have also been identified on DI and DII (Oliphant et al., 2006, Crill and Chang, 2004; Goncalvez et al., 2004; Ledizet et al., 2007).
A pseudo-atomic structure of the neutralizing monoclonal antibody E16 Fab in complex with WNV was recently determined (Nybakken et al., 2005; Kaufmann et al., 2006). This structure suggests that E16 interferes with virus entry by blocking conformational rearrangement of E at a step following receptor attachment. In this structure, E16 only partially obscures the surface of the particle by binding to 120 of 180 DIII domains, leaving those on the five-fold axes of the particle available for receptor binding. This preferential binding has been attributed to steric hindrance that prevents complete occupancy of all DIII epitopes. These observations were predicted from previous in vitro studies that indicated that E16 only partially prevents attachment of the virus to cell surface receptors and blocks infection at a step following receptor binding (Nybakkan et al., 2005; Oliphant et al., 2005). It is plausible that E16 may function at a step following receptor binding by inhibiting these rearrangements necessary for the mature virus to transform to its fusion-active state. This structure also suggests a combination therapeutic strategy that could utilize both E16 antibodies and antivirals designed against the co-receptors (exposed at the five fold axes) to block flavivirus entry. Studies of mice exposed to WNV showed that treatment with E16 five days postexposure to WNV resulted in a 90% survival rate, with complete clearance of the virus in the brains of 68% of the treated mice (Oliphant et al., 2005). Interestingly, the interaction of E16 with WNV seems to be quite different from the interaction the 1A1D-2 Fab with DENV (Lok et al., 2008).
In DENV infections, the monoclonal antibody 1A1D2 strongly neutralizes DENV serotypes 1, 2 and 3 by inhibiting attachment of virus to host cells, but it does not bind to serotype 4 (Roehrig et al., 1998). A pseudo-atomic structure of the Fab fragment of 1A1D2 in complex with the virus has recently been determined and suggests a mechanism for virus neutralization (Lok et al., 2008). In this structure, it was observed that the Fab bound to an epitope on DIII of the E protein that was normally occluded in the mature virus. Fab binding required higher temperatures, suggesting that “breathing” of the virus was a prerequisite for antibody recognition. Based on these observations, structural transitions that might require breathing of the viral proteins could be trapped through the use of antibodies. It has already been demonstrated that such trapped complexes are incapable of efficient infection (as demonstrated by the neutralization efficacy of 1A1D2). This structure also provides insight into cryptic epitopes in E that could be targeted by antivirals. As discussed previously, this phenomenon has been observed for nodavirus (Bothner et al., 1998) and rhinovirus (Lewis etal., 1998). For instance, the breathing of rhinovirus exposed internal regions of the VP4 structural protein making this protein sensitive to proteolysis. These breathing motions were inhibited however, in the presence of antiviral compounds that bound in a cavity within VP1 that stabilized the virion (Lewis et al., 1998; Reisdorph et al., 2003; Smith et al., 1986; Badger et al., 1988; Kim et al., 1993). Further evidence that hidden epitopes might function as immunogenic sites and potential targets for antivirals stems from a recent report that immunoglobulins raised against linear epitopes of all three domains of the E protein protected mice against lethal WNV challenge (Ledizet et al., 2007).
An alternate scenario is possible for MAbs that bind DI and DII epitopes of the E protein. As mentioned previously, ADE is a phenomenon with devastating consequences, caused by a weakly neutralizing immune response to a prior DENV infection and is a serious concern for flavivirus vaccine development (Halstead, 1979). Several MAbs directed against DI and DII of the E proteins demonstrated more enhancing effects (characteristic of ADE) than neutralization properties during virus challenge, in contrast to antibodies against DIII, which were strongly neutralizing (Oliphant et al., 2006). The authors therefore suggested that productive neutralization of DENV would require redirecting the antibody response from the enhancing effects of the DI and DII epitopes towards the more protective DIII epitopes. Structure-based antiviral design could allow this by eliminating or inhibiting the availability of epitopes of DI and DII to the immune response, preventing their ADE effects, and prove to be a potential strategy to prevent flavivirus infection.
V. Concluding remarks
Given all of these possible strategies, it is remarkable that few successful antivirals have been developed against the flaviviruses, but the challenges ahead are clear. New structural insights into the flavivirus life cycle and viral interactions with cellular molecules and antibodies provide great opportunities for identifying new classes of inhibitors. The ability to obtain high-resolution structures of viral components and inhibitory compounds suggests that powerful structure-based approaches could rapidly focus the development of highly efficacious compounds. The same techniques could be used to design compounds that evade virus resistance and demonstrate broad anti-flaviviral activity. However, it is important to recognize that rapid and precise diagnosis will be essential to the effectiveness of anti-dengue drugs. Therefore, improvement in diagnostic tests for dengue must proceed in parallel with new therapies. The severity and duration of dengue fever might be effectively controlled by antivirals upon early diagnosis, however, these compounds may not be as effective if the infection has progressed to DHF. In this form of the disease, due to complications caused by an active immune response, immunomodulatory compounds and strategies may have greater impact on disease intervention.
