Abstract
Cocaine is recognized as the most reinforcing of all drugs of abuse. There is no anti-cocaine medication available. The disastrous medical and social consequences of cocaine addiction have made the development of an anti-cocaine medication a high priority. It has been recognized as an ideal anti-cocaine medication to accelerate cocaine metabolism producing biologically inactive metabolites via a route similar to the primary cocaine-metabolizing pathway, i.e. cocaine hydrolysis catalyzed by plasma enzyme butyrylcholinesterase (BChE). However, wild-type BChE has a low catalytic efficiency against the abused cocaine. Design of a high-activity enzyme mutant is extremely challenging, particularly when the chemical reaction process is rate determining for the enzymatic reaction. Here we report the design and discovery of a high-activity mutant of human BChE by using a novel, systematic computational design approach based on transition-state simulations and activation energy calculations. The novel computational design approach has led to discovery of the most efficient cocaine hydrolase, i.e. a human BChE mutant with a ∼2000-fold improved catalytic efficiency, promising for therapeutic treatment of cocaine overdose and addiction as an exogenous enzyme in human. The encouraging discovery resulted from the computational design not only provides a promising anti-cocaine medication, but also demonstrates that the novel, generally applicable computational design approach is promising for rational enzyme redesign and drug discovery.
Introduction
Cocaine is recognized as the most reinforcing of all drugs of abuse.1,2,3 There is no anti-cocaine medication available. The disastrous medical and social consequences of cocaine addiction have made the development of an anti-cocaine medication a high priority.4,5 An ideal anti-cocaine medication would be to accelerate cocaine metabolism producing biologically inactive metabolites via a route similar to the primary cocaine-metabolizing pathway, i.e. cocaine hydrolysis catalyzed by plasma enzyme butyrylcholinesterase (BChE).4,6,7,8,9,10 However, wild-type BChE has a low catalytic efficiency against naturally occurring (-)-cocaine.11,12,13,14,15 Here we report a unique computational design leading to discovery of a human BChE mutant with a ∼2000-fold improved catalytic efficiency, which is sufficient for use as an exogenous enzyme in human to prevent (-)-cocaine reaching central nervous system (CNS). The encouraging outcome in vitro and in vivo not only provides a promising anti-cocaine medication, but also demonstrates a novel, generally applicable approach for rational enzyme redesign and drug discovery.
The primary pathway for cocaine metabolism in primates is hydrolysis at the benzoyl ester or methyl ester group.4,10 Benzoyl ester hydrolysis generates ecgonine methyl ester (EME), whereas the methyl ester hydrolysis yields benzoylecgonine (BE). The major cocaine-metabolizing enzymes in humans are butyrylcholinesterase (BChE) which catalyzes benzoyl ester hydrolysis (Figure 1) and two liver carboxylesterases (denoted by hCE-1 and hCE-2) which catalyze hydrolysis at the methyl ester and the benzoyl ester, respectively. Among the three, BChE is the principal cocaine hydrolase in human serum. Hydrolysis accounts for about 95% of cocaine metabolism in humans. The remaining 5% is deactivated through oxidation by the liver microsomal cytochrome P450 system, producing norcocaine.4,16 EME appears the least pharmacologically active of the cocaine metabolites and may even cause vasodilation,4 whereas both BE and norcocaine appear to cause vasoconstriction and lower the seizure threshold, similar to cocaine itself. Norcocaine is hepatotoxic and a local anesthetic.17 Thus, hydrolysis of cocaine at the benzoyl ester by an enzyme is the pathway most suitable for amplification. The use of an exogenous enzyme has some potential advantages over active immunization since the enzyme administration would immediately enhance cocaine metabolism and would not require an immune response to be effective.4
Figure 1.