Acknowledgements
We would like to thank Joshua Yoder, Mayuri and Michael Owsten for critical reading of the manuscript. We also thank Mark Cushman and Carol Post for extensive discussions. This work is sponsored by the NIH/NIAID Regional Center of Excellence for Bio-defense and Emerging Infectious Dieseases Research (RCE) program. The authors wish to acknowledge membership within and support from the Region V ‘Great Lakes’ RCE (NIH award 1-U54-AI-057153).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allison SL, Schalich J, Stiasny K, Mandl CW, Heinz FX. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J. Virol. 2001;75:4268–4275. doi: 10.1128/JVI.75.9.4268-4275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison SL, Stiasny K, Stadler K, Mandl CW, Heinz FX. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J. Virol. 1999;73:5605–5612. doi: 10.1128/jvi.73.7.5605-5612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison SL, Schalich J, Stiasny K, Mandl CW, Kunz C, Heinz FX. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 1995;69:695–700. doi: 10.1128/jvi.69.2.695-700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger J, Minor I, Kremer MJ, Oliveira MA, Smith TJ, Griffith JP, Guerin DMA, Krishnaswamy S, Luo M, Rossmann MG, McKinlay MA, Diana GD, Dutko FJ, Fancher M, Rueckert RR, Heinz BA. Structural analysis of a series of antiviral agents complexed with human rhinovirus 14. Proc. Natl. Acad. Sci. USA. 1988;85:3304–3308. doi: 10.1073/pnas.85.10.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F, Town T, Pradhan D, Cox J, Ashish, Ledizet M, Anderson JF, Flavell RA, Krueger JK, Koski RA, Fikrig E. Antiviral peptides targeting the west nile virus envelope protein. J. Virol. 2007;81:2047–2055. doi: 10.1128/JVI.01840-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M, Harfe B, Freimuth P. Mutations that alter an Arg-Gly-Asp (RGD) sequence in the adenovirus type 2 penton base protein abolish its cell-rounding activity and delay virus reproduction in flat cells. J. Virol. 1993;67:5198–5205. doi: 10.1128/jvi.67.9.5198-5205.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barba-Spaeth G, Longman RS, Albert ML, Rice CM. Live attenuated yellow fever 17D infects human DCs and allows for presentation of endogenous and recombinant T cell epitopes. J. Exp. Med. 2005;202:1179–1184. doi: 10.1084/jem.20051352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley DW, Barrett AD. Identification of neutralizing epitopes within structural domain III of the West Nile virus envelope protein. J. Virol. 2002;76:13097–13100. doi: 10.1128/JVI.76.24.13097-13100.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj S, Holbrook M, Shope RE, Barrett AD, Watowich SJ. Biophysical characterization and vector-specific antagonist activity of domain III of the tick-borne flavivirus envelope protein. J. Virol. 2001;75:4002–4007. doi: 10.1128/JVI.75.8.4002-4007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothner B, Dong XF, Bibbs L, Johnson JE, Siuzdak G. Evidence of viral capsid dynamics using limited proteolysis and mass spectrometry. J. Biol. Chem. 1998;273:673–676. doi: 10.1074/jbc.273.2.673. [DOI] [PubMed] [Google Scholar]
- Bressanelli S, Stiasny K, Allison SL, Stura EA, Duquerroy S, Lescar J, Heinz FX, Rey FA. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 2004;23:1–11. doi: 10.1038/sj.emboj.7600064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant JE, Calvert AE, Mesesan K, Crabtree MB, Volpe KE, Silengo S, Kinney RM, Huang CY, Miller BR, Roehrig JT. Glycosylation of the dengue 2 virus E protein at N67 is critical for virus growth in vitro but not for growth in intrathoracically inoculated Aedes aegypti mosquitoes. Virol. 2007;366:415–423. doi: 10.1016/j.virol.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Chang DK, Cheng SF, Kantchev EA, Lin CH, Liu YT. Membrane interaction and structure of the transmembrane domain of influenza hemagglutinin and its fusion peptide complex. BMC Biol. 2008;15(6):2. doi: 10.1186/1741-7007-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Maguire T, Hileman RE, Fromm JR, Esko JD, Linhardt RJ, Marks RM. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat. Med. 1997;3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- Chin JFL, Chu JJH, Ng ML. Tee envelope glycoprotein domain III of dengue virus serotypes 1 and 2 inhibit virus entry. Microb. Infect. 2007;9:1–6. doi: 10.1016/j.micinf.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Chu CL, Buczek-Thomas JA, Nugent MA. Heparan sulphate proteoglycans modulate fibroblast growth factor-2 binding through a lipid raft-mediated mechanism. Biochem. J. 2004;379(Pt 2):331–341. doi: 10.1042/BJ20031082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu JJ, Rajamanonmani R, Li J, Bhuvanakantham R, Lescar J, Ng ML. Inhibition of West Nile virus entry by using a recombinant domain III from the envelope glycoprotein. J. Gen. Virol. 2005;86(Pt 2):405–412. doi: 10.1099/vir.0.80411-0. [DOI] [PubMed] [Google Scholar]
- Chu JJ, Ng ML. Interaction of West Nile virus with alpha v beta 3 integrin mediates virus entry into cells. J. Biol. Chem. 2004;279:54533–54541. doi: 10.1074/jbc.M410208200. [DOI] [PubMed] [Google Scholar]
- Cohen FS, Melikyan GB. The energetics of membrane fusion from binding, through hemifusion, pore formation, and pore enlargement. J. Membr. Biol. 2004;199:1–14. doi: 10.1007/s00232-004-0669-8. [DOI] [PubMed] [Google Scholar]