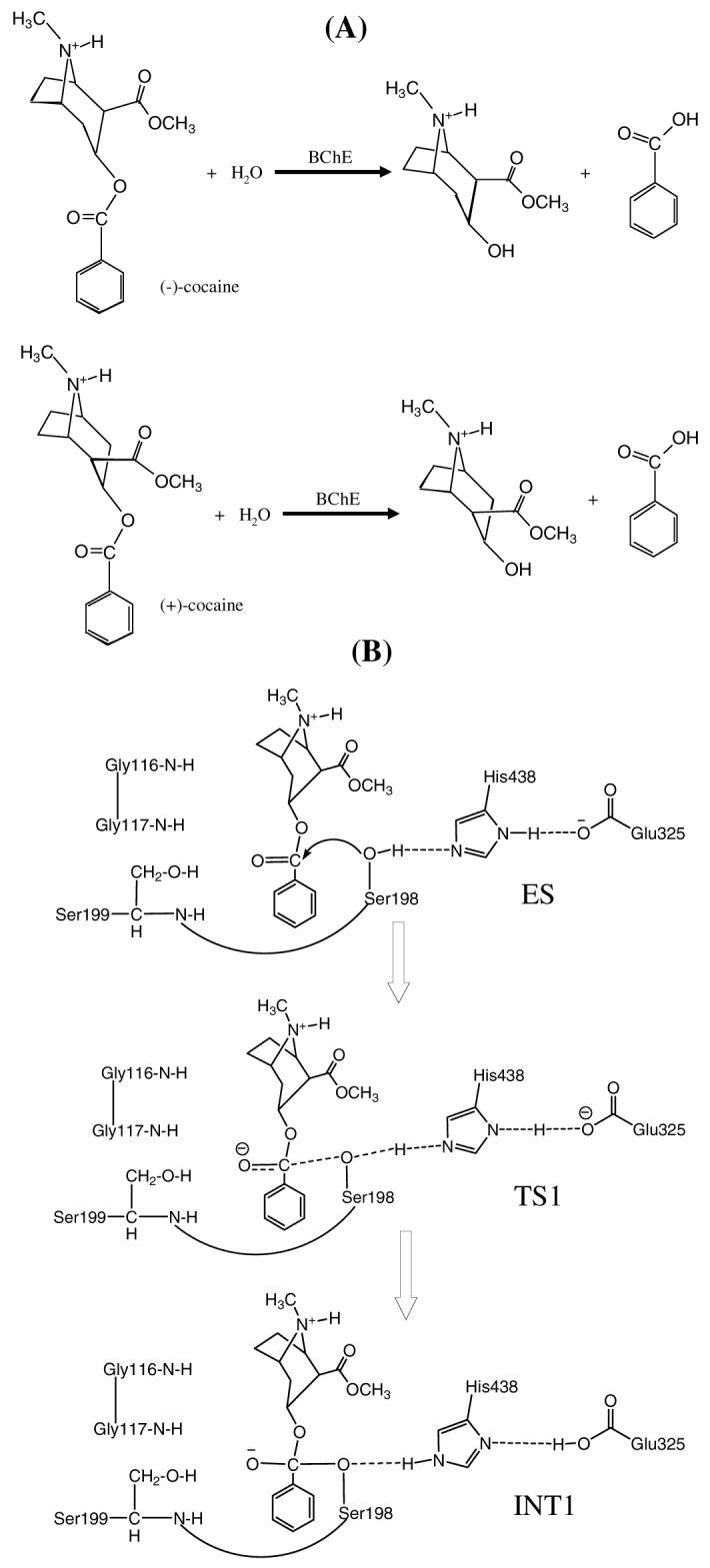
Hydrolysis reactions of (-)-cocaine and (+)-cocaine: (a) reactants and products of the reactions; (b) schematic representation of the first reaction step for (-)-cocaine hydrolysis catalyzed by a BChE mutant including A199S mutation.
Ignoring hCE-2, so far, two types of known native enzymes may be used to catalyze hydrolysis of cocaine at the benzoyl ester: human BChE and bacterial cocaine esterase (CocE).18,19 BChE from human source can be tolerated perfectly in human body and, in fact, BChE has a long history of successful clinic use for other purposes.4 However, the catalytic activity of this plasma enzyme (kcat = 4.1 min-1 and KM = 4.5 μM)11 is significantly lower against the naturally occurring (-)-cocaine than CocE (kcat = 468 min-1 and KM = 0.64 μM).19 Due to the low activity of BChE, (-)-cocaine has a plasma half-life of ∼ 45 - 90 min (before BChE saturation), long enough for manifestation of the effects on CNS which peak in minutes.13 Hence, BChE mutants with a significantly higher activity against (-)-cocaine are highly desired for use as an exogenous enzyme in humans.
In general, for rational design of an enzyme mutant with a higher catalytic activity for a given substrate, one needs to design a mutation that can accelerate the rate-determining step of the entire catalytic reaction process while the other steps are not slowed down by the mutation. Reported computational modeling and experimental data indicated that the formation of the prereactive BChE-(-)-cocaine complex (ES) is the rate-determining step of (-)-cocaine hydrolysis (Figure 1) catalyzed by wild-type BChE,11,12,20,21 whereas the rate-determining step of the unnatural, biologically inactive (+)-cocaine hydrolysis catalyzed by the same enzyme is the chemical reaction process consisting of four individual reaction steps.21 Based on this mechanistic understanding, previous efforts reported by other researchers for rational design of BChE mutants were focused on how to improve the ES formation process and several BChE mutants11,12,22 were found to have a ∼9 to 34-fold improved catalytic efficiency (kcat/KM) against (-)-cocaine. Recently reported computational modeling also suggests that the formation of the prereactive BChE-(-)-cocaine complex (ES) is hindered mainly by the bulky side chain of Y332 residue in wild-type BChE, but the hindering can be removed by the Y332A mutation and the Y332G mutation can produce a more significant improvement.12 The combined computational and experimental data11,20,23,24,25,26 have revealed that the rate-determining step of (-)-cocaine hydrolysis catalyzed by the A328W/Y332A and A328W/Y332G mutants is the first step of the chemical reaction process. Therefore, starting from the A328W/Y332A or A328W/Y332G mutant, our work for further improving the catalytic efficiency of BChE against (-)-cocaine aimed to decrease the energy barrier for the first reaction step without significantly affecting the ES formation and other chemical reaction steps.23,25,27,28
Our recently reported computational studies suggest that the hydrogen bonding between the carbonyl oxygen of (-)-cocaine benzoyl ester and the oxyanion hole (residues #116, #117, and #199) of the enzyme in the rate-determining transition state (TS1, i.e. the transition state for the first step of the chemical reaction process) can be enhanced by some mutations.23,25,26 The enhanced hydrogen bonding (HB) may potentially stabilize the TS1 structure and thus possibly lower the energy barrier for the first reaction step of (-)-cocaine hydrolysis. The HB energy-based virtual screening led to discovery of some high-activity mutants of BChE; the most active one is A199S/S287G/A328W/Y332G BChE25 whose catalytic efficiency (kcat/KM) against (-)-cocaine is still lower than bacterial CocE. As expected, A199S/S287G/A328W/Y332G BChE indeed can selectively block cocaine toxicity and reinstatement of drug seeking in rats.29
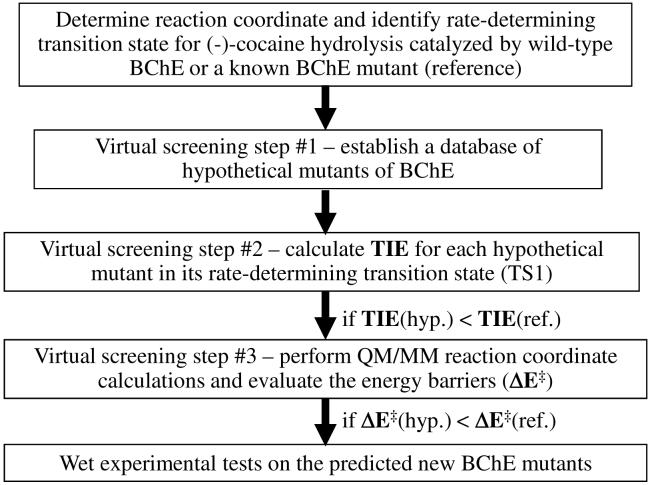
Concerning the computational design methodology, our previous HB energy calculations23,25,26 were simply based on an empirical HB energy equation which does not account for any nonbonding interaction. So, nonbonding interactions between the (-)-cocaine atoms and the oxyanion hole of the enzyme and their effects on the energy barrier of the enzymatic reaction remain to be evaluated in computational modeling. In the present study, we first aimed to develop and test a novel, systematic design protocol (depicted in Figure 2) for a virtual screening of various BChE mutants. The virtual screening starts by first estimating the total interaction energy (TIE) between the whole oxyanion hole of the enzyme and all (-)-cocaine atoms in the TS1 structure for each possible mutant. The practical effects of the interactions on the energy barrier for the rate-determining reaction step are evaluated by performing more sophisticated, hybrid quantum mechanical/molecular mechanical (QM/MM) calculations, followed by wet experimental tests in vitro and in vivo.
Figure 2.
Schematic representation of structure-and-mechanism-based computational design and discovery of high-activity mutants of human BChE. TIE refers to the total interaction energy between the whole oxyanion hole of the enzyme and all (-)-cocaine atoms in the TS1 structure for each possible mutant. ΔE‡ represents the energy barrier for the first step of the enzymatic hydrolysis of (-)-cocaine. The approach depicted here can be used for any other enzyme, as TIE can be the total interaction energy between all (or some key) atoms of the substrate and all (or some key) atoms of the enzyme.
Results and Discussion
To carry out the TIE-based initial screening, we first examined a variety of possible BChE mutants by performing molecular dynamics (MD) simulation on the TS1 structure for each mutant. The general strategy of performing a classical MD simulation on a transition state structure of the enzymatic reaction has been described and justified in our recently reported studies.23,24,25,26,28 Each MD simulation in water was performed for 1 ns or longer to make sure that we obtained a stable MD trajectory for each TS1 structure simulated. Within the stable MD trajectory obtained for a dynamically stable TS1 structure, we collected a large number of snapshots of the simulated TS1 structure (one snapshot for each 1 ps). One can estimate a TIE value for each snapshot of the simulated TS1 structure. In principle, a TIE value includes energetic contributions from both the hydrogen bonding and nonbonding interactions. Technically, based on the popularly used Amber force field,30 the TIE value can be evaluated as the sum of the electrostatic and van der Waals interaction energies between all atoms of (-)-cocaine and all atoms of residues #116, #117, and #199. The final TIE value is the average of the TIE values calculated for all of the snapshots of the MD-simulated TS1 structure.
The virtual screening based on extensive transition state simulations and subsequent TIE calculations reveals that the most stable TS1 structure is associated with the A199S/F227A/S287G/A328W/Y332G mutant. Summarized in Table 1 are important numerical results concerning the MD-simulated TS1 structures and energetics for wild-type BChE and for representative mutants of BChE. The simulated H···O distance D1 (Table 1) is always too long for the peptidic NH of G116 to form an N-H···O hydrogen bond with the carbonyl oxygen of (-)-cocaine in all of the simulated TS1 structures. The interaction energy (IE116) of all (-)-cocaine atoms with all atoms of G116 is purely nonbonding, as seen in Table 1. All of the calculated IE116 values are positive, except for the A199S/F227A/S287G/A328W/Y332G mutant. The positive interaction energy (IE) value means to destabilize the TS1 structure, whereas the negative IE value means to stabilize the TS1 structure. In the simulated TS1 structure for wild-type BChE, the carbonyl oxygen of (-)-cocaine formed an N-H···O hydrogen bond with the peptidic NH hydrogen atom of A199 residue; the simulated H···O distance (D3) was 1.61 to 2.35 Å, with an average D3 value of 1.92 Å. Meanwhile, the carbonyl oxygen of (-)-cocaine also had a partial N-H···O hydrogen bond with the peptidic NH hydrogen atom of G117 residue; the simulated H···O distance (D2) was 1.97 to 4.14 Å (the average D2 value: 2.91 Å). Due to the hydrogen bonding, all of the calculated interaction energies IE117 and IE199 (including both the hydrogen bonding and nonbonding interaction energies) in Table 1 are negative values.