- Copeland KF. Inhibition of HIV-1 entry into cells. Recent Patents Anti-Infect. Drug Disc. 2006;1(1):107–112. doi: 10.2174/157489106775244118. [DOI] [PubMed] [Google Scholar]
- Corver J, Ortiz A, Allison SL, Schalich J, Heinz FX, Wilschut J. Membrane fusion activity of tick-borne encephalitis virus and recombinant subviral particles in a liposomal model system. Virol. 2000;269:37–46. doi: 10.1006/viro.1999.0172. [DOI] [PubMed] [Google Scholar]
- Crill WD, Chang GJ. Localization and characterization of flavivirus envelope glycoprotein cross-reactive epitopes. J. Virol. 2004;78:13975–13986. doi: 10.1128/JVI.78.24.13975-13986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crill WD, Roehrig JT. Monoclonal antibodies that bind to domain III of Dengue E glycoprotein are the most efficient blockers of virus adsorption to vero cells. J. Virol. 2001;75:7769–7773. doi: 10.1128/JVI.75.16.7769-7773.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CW, Nguyen HY, Hanna SL, Sanchez MD, Doms RW, Pierson TC. West Nile virus discriminates between DC-SIGN and DC-SIGNR for cellular attachment and infection. J. Virol. 2006;80:1290–1301. doi: 10.1128/JVI.80.3.1290-1301.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres P, Frenkiel MP, Deubel V. Differences between cell membrane fusion activities of two dengue type-1 isolates reflect modifications of viral structure. Virol. 1993;196:209–219. doi: 10.1006/viro.1993.1469. [DOI] [PubMed] [Google Scholar]
- Diamond MS, Sitati EM, Friend LD, Higgs S, Shrestha B, Engle M. A critical role for induced IgM in the protection against West Nile virus infection. J. Exp. Med. 2003;198:1853–1862. doi: 10.1084/jem.20031223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokland T, Walsh M, Mackenzie JM, Khromykh AA, Ee KH, Wang S. West Nile virus core protein; tetramer structure and ribbon formation. Structure. 2004;12:1157–1163. doi: 10.1016/j.str.2004.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earp LJ, Delos SE, Park HE, White JM. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 2005;285:25–66. doi: 10.1007/3-540-26764-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Ann. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- Elshuber S, Allison SL, Heinz FX, Mandl CW. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J. Gen. Virol. 2003;84(Pt 1):183–191. doi: 10.1099/vir.0.18723-0. [DOI] [PubMed] [Google Scholar]
- Esté JA, Telenti A. HIV entry inhibitors. Lancet. 2007;370:81–88. doi: 10.1016/S0140-6736(07)61052-6. [DOI] [PubMed] [Google Scholar]
- Ferlenghi I, Clarke M, Ruttan T, Allison SL, Schalich J, Heinz FX, Harrison SC, Rey FA, Fuller SD. Molecular organization of a recombinant subviral particle from tick-borne encephalitis. Mol. Cell. 2001;7:593–602. doi: 10.1016/s1097-2765(01)00206-4. [DOI] [PubMed] [Google Scholar]
- Fox MP, Otto MJ, McKinlay MA. The prevention of rhinovirus and poliovirus uncoating by WIN 51711: a new antiviral drug. Antimicrob. Agents Chemother. 1986;30:110–116. doi: 10.1128/aac.30.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollins SW, Porterfield JS. Flavivirus infection enhancement in macrophages: radioactive and biological studies on the effect of antibody on viral fate. J. Gen. Virol. 1984;65:1261–1272. doi: 10.1099/0022-1317-65-8-1261. [DOI] [PubMed] [Google Scholar]
- Gollins SW, Porterfield JS. pH-dependent fusion between the flavivirus West Nile and liposomal model membranes. J. Gen. Virol. 1986;67:157–166. doi: 10.1099/0022-1317-67-1-157. [DOI] [PubMed] [Google Scholar]
- Goncalvez AP, Purcell RH, Lai CJ. Epitope determinants of a chimpanzee Fab antibody that efficiently cross-neutralizes dengue type 1 and type 2 viruses map to inside and in close proximity to fusion loop of the dengue type 2 virus envelope glycoprotein. J. Virol. 2004;78:12919–12928. doi: 10.1128/JVI.78.23.12919-12928.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto A, Hayasaka D, Yoshii K, Mizutani T, Kariwa H, Takashima I. A BHK-21 cell culture-adapted tick-borne encephalitis virus mutant is attenuated for neuroinvasiveness. Vaccine. 2003;21:4043–4051. doi: 10.1016/s0264-410x(03)00269-x. [DOI] [PubMed] [Google Scholar]
- Gould EA, Solomon T. Pathogenic flaviviruses. Lancet. 2008;371:500–509. doi: 10.1016/S0140-6736(08)60238-X. [DOI] [PubMed] [Google Scholar]
- Guirakhoo F, Bolin RA, Roehrig JT. The Murray Valley encephalitis virus prM protein confers acid resistance to virus particles and alters the expression of epitopes within the R2 domain of E glycoprotein. Virol. 1992;191:921–931. doi: 10.1016/0042-6822(92)90267-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirakhoo F, Heinz FX, Mandl CW, Holzmann H, Kunz C. Fusion activity of flaviviruses: comparison of mature and immature (prM-containing) tick-borne encephalitis virions. J. Gen. Virol. 1991;72:1323–1329. doi: 10.1099/0022-1317-72-6-1323. [DOI] [PubMed] [Google Scholar]
- Guirakhoo F, Hunt AR, Lewis JG, Roehrig JT. Selection and partial characterization of dengue 2 virus mutants that induce fusion at elevated pH. Virol. 1993;194:219–223. doi: 10.1006/viro.1993.1252. [DOI] [PubMed] [Google Scholar]