Table 1.
MD-simulated key distances and the calculated interaction energies (IEs) between all atoms of (-)-cocaine and all atoms of the oxyanion hole (residues #116, #117, and #199) in the simulated first transition state (TS1).
| Transition State | Distances (D1 to D4)a and interaction energies (IEs)b | ||||
|---|---|---|---|---|---|
| D1 or IE116 | D2 or IE117 | D3/D4 or IE199 | Total IE (i.e. TIE)c | ||
| TS1 structure for (-)-cocaine hydrolysis catalyzed by wild-type BChE | Average (Å) | 4.59 | 2.91 | 1.92 | |
| Maximum (Å) | 5.73 | 4.14 | 2.35 | ||
| Minimum (Å) | 3.35 | 1.97 | 1.61 | ||
| Fluctuation (Å) | 0.35 | 0.35 | 0.12 | ||
| IE (kcal/mol) | 3.18 | -4.52 | -4.23 | -5.57 | |
| TS1 structure for (-)-cocaine hydrolysis catalyzed by A328W/Y332A mutant of BChE | Average (Å) | 3.62 | 2.35 | 1.95 | |
| Maximum (Å) | 4.35 | 3.37 | 3.02 | ||
| Minimum (Å) | 2.92 | 1.78 | 1.61 | ||
| Fluctuation (Å) | 0.23 | 0.27 | 0.17 | ||
| IE (kcal/mol) | 0.84 | -4.25 | -3.03 | -6.44 | |
| TS1 structure for (-)-cocaine hydrolysis catalyzed by A328W/Y332G mutant of BChE | Average (Å) | 3.60 | 2.25 | 1.97 | |
| Maximum (Å) | 4.24 | 3.17 | 2.76 | ||
| Minimum (Å) | 2.89 | 1.77 | 1.62 | ||
| Fluctuation (Å) | 0.23 | 0.24 | 0.17 | ||
| IE (kcal/mol) | 0.04 | -4.94 | -2.67 | -7.57 | |
| TS1 structure for (-)-cocaine hydrolysis catalyzed by A199S/S287G/A328W/Y332G mutant of BChE | Average (Å) | 4.39 | 2.60 | 2.01/1.76 | |
| Maximum (Å) | 5.72 | 4.42 | 2.68/2.50 | ||
| Minimum (Å) | 2.87 | 1.76 | 1.62/1.48 | ||
| Fluctuation (Å) | 0.48 | 0.36 | 0.17/0.12 | ||
| IE (kcal/mol) | 2.96 | -5.12 | -9.92 | -12.08 | |
| TS1 structure for (-)-cocaine hydrolysis catalyzed by A199S/F227A/S287G/A328W/Y332G mutant of BChE | Average (Å) | 5.28 | 2.15 | 1.96/2.20 | |
| Maximum (Å) | 5.98 | 2.83 | 2.40/3.28 | ||
| Minimum (Å) | 2.72 | 1.68 | 1.65/1.59 | ||
| Fluctuation (Å) | 0.25 | 0.18 | 0.12/0.30 | ||
| IE (kcal/mol) | -0.97 | -5.59 | -8.22 | -14.78 | |
D1, D2, and D3 represent the internuclear distances between the carbonyl oxygen of cocaine benzoyl ester and the NH hydrogen of residues #116 (i.e. G116), #117 (i.e. G117), and #199 (i.e. A199 or S199) of BChE, respectively. D4 is the internuclear distance between the carbonyl oxygen of cocaine benzoyl ester and the hydroxyl hydrogen of S199 side chain in the A199S/S287G/A328W/Y332G and A199S/F227A/S287G/A328W/Y332G mutants.
IE116, IE117, and IE199 represent the average interaction energies (IEs) between all atoms of (-)-cocaine and all atoms of amino acid residues #116, #117, and #199 of BChE, respectively, in the MD-simulated TS1 structure.
TIE = IE116 + IE117 + IE199.
In going from wild-type BChE to the A328W/Y332A and A328W/Y332G mutants, the largest IE change in the TS1 structure is associated with the decrease of the IE116 value, as seen in Table 1. Hence, the total IE (i.e. TIE = IE116 + IE117 + IE199) value between all atoms of (-)-cocaine and all atoms of the oxyanion hole only slightly decreases when wild-type BChE is replaced by the A328W/Y332A or A328W/Y332G mutant, as seen from the calculated TIE values in Table 1. In the simulated TS1 structures for A199S/S287G/A328W/Y332G and A199S/F227A/S287G/A328W/Y332G BChE’s, an O-H···O hydrogen bond formed between the hydroxyl group on the S199 side chain and the carbonyl oxygen of (-)-cocaine, in addition to the two N-H···O hydrogen bonds with the peptidic NH of G117 and S199, as seen in Table 1. Due to additional O-H···O hydrogen bond with the hydroxyl oxygen of residue #199, the IE199 values calculated for A199S/S287G/A328W/Y332G and A199S/F227A/S287G/A328W/Y332G BChE’s are remarkably lower than those calculated for wild-type, A328W/Y332A, and A328W/Y332G BChE’s, as seen in Table 1. Thus, the TIE values calculated for the A199S/S287G/A328W/Y332G and A199S/F227A/S287G/A328W/Y332G mutants are remarkably lower than the TIE values for wild-type, A328W/Y332A, and A328W/Y332G BChE’s. The lowest TIE value is associated with the A199S/F227A/S287G/A328W/Y332G mutant, due to the contributions from both the additional hydrogen bond with S199 side chain and the nonbonding interactions with residue G116.
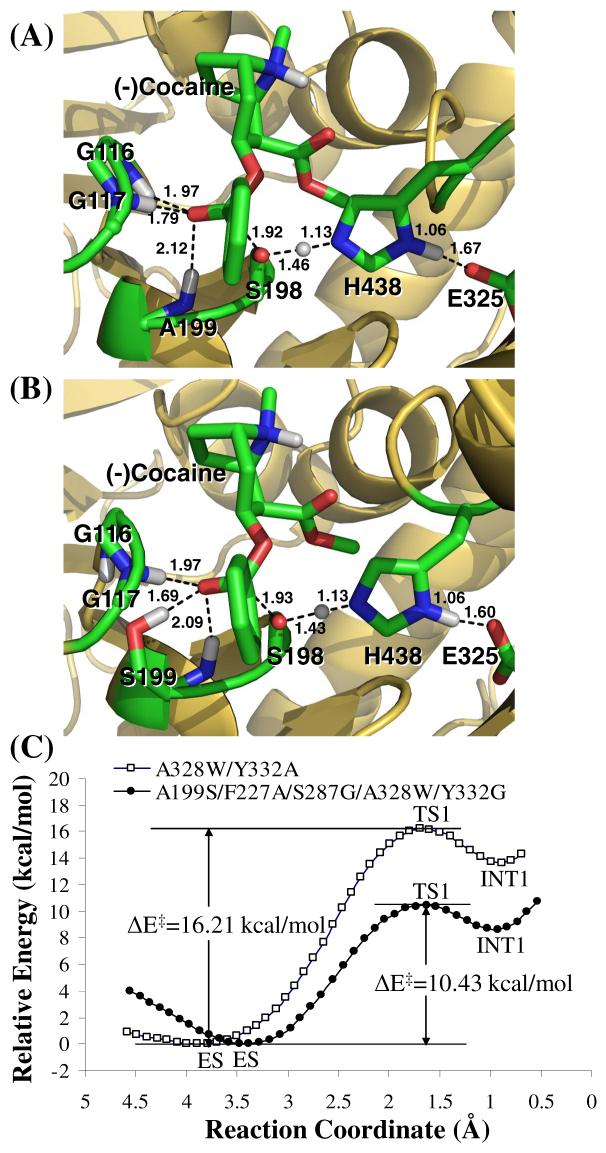
The calculated relative TIE values clearly suggest that the MD-simulated TS1 structure for (-)-cocaine hydrolysis catalyzed by A199S/F227A/S287G/A328W/Y332G BChE should be more stable than the TS1 structures corresponding to wild-type BChE and other mutants. To examine whether the TS1 stabilization can decrease the energy barrier for the rate-determining reaction step, we further performed QM/MM reaction coordinate calculations (at the B3LYP/6-31G*:Amber level) on the first step of (-)-cocaine hydrolysis catalyzed by the A199S/A328W and A199S/F227A/S287G/A328W/Y332G mutants. The QM/MM calculations were performed to optimize the ES, TS1, and INT1 geometries and determine the corresponding energy changes from ES to TS1 and INT1. Depicted in Figure 3 are the QM/MM-optimized TS1 geometries and the calculated energy profiles along the reaction coordinate for the (-)-cocaine hydrolysis catalyzed by the two mutants. As seen in Figure 3(C), the energy barrier calculated for the first step of (-)-cocaine hydrolysis catalyzed by A199S/A328W BChE is ∼16.2 kcal/mol, whereas the energy barrier calculated for the first step of (-)-cocaine hydrolysis catalyzed by A199S/F227A/S287G/A328W/Y332G BChE is only ∼10.4 kcal/mol. According to the QM/MM results, the energy barrier decreases by ∼6 kcal/mol in going from A199S/A328W BChE to A199S/F227A/S287G/A328W/Y332G BChE. The QM/MM-calculated energy barrier decrease of ∼6 kcal/mol correlates very well with the TIE decrease of ∼8 kcal/mol estimated from the initial interaction energy calculations. The QM/MM calculations qualitatively support the results obtained from the TIE-based virtual screening. All of the computational results consistently suggest that A199S/F227A/S287G/A328W/Y332G BChE should be a very active cocaine hydrolase catalyzing hydrolysis of (-)-cocaine.