- Hadfield AT, Diana GD, Rossmann MG. Analysis of three structurally related antiviral compounds in complex with human rhinovirus 16. Proc. Natl. Acad. Sci. USA. 1999;96:14730–14735. doi: 10.1073/pnas.96.26.14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead SB. In vivo enhancement of dengue virus infection in rhesus monkeys by passively transferred antibody. J. Infect. Dis. 1979;140:527–533. doi: 10.1093/infdis/140.4.527. [DOI] [PubMed] [Google Scholar]
- Halstead SB. Antibody, macrophages, Dengue virus infection, shock, and hemorrhage: a pathogenic cascade. Rev. Infect. Dis. 1989;11:S830–S839. doi: 10.1093/clinids/11.supplement_4.s830. [DOI] [PubMed] [Google Scholar]
- Halstead SB, Porterfield JS, O'Rourke EJ. Enhancement of dengue virus infection in monocytes by flavivirus antisera. Am. J. Trop. Med. Hyg. 1980;29:638–642. doi: 10.4269/ajtmh.1980.29.638. [DOI] [PubMed] [Google Scholar]
- Halstead SB, Heinz FX, Barrett AD, Roehrig JT. Dengue virus: molecular basis of cell entry and pathogenesis, 25–27 June 2003, Vienna, Austria. Vaccine. 2005;23:849–856. doi: 10.1016/j.vaccine.2004.03.069. [DOI] [PubMed] [Google Scholar]
- Harrison SC. Mechanism of membrane fusion by viral envelope proteins. Adv. Virus Res. 2005;64:231–261. doi: 10.1016/S0065-3527(05)64007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz BA, Rueckert RR, Shepard DA, Dutko FJ, McKinlay MA, Fancher M, Rossmann MG, Badger J, Smith TJ. Genetic and molecular analyses of spontaneous mutants of human rhinovirus 14 that are resistant to an antiviral compound. J. Virol. 1989;63:2476–2485. doi: 10.1128/jvi.63.6.2476-2485.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz FX, Allison SL. The machinery for flavivirus fusion with host cell membranes. Curr. Op. Microbiol. 2001;4:450–455. doi: 10.1016/s1369-5274(00)00234-4. [DOI] [PubMed] [Google Scholar]
- Heinz FX, Mandl CW, Holzmann H, Kunz C, Harris BA, Rey F, Harrison SC. The flavivirus envelope protein E: isolation of a soluble form from tick-borne encephalitis virus and its crystallization. J. Virol. 1991;65:5579–5583. doi: 10.1128/jvi.65.10.5579-5583.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh HP, Hsu JT. Strategies of development of antiviral agents directed against influenza virus replication. Curr. Pharm. Des. 2007;13:3531–3542. doi: 10.2174/138161207782794248. [DOI] [PubMed] [Google Scholar]
- Hung SL, Lee PL, Chen HW, Chen LK, Kao CL, King CC. Analysis of the steps involved in Dengue virus entry into host cells. Virol. 1999;257:156–167. doi: 10.1006/viro.1999.9633. [DOI] [PubMed] [Google Scholar]
- Ishak R, Tovey DG, Howard CR. Morphogenesis of yellow fever virus 17D in infected cell cultures. J. Gen. Virol. 1988;69:325–335. doi: 10.1099/0022-1317-69-2-325. [DOI] [PubMed] [Google Scholar]
- Jardetzky TS, Lamb RA. Virology: a class act. Nature. 2004;427:307–308. doi: 10.1038/427307a. [DOI] [PubMed] [Google Scholar]
- Jindadamrongwech S, Thepparit C, Smith DR. Identification of GRP 78 (BiP) as a liver cell expressed receptor element for dengue virus serotype 2. Arch. Virol. 2004;149:915–927. doi: 10.1007/s00705-003-0263-x. [DOI] [PubMed] [Google Scholar]
- Johnson ER, McKay DB. Mapping the role of active site residues for transducing an ATP-induced conformational change in the bovine 70-kDa heat shock cognate protein. Biochem. 1999;38:10823–10830. doi: 10.1021/bi990816g. [DOI] [PubMed] [Google Scholar]
- Johnson AJ, Roehrig JT. New mouse model for dengue virus vaccine testing. J. Virol. 1999;73:783–786. doi: 10.1128/jvi.73.1.783-786.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CT, Patkar CG, Kuhn RJ. Construction and applications of yellow fever virus replicons. Virol. 2005;331:247–259. doi: 10.1016/j.virol.2004.10.034. [DOI] [PubMed] [Google Scholar]
- Kanai R, Kar K, Anthony K, Gould LH, Ledizet M, Fikrig E, Marasco WA, Koski RA, Modis Y. Crystal structure of west nile virus envelope glycoprotein reveals viral surface epitopes. J. Virol. 2006;80:11000–11008. doi: 10.1128/JVI.01735-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann BM, Summers PL, Dubois DR, Eckels KH. Monoclonal antibodies against dengue 2 virus E-glycoprotein protect mice against lethal dengue infection. Am. J. Trop. Med. Hyg. 1987;36:427–434. doi: 10.4269/ajtmh.1987.36.427. [DOI] [PubMed] [Google Scholar]
- Kaufman BM, Summers PL, Dubois DR, Cohen WH, Gentry MK, Timchak RL, Burke DS, Eckels KH. Monoclonal antibodies for dengue virus prM glycoprotein protect mice against lethal dengue infection. Am. J. Trop. Med. Hyg. 1989;41:576–580. doi: 10.4269/ajtmh.1989.41.576. [DOI] [PubMed] [Google Scholar]
- Kaufmann B, Nybakken GE, Chipman PR, Zhang W, Diamond MS, Fremont DH, Kuhn RJ, Rossmann MG. West Nile virus in complex with the Fab fragment of a neutralizing monoclonal antibody. Proc. Natl. Acad. Sci. USA. 2006;103:12400–12404. doi: 10.1073/pnas.0603488103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielian M, Chatterjee PK, Gibbons DL, Lu YE. Specific roles for lipids in virus fusion and exit. Examples from the alphaviruses. Sub-Cell. Biochem. 2000;34:409–455. doi: 10.1007/0-306-46824-7_11. [DOI] [PubMed] [Google Scholar]
- Kim KH, Willingmann P, Gong ZX, Kremer MJ, Chapman MS, Minor I, Oliveira MA, Rossmann MG, Andries K, Diana GD, Dutko FJ, McKinlay MA, Pevear DC. A comparison of the anti-rhinoviral drug binding pocket in HRV14 and HRV1A. J. Mol. Biol. 1993;230:206–226. doi: 10.1006/jmbi.1993.1137. [DOI] [PubMed] [Google Scholar]