Figure 3.
Results from the QM/MM reaction coordinate calculations at the B3LYP/6-31G*:Amber level. (A) Optimized geometry of the transition state (TS1) for the first reaction step of (-)-cocaine hydrolysis catalyzed by A328W/Y332G BChE. (B) Optimized geometry of the transition state (TS1) for the first reaction step of (-)-cocaine hydrolysis catalyzed by A199S/F227A/S287G/A328W/Y332G BChE. (C) Calculated potential energy surface along the reaction coordinate (R1 - R2 + R3) of the enzymatic hydrolysis of (-)-cocaine. R1 is the length of the C···O transition bond between the hydroxyl oxygen of S198 and carbonyl carbon of (-)-cocaine benzoyl ester. R2 refers to the length of the O···H transition bond in the S198 hydroxyl group. R3 represents the length of the N···H transition bond between the hydroxyl hydrogen of S198 and the nitrogen of H438 side chain.
To examine the theoretical prediction of high catalytic efficiency for A199S/F227A/S287G/A328W/Y332G BChE against (-)-cocaine, we produced the A199S/F227A/S287G/A328W/Y332G mutant through site-directed mutagenesis, protein expression, and enzyme activity assays in vitro and in vivo. To minimize the possible systematic experimental errors of the in vitro kinetic data, we expressed the enzymes and performed kinetic studies with wild-type BChE and the A199S/F227A/S287G/A328W/Y332G mutant under the same condition and compared the catalytic efficiency of A199S/F227A/S287G/A328W/Y332G BChE to that of wild-type BChE for (-)-cocaine hydrolysis at benzoyl ester group. Michaelis-Menten kinetics of the enzymatic (-)-cocaine hydrolysis at benzoyl ester group was determined by performing the sensitive radiometric assays using [3H](-)-cocaine (labeled on its benzene ring) with varying concentrations of the substrate in combination with an enzyme-linked immunosorbent assay (ELISA). The kinetic analysis revealed that kcat = 5700 ± 400 min-1 and KM = 3.1 ± 0.2 μM for (-)-cocaine hydrolysis catalyzed by A199S/F227A/S287G/A328W/Y332G BChE while the known kinetic parameters (kcat = 4.1 min-1 and KM = 4.5 μM)11 for (-)-cocaine hydrolysis catalyzed by wild-type BChE were reproduced by the same in vitro activity assays. Thus, A199S/F227A/S287G/A328W/Y332G BChE has a ∼2020-fold improved catalytic efficiency (kcat/KM = ∼1.84 × 109 M min-1) compared to wild-type BChE (kcat/KM = ∼9.1 × 105 M min-1) against (-)-cocaine.
What does the ∼2020-fold improvement of the catalytic efficiency mean for decreasing the (-)-cocaine half-life in plasma? Using the designed A199S/F227A/S287G/A328W/Y332G BChE as an exogenous enzyme in human, when the concentration of this mutant is kept the same as that of wild-type BChE in plasma, the half-life of (-)-cocaine in plasma should be reduced from the ∼45 - 90 min to only ∼1.3 - 2.7 sec. It is interesting to note that the catalytic efficiency of A199S/F227A/S287G/A328W/Y332G BChE is even significantly higher than that of the most active native enzyme yet identified, i.e. bacterial CocE with kcat/KM = ∼7.2 × 108 M min-1, against (-)-cocaine. More importantly, the catalytic rate constant (kcat = 5700 min-1) of A199S/F227A/S287G/A328W/Y332G BChE against (-)-cocaine is higher than that (kcat = 468 min-1)19 of bacterial CocE by ∼12-fold.
Assessment of the catalytic activity using an in vivo model based on protection of A199S/F227A/S287G/A328W/Y332G BChE against cocaine-induced lethality correlated well with the in vitro activity data. Our in vivo activity assays on the BChE mutant were performed to compare with the in vivo activity of CocE under the same experimental condition. It has been demonstrated31 that the minimum effective dose of CocE to protect mice from cocaine-induced lethality is 0.1 mg (per mouse). Our theoretical estimation of the minimum effective dose of A199S/F227A/S287G/A328W/Y332G BChE to protect mice from cocaine-induced lethality was based on the relative kcat values of CocE and A199S/F227A/S287G/A328W/Y332G BChE (kcat = 468 min-1 for CocE versus kcat = 5700 min-1 for the BChE mutant) and the relative molecular weights (∼65 kDa for CocE versus ∼84 kDa for the BChE mutant). This is because in the treatment of cocaine overdose, the cocaine concentration in the body is significantly higher than KM of the enzyme and the overall velocity (V) of cocaine hydrolysis is expected to reach the maximum (Vmax), i.e. Vmax = [E]kcat in which [E] is the concentration of the enzyme. Based on the relative molecular weights and the relative catalytic rate constants, the minimum effective dose of A199S/F227A/S287G/A328W/Y332G BChE to protect mice from cocaine-induced lethality was theoretically estimated to be 0.01 mg. Figure 4 shows that the A199S/F227A/S287G/A328W/Y332G mutant dose-dependently protected mice from cocaine-induced convulsions and lethality. Intraperitoneal administration of cocaine 180 mg/kg produced convulsions and lethality in all tested mice (n = 6). Pretreatment with the BChE mutant (i.e. 1 min prior to cocaine administration) dose-dependently protected mice against cocaine-induced convulsions and lethality. In particular, the BChE mutant 0.01 mg and 0.03 mg produced full protection in mice from cocaine overdose induced by a lethal dose of cocaine 180 mg/kg (p < 0.05). The in vivo data show that A199S/F227A/S287G/A328W/Y332G BChE is indeed roughly 10-fold more potent than CocE for the in vivo protection of mice from cocaine-induced lethality under the same experimental condition.
Figure 4.
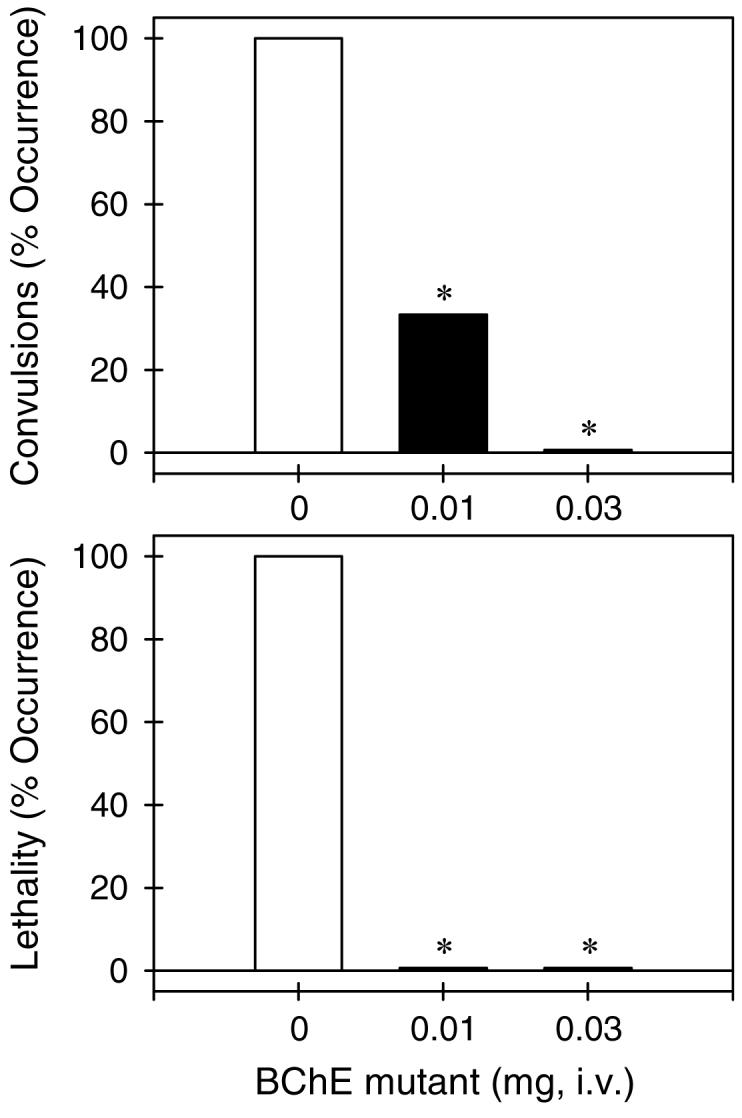
Protective effects of A199S/F227A/S287G/A328W/Y332G BChE against cocaine-induced toxicity. The BChE mutant (mg/mouse) was administered intravenously 1 min before intraperitoneal administration of cocaine 180 mg/kg. Each data point represents the percentage of mice (n = 6 for each dosing condition) exhibiting cocaine-induced convulsions or lethality. The asterisks represent a significant difference from the condition of mice pretreated with phosphate buffered saline (p < 0.05). See supporting materials section for other details.
The encouraging outcome of this study suggests that the unique virtual screening approach (depicted in Figure 2) based on the transition-state simulations and subsequent energetic calculations is promising for rational enzyme redesign and drug discovery. The general design approach can be used to rationally design mutants of any enzyme. When BChE and cocaine in Figure 2 are replaced by another enzyme and its own substrate, one still can carry out the similar QM/MM reaction coordinate calculations on the enzymatic reaction to identify the rate-determining transition state and calculate the energy barriers associated with the wild-type and mutant enzymes. The TIE to be calculated in virtual screening step #2 can be the total interaction energy between all (or some key) atoms of the substrate and all (or some key) atoms of the enzyme. For example, the similar computational design strategy integrated with appropriate wet experiments may also be valuable in rational redesign of other metabolic enzymes for therapeutic treatment of metabolic diseases or for detoxification of toxic compounds (including chemical warfare nerve agents).
Conclusion
We have developed a novel, generally applicable computational design approach based on a systematic virtual screening of transition-states of enzymatic reaction and activation energy calculations. The novel design approach has led to discovery of a human BChE mutant with a ∼2000-fold improved catalytic efficiency, sufficient for therapeutic use as an exogenous enzyme in human to treat cocaine overdose and addiction. The encouraging discovery resulted from the computational design not only provides a promising anti-cocaine medication, but also demonstrates that the novel, generally applicable computational design approach is promising for rational enzyme redesign and drug discovery.