- Krishnan MN, Sukumaran B, Pal U, Agaisse H, Murray JL, Hodge TW, Fikrig E. Rab 5 is required for the cellular entry of dengue and West Nile viruses. J. Virol. 2007;81:4881–4885. doi: 10.1128/JVI.02210-06. Epub 2007 Feb 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschewski H, Allison SL, Heinz FX, Mandl CW. Role of heparan sulfate for attachment and entry of tick-borne encephalitis virus. Virol. 2003;308:92–100. doi: 10.1016/s0042-6822(02)00097-1. [DOI] [PubMed] [Google Scholar]
- Kuhn RJ. Flaviviruses. In: Acheson NA, editor. Fundamentals of Molecular Virology. John Wiley & Sons, Inc.; 2006. pp. 181–190. [Google Scholar]
- Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledizet M, Kar K, Foellmer HG, Bonafe N, Anthony KG, Gould LH, Bushmich SL, Fikrig E, Koski RA. Antibodies targeting linear determinants of the envelope protein protect mice against West Nile virus. J. Infect. Dis. 2007;196:1741–1748. doi: 10.1086/523654. [DOI] [PubMed] [Google Scholar]
- Lee E, Lobigs M. Substitutions at the putative receptor-binding site of an encephalitic flavivirus alter virulence and host cell tropism and reveal a role for glycosaminoglycans in entry. J. Virol. 2000;74:8867–8875. doi: 10.1128/jvi.74.19.8867-8875.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Lobigs M. Mechanism of virulence attenuation of glycosaminoglycan-binding variants of Japanese encephalitis virus and Murray Valley encephalitis virus. J. Virol. 2002;76:4901–4911. doi: 10.1128/JVI.76.10.4901-4911.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Pavy M, Young N, Freeman C, Lobigs M. Antiviral effect of the heparan sulfate mimetic, PI-88, against dengue and encephalitic flaviviruses. Antiviral Res. 2006;69:31–38. doi: 10.1016/j.antiviral.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Lewis JK, Bothner B, Smith TJ, Siuzdak G. Antiviral agent blocks breathing of the common cold virus. Proc. Natl. Acad. Sci. USA. 1998;95:6774–6778. doi: 10.1073/pnas.95.12.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lok S, Yu I, Zhang Y, Kuhn RJ, Chen J, Rossmann MG. The flavivirus precursor membrane-envelope protein complex: Structure and maturation. Science. 2008;319:1830–1834. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- Li L, Barrett AD, Beasley DW. Differential expression of domain III neutralizing epitopes on the envelope proteins of West Nile virus strains. Virol. 2005;335:99–105. doi: 10.1016/j.virol.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Liao M, Kielian M. Domain III from class II fusion proteins functions as a dominant-negative inhibitor of virus membrane fusion. J. Cell. Biol. 2005;171:111–120. doi: 10.1083/jcb.200507075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-L, Lei H-Y, Lin Y-S, Yeh T-M, Chen S-H, Liu H-S. Heparin inhibits Dengue-2 virus infection of five human liver cell lines. Antiviral. Res. 2002;56:93–96. doi: 10.1016/s0166-3542(02)00095-5. [DOI] [PubMed] [Google Scholar]
- Lin B, Parrish CR, Murray JM, Wright PJ. Localization of a neutralizing epitope on the envelope protein of dengue virus type 2. Virol. 1994;202:885–890. doi: 10.1006/viro.1994.1410. [DOI] [PubMed] [Google Scholar]
- Lin CW, Wu SC. A functional epitope determinant on domain III of the Japanese encephalitis virus envelope protein interacted with neutralizing-antibody combining sites. J. Virol. 2003;77:2600–2606. doi: 10.1128/JVI.77.4.2600-2606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Flaviviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Fields Virology. 4th edn. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 991–1041. [Google Scholar]
- Liu H, Chiou SS, Chen WJ. Differential binding efficiency between the envelope protein of Japanese encephalitis virus variants and heparan sulfate on the cell surface. J. Med. Virol. 2004;72:618–624. doi: 10.1002/jmv.20025. [DOI] [PubMed] [Google Scholar]
- Lok S-M, Kostyuchenko VA, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S, Sedlak D, Fremont H, Chipman PR, Roehrig JT, Diamond MS, Kuhn RJ, Rossmann MG. Binding of a neutralizing antibody to dengue virus resulted in an altered surface glycoproteins arrangement. Nat. Struct. Mol. Biol. 2007 doi: 10.1038/nsmb.1382. [DOI] [PubMed] [Google Scholar]
- Lozach PY, Burleigh L, Staropoli I, Navarro-Sanchez E, Harriague J, Virelizier JL, Rey FA, Despres P, Arenzana-Seisdedos F, Amara A. Dendritic cell-specific intercellular adhesion molecule 3-grabbing non-integrin (DC-SIGN)-mediated enhancement of dengue virus infection is independent of DC-SIGN internalization signals. J. Biol. Chem. 2005;280:23698–23708. doi: 10.1074/jbc.M504337200. [DOI] [PubMed] [Google Scholar]
- Ma L, Jones CT, Groesch TD, Kuhn RJ, Post CB. Solution structure of dengue virus capsid protein reveals another fold. Proc. Natl. Acad. Sci. USA. 2004;101:3414–3419. doi: 10.1073/pnas.0305892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JM, Khromykh AA, Westaway EG. Stable expression of noncytopathic Kunjin replicons simulates both ultrastructural and biochemical characteristics observed during replication of Kunjin virus. Virol. 2001;279:161–172. doi: 10.1006/viro.2000.0691. [DOI] [PubMed] [Google Scholar]
- Mandl CW, Kroschewski H, Allison SL, Kofler R, Holzmann H, Meixner T, Heinz FX. Adaptation of tick-borne encephalitis virus to BHK-21 cells results in the formation of multiple heparan sulfate binding sites in the envelope protein and attenuation in vivo. J. Virol. 2001;75:5627–5637. doi: 10.1128/JVI.75.12.5627-5637.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks RM, Lu H, Sundaresan R, Toida T, Suzuki A, Imanari T, Hernaiz MJ, Linhardt RJ. Probing the interaction of dengue virus envelope protein with heparin: assessment of glycosaminoglycan-derived inhibitors. J. Med. Chem. 2001;44:2178–2187. doi: 10.1021/jm000412i. [DOI] [PubMed] [Google Scholar]
- Martin JE, Pierson TC, Hubka S, Rucker S, Gordon IJ, Enama ME, Andrews CA, Xu Q, Davis BS, Nason M, Fay M, Koup RA, Roederer M, Bailer RT, Gomez PL, Mascola JR, Chang GJ, Nabel GJ, Graham BS. A West Nile virus DNA vaccine induces neutralizing antibody in healthy adults during a phase 1 clinical trial. J. Infect. Dis. 2007;196:1732–1740. doi: 10.1086/523650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews JH, Roehrig JT. Elucidation of the topography and determination of the protective epitopes on the E glycoprotein of Saint Louis encephalitis virus by passive transfer with monoclonal antibodies. J Immunol. 1984;132:1533–1537. [PubMed] [Google Scholar]
- McMinn PC, Weir RC, Dalgarno L. A mouse-attenuated envelope protein variant of Murray Valley encephalitis virus with altered fusion activity. J. Gen. Virol. 1996;77:2085–2088. doi: 10.1099/0022-1317-77-9-2085. [DOI] [PubMed] [Google Scholar]
- Melikyan GB, Markosyan RM, Roth MG, Cohen FS. A point mutation in the transmembrane domain of the hemagglutinin of influenza virus stabilizes a hemifusion intermediate that can transit to fusion. Mol. Biol. Cell. 2000;11:3765–3775. doi: 10.1091/mbc.11.11.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JL, Dewet BJ, Martinez-Pomares L, Radcliffe CM, Dwek RA, Rudd PM, Gordon S. The Mannose Receptor Mediates Dengue Virus Infection of Macrophages. PLoS Pathog. 2008;4:e17. doi: 10.1371/journal.ppat.0040017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. Structure of the Dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J. Virol. 2005;79:1223–1231. doi: 10.1128/JVI.79.2.1223-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monath TP, Liu J, Kanesa-Thasan N, Myers GA, Nichols R, Deary A, McCarthy K, Johnson C, Ermak T, Shin S, Arroyo J, Guirakhoo F, Kennedy JS, Ennis FA, Green S, Bedford P. A live, attenuated recombinant West Nile virus vaccine. Proc. Natl. Acad. Sci. USA. 2006;103:6694–6699. doi: 10.1073/pnas.0601932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monath TP, Liu J, Kanesa-Thasan N, Myers GA, Nichols R, Deary A, McCarthy K, Johnson C, Ermak T, Shin S, Arroyo J, Guirakhoo F, Kennedy JS, Ennis FA, Green S, Bedford P. A live, attenuated recombinant West Nile virus vaccine. Proc. Natl. Acad. Sci. USA. 2006;103:6694–6699. doi: 10.1073/pnas.0601932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondotte JA, Lozach PY, Amara A, Gamarnik AV. Essential role of dengue virus envelope protein N glycosylation at asparagine-67 during viral propagation. J. Virol. 2007;81:7136–7148. doi: 10.1128/JVI.00116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Kim B-S, Chipman PR, Rossmann MG, Kuhn RJ. Structure of West Nile virus. Science. 2003;303:248. doi: 10.1126/science.1089316. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the Flavivirus life cycle. Nat. Rev. Microbiol. 2005;3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- Navarro-Sanchez E, Altmeyer R, Amara A, Schwartz O, Fieschi F, Virelizier JL, Arenzana-Seisdedos F, Despres P. Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep. 2003;4:723–728. doi: 10.1038/sj.embor.embor866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybakken GE, Oliphant T, Johnson S, Burke S, Diamond MS, Fremont DH. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature. 2005;437:764–769. doi: 10.1038/nature03956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybakken GE, Nelson CA, Chen BR, Diamond MS, Fremont DH. Crystal structure of the West Nile virus envelope glycoprotein. J. Virol. 2006;80:11467–11474. doi: 10.1128/JVI.01125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant T, Engle M, Nybakken GE, Doane C, Johnson S, Huang L, Gorlatov S, Mehlhop E, Marri A, Chung KM, Ebel GD, Kramer LD, Fremont DH, Diamond MS. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat. Med. 2005;11:522–530. doi: 10.1038/nm1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant T, Nybakken GE, Engle M, Xu Q, Nelson CA, Sukupolvi-Petty S, Marri A, Lachmi BE, Olshevsky U, Fremont DH, Pierson TC, Diamond MS. Antibody recognition and neutralization determinants on domains I and II of West Nile Virus envelope protein. J. Virol. 2006;80:12149–12159. doi: 10.1128/JVI.01732-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono L, Wollinger W, Rocco IM, Coimbra TL, Gorin PA, Sierakowski MR. In vitro and in vivo antiviral properties of sulfated galactomannans against yellow fever virus (BeH111 strain) and dengue 1 virus (Hawaii strain) Antiviral Res. 2003;60:201–208. doi: 10.1016/s0166-3542(03)00175-x. [DOI] [PubMed] [Google Scholar]