Materials and Methods
Molecular dynamics simulations
The general strategy of performing a classical MD simulation on a transition state structure of an enzymatic reaction has been described and justified in our recent studies.23,24,25,26 In principle, MD simulation using a classical force field (molecular mechanics) can only simulate a stable structure corresponding to a local minimum on the potential energy surface, whereas a transition state during a reaction process is always associated with a first-order saddle point on the potential energy surface. Hence, MD simulation using a classical force field cannot directly simulate a transition state without any restraint on the geometry of the transition state. Nevertheless, if we can technically remove the freedom of imaginary vibration in the transition state structure, then the number of the degrees of vibrational freedom (normal vibration modes) for a nonlinear molecule will decrease from 3N - 6. The transition state structure is associated with a local minimum on the potential energy surface within a subspace of the reduced degrees of vibrational freedom, although it is associated with a first-order saddle point on the potential energy surface with all of the 3N - 6 degrees of vibrational freedom. Theoretically, the degree of vibrational freedom associated with the imaginary vibrational frequency in the transition state structure can be removed by appropriately freezing the reaction coordinate. The reaction coordinate corresponding to the imaginary vibration of the transition state is generally characterized by a combination of some key geometric parameters. These key geometric parameters are bond lengths of the transition bonds, i.e. the forming and breaking covalent bonds during this reaction step of BChE-catalyzed hydrolysis of cocaine, as seen in Figure 1 of the text. Thus, we just need to maintain the lengths of the transition bonds during the MD simulation on a transition state. Technically, we can maintain the lengths of the transition bonds by simply fixing all atoms within the reaction center, by using some constraints on the transition bonds, or by redefining the transition bonds. It should be pointed out that the only purpose of performing such type of MD simulation on a transition state is to examine the dynamic change of the protein environment surrounding the reaction center and the interaction between the reaction center and the protein environment. We are only interested in the simulated structures, as the total energies calculated in this way are meaningless.
The initial BChE structures used in the MD simulations were prepared based on our previous MD simulation12 on the prereactive ES complex for wild-type BChE with (-)-cocaine in water by using Amber7 program package.30 Our previous MD simulations12 on the prereactive BChE-(-)-cocaine complex (ES) started from the X-ray crystal structure32 deposited in the Protein Data Bank (pdb code: 1POP). The present MD simulations on the transition state for the first step (TS1) were performed in such a way that bond lengths of the transition bonds in the transition state were all constrained to be the same as those obtained from our previous ab initio reaction coordinate calculations on the model reaction system of wild-type BChE.21 A sufficiently long MD simulation with the transition bonds constrained should lead to a reasonable protein environment stabilizing the reaction center in the transition-state structure simulated. Further, the simulated TS1 structure for wild-type BChE with (-)-cocaine was used to build the initial structures of TS1 for the examined BChE mutants with (-)-cocaine; only the side chains of mutated residues needed to be changed.
The partial atomic charges for the non-standard residue atoms, including cocaine atoms, in the TS1 structures were calculated by using the RESP protocol implemented in the Antechamber module of the Amber7 package following electrostatic potential (ESP) calculations at ab initio HF/6-31G* level using Gaussian03 program.33 The geometries used in the ESP calculations came from those obtained from the previous ab initio reaction coordinate calculations,21 but the functional groups representing the oxyanion hole were removed. Thus, residues G116, G117, and A199 were the standard residues as supplied by Amber7 in the MD simulations. The general procedure for carrying out the MD simulations in water is essentially the same as that used in our previously reported other computational studies.12,21,30,32 Each aforementioned starting TS1 structure was neutralized by adding chloride counterions and was solvated in a rectangular box of TIP3P water molecules with a minimum solute-wall distance of 10 Å. The total numbers of atoms in the solvated protein structures for the MD simulations are nearly 70,000, although the total number of atoms of BChE and (-)-cocaine is only 8417 (for the wild-type BChE). All of the MD simulations were performed by using the Sander module of Amber7 package. The solvated systems were carefully equilibrated and fully energy minimized. These systems were gradually heated from T = 10 K to T = 298.15 K in 30 ps before running the MD simulation at T = 298.15 K for 1 ns or longer, making sure that we obtained a stable MD trajectory for each of the simulated TS1 structures. The time step used for the MD simulations was 2 fs. Periodic boundary conditions in the NPT ensemble at T = 298.15 K with Berendsen temperature coupling and P = 1 atm with isotropic molecule-based scaling were applied. The SHAKE algorithm was used to fix all covalent bonds containing hydrogen atoms. The non-bonded pair list was updated every 10 steps. The particle mesh Ewald (PME) method was used to treat long-range electrostatic interactions. A residue-based cutoff of 10 Å was utilized to the non-covalent interactions. The coordinates of the simulated systems were collected every 1 ps during the production MD stages.
QM/MM calculations
All of the QM/MM calculations were performed by a pseudobond QM/MM method.34,35 The pseudobond QM/MM method was initially implemented in revised Gaussian03 and Tinker programs.34,35 The revised Gaussian03 and Tinker programs can be used to carry out the QM and MM parts of the QM/MM calculation iteratively until the full self-consistency is achieved. The pseudobond QM/MM method uses a seven