- Padalko E, Ohnishi T, Matsushita K, Sun H, Fox-Talbot K, Bao C, Baldwin WM, 3rd, Lowenstein CJ. Peroxynitrite inhibition of Coxsackievirus infection by prevention of viral RNA entry. Proc. Natl. Acad. Sci. USA. 2004;101:11731–11736. doi: 10.1073/pnas.0400518101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson TC, Xu Q, Nelson S, Oliphant T, Nybakken GE, Fremont DH, Diamond MS. The stoichiometry of antibody-mediated neutralization and enhancement of West Nile virus infection. Cell Host Microbe. 2007;1:135–145. doi: 10.1016/j.chom.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevear DC, Fancher FJ, Feloc PJ, Rossmann MG, Miller MS, Diana G, Treasurywala AM, McKinlay MA, F.J D. Conformational change in the floor of the human rhinovirus canyon blocks adsorption to HeLa cell receptors. J. Virol. 1989;63:2002–2007. doi: 10.1128/jvi.63.5.2002-2007.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillpotts RJ, Stephenson JR, Porterfield JS. Passive immunization of mice with monoclonal antibodies raised against tick-borne encephalitis virus. Brief report. Arch. Virol. 1987;93:295–301. doi: 10.1007/BF01310983. [DOI] [PubMed] [Google Scholar]
- Pokidysheva E, Zhang Y, Battisti AJ, Bator-Kelly CM, Chipman PR, Xiao C, Gregorio GG, Hendrickson A, Kuhn RJ, Rossmann MG. Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN. Cell. 2006;124:485–493. doi: 10.1016/j.cell.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Pujol CA, Estevez JM, Carlucci MJ, Ciancia M, Cerezo AS, Damonte EB. Novel DL-galactan hybrids from the red seaweed Gymnogongrus torulosus are potent inhibitors of herpes simplex virus and dengue virus. Antivir. Chem. Chemother. 2002;13:83–89. doi: 10.1177/095632020201300202. [DOI] [PubMed] [Google Scholar]
- Randolph VB, Stollar V. Low pH-induced cell fusion in flavivirus-infected Aedes albopictus cell cultures. J. Gen. Virol. 1990;71:1845–1850. doi: 10.1099/0022-1317-71-8-1845. [DOI] [PubMed] [Google Scholar]
- Ray D, Shi PY. Recent advances in flavivirus antiviral drug discovery and vaccine development. Rec. Pat. Anti-Infect Drug Disc. 2006;1:45–55. doi: 10.2174/157489106775244055. [DOI] [PubMed] [Google Scholar]
- Rieder E, Berinstein A, Baxt B, Kang A, Mason PW. Propagation of an attenuated virus by design: engineering a novel receptor for a noninfectious foot-and-mouth disease virus. Proc. Natl. Acad. Sci. USA. 1996;93:10428–10433. doi: 10.1073/pnas.93.19.10428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisdorph N, Thomas JJ, Katpally U, Chase E, Harris K, Siuzdak G, Smith TJ. Human rhinovirus capsid dynamics is controlled by canyon flexibility. Virol. 2003;314:34–44. doi: 10.1016/s0042-6822(03)00452-5. [DOI] [PubMed] [Google Scholar]
- Ren J, Ding T, Zhang W, Song J, Ma W. Does Japanese encephalitis virus share the same cellular receptor with other mosquito-borne flaviviruses on the C6/36 mosquito cells? Virol. 2007;4:83. doi: 10.1186/1743-422X-4-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey FA. Dengue virus envelope glycoprotein structure: new insight into its interactions during viral entry. Proc. Natl. Acad. Sci. USA. 2003;100:6899–6901. doi: 10.1073/pnas.1332695100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- Roehrig JT, Bolin RA, Kelly RG. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus, Jamaica. Virol. 1998;246:317–328. doi: 10.1006/viro.1998.9200. [DOI] [PubMed] [Google Scholar]
- Roehrig JT, Staudinger LA, Hunt AR, Mathews JH, Blair CD. Antibody prophylaxis and therapy for flavivirus encephalitis infections. Ann N Y Acad Sci. 2001;951:286–297. doi: 10.1111/j.1749-6632.2001.tb02704.x. [DOI] [PubMed] [Google Scholar]
- Roehrig JT. Antigenic structure of flavivirus proteins. Adv Virus Res. 2003;59:141–175. doi: 10.1016/s0065-3527(03)59005-4. [DOI] [PubMed] [Google Scholar]
- Roivainen M, Hyypiä T, Piirainen L, Kalkkinen N, Stanway G, Hovi T. RGD-dependent entry of coxsackievirus A9 into host cells and its bypass after cleavage of VP1 protein by intestinal proteases. J. Virol. 1991;65:4735–4740. doi: 10.1128/jvi.65.9.4735-4740.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG, Arnold E, Erickson JW, Frankenberger EA, Griffith JP, Hecht HJ, Johnson JE, Kamer G, Luo M, et al. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- Rossmann MG. Viral cell recognition and entry. Prot. Sci. 1994;3:1712–1725. doi: 10.1002/pro.5560031010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG, Bella J, Kolatkar PR, He YN, Wimmer E, Kuhn RJ, Baker TS. Cell recognition and entry by rhino- and enteroviruses. Virol. 2000;269:239–247. doi: 10.1006/viro.2000.0258. [DOI] [PubMed] [Google Scholar]
- Rusconi S, Scozzafava A, Mastrolorenzo A, Supuran CT. An update in the development of HIV entry inhibitors. Curr. Top. Med. Chem. 2007;7:1273–1289. doi: 10.2174/156802607781212239. [DOI] [PubMed] [Google Scholar]
- Russell CJ, Jardetzky TS, Lamb RA. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. Embo J. 2001;20:4024–4034. doi: 10.1093/emboj/20.15.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez MD, Pierson TC, McAllister D, Hanna SL, Puffer BA, Valentine LE, Murtadha MM, Hoxie JA, Doms RW. Characterization of neutralizing antibodies to West Nile virus. Virol. 2005;336:70–82. doi: 10.1016/j.virol.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Schibli DJ, Weissenhorn W. Class I and class II viral fusion protein structures reveal similar principles in membrane fusion. Mol. Membr. Biol. 2004;21:361–371. doi: 10.1080/09687860400017784. [DOI] [PubMed] [Google Scholar]
- Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- Smith TJ, Kremer MJ, Luo M, Vriend G, Arnold E, Kamer G, Rossmann MG, McKinlay MA, Diana GD, Otto MJ. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science. 1986;233:1286–1293. doi: 10.1126/science.3018924. [DOI] [PubMed] [Google Scholar]
- Stadler K, Allison SL, Schalich J, Heinz FX. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 1997;71:8475–8481. doi: 10.1128/jvi.71.11.8475-8481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J, Donis RO. Influenza virus hemagglutinin--structural studies and their implications for the development of therapeutic approaches. Infect. Disord. Drug Targ. 2007;7:329–335. doi: 10.2174/187152607783018727. [DOI] [PubMed] [Google Scholar]
- Stiasny K, Allison SL, Marchler-Bauer A, Kunz C, Heinz FX. Structural requirements for low-pH-induced rearrangements in the envelope glycoprotein of tick-borne encephalitis virus. J. Virol. 1996;70:8142–8147. doi: 10.1128/jvi.70.11.8142-8147.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiasny K, Kiermayr S, Holzmann H, Heinz FX. Cryptic properties of a cluster of dominant flavivirus cross-reactive antigenic sites. J. Virol. 2006;80:9557–9568. doi: 10.1128/JVI.00080-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiasny K, Allison SL, Schalich J, Heinz FX. Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J. Virol. 2002;76:3784–3790. doi: 10.1128/JVI.76.8.3784-3790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiasny K, Bressanelli S, Lepault J, Rey FA, Heinz FX. Characterization of a membrane-associated trimeric low-pH-induced Form of the class II viral fusion protein E from tick-borne encephalitis virus and its crystallization. J. Virol. 2004;78:3178–3183. doi: 10.1128/JVI.78.6.3178-3183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiasny K, Koessl C, Heinz FX. Involvement of lipids in different steps of the Flavivirus fusion mechanism. J. Virol. 2003;77:7856–7862. doi: 10.1128/JVI.77.14.7856-7862.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiasny K, Kossl C, Lepault J, Rey FA, Heinz FX. Characterization of a structural intermediate of flavivirus membrane fusion. PLoS Pathog. 2007;3:e20. doi: 10.1371/journal.ppat.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukupolvi-Petty S, Austin SK, Purtha WE, Oliphant T, Nybakken GE, Schlesinger JJ, Roehrig JT, Gromowski GD, Barrett AD, Fremont DH, Diamond MS. Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. J. Virol. 2007;81:12816–12826. doi: 10.1128/JVI.00432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers PL, Cohen WH, Ruiz MM, Hase T, Eckels KH. Flaviviruses can mediate fusion from without in Aedes albopictus mosquito cell cultures. Virus Res. 1989;12:383–392. doi: 10.1016/0168-1702(89)90095-6. [DOI] [PubMed] [Google Scholar]
- Talarico LB, Pujol CA, Zibetti RG, Faria PC, Noseda MD, Duarte ME, Damonte EB. The antiviral activity of sulfated polysaccharides against dengue virus is dependent on virus serotype and host cell. Antiviral Res. 2005;66:103–110. doi: 10.1016/j.antiviral.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Tassaneetrithep B, Burgess TH, Granelli-Piperno A, Trumpfheller C, Finke J, Sun W, Eller MA, Pattanapanyasat K, Sarasombath S, Birx DL, Steinman RM, Schlesinger S, Marovich MA. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J. Exp. Med. 2003;197:823–829. doi: 10.1084/jem.20021840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueba N, Kimura T. Polykaryocytosis induced by certain arboviruses in monolayers of BHK-21-528 cells. J. Gen. Virol. 1977;34:369–373. doi: 10.1099/0022-1317-34-2-369. [DOI] [PubMed] [Google Scholar]
- van der Most RG, Corver J, Strauss JH. Mutagenesis of the RGD motif in the yellow fever virus 17D envelope protein. Virology. 1999;265(1):83–95. doi: 10.1006/viro.1999.0026. [DOI] [PubMed] [Google Scholar]
- van der Schaar HM, Rust MJ, Waarts BL, van der Ende-Metselaar H, Kuhn RJ, Wilschut J, Zhuang X, Smit JM. Characterization of the early events in dengue virus cell entry by biochemical assays and single-virus tracking. J. Virol. 2007;81:12019–12028. doi: 10.1128/JVI.00300-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga AS, Santos NC, Castanho MA. An insight on the leading HIV entry inhibitors. Rec. Pat. Anti-Infect. Drug Disc. 2006;1:67–73. doi: 10.2174/157489106775244046. [DOI] [PubMed] [Google Scholar]
- Wang T, Duan Y. Binding modes of CCR5-targetting HIV entry inhibitors: Partial and full antagonists. J. Mol. Graph. Model. 2007 doi: 10.1016/j.jmgm.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu GL, Hasson M, Blackburn EH. Circular ribosomal DNA plasmids transform Tetrahymena thermophila by homologous recombination with endogenous macronuclear ribosomal DNA. Proc. Natl. Acad. Sci. USA. 1988;85:5151–5155. doi: 10.1073/pnas.85.14.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319:1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- Zhang W, Chipman PR, Corver J, Johnson PR, Zhang Y, Mukhopadhyay S, Baker TS, Strauss JH, Rossmann MG, Kuhn RJ. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 2003a;10:907–912. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Corver J, Chipman PR, Pletnev SV, Sedlak D, Baker TS, Strauss JH, Kuhn RJ, Rossmann MG. Structures of immature flavivirus particles. J. EMBO. 2003b;22:2604–2613. doi: 10.1093/emboj/cdg270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG. Conformational changes of the flavivirus E glycoprotein. Structure. 2004;12:1607–1618. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kaufmann B, Chipman PR, Kuhn RJ, Rossmann MG. Structure of immature West Nile Virus. J. Virol. 2007;81:6141–6145. doi: 10.1128/JVI.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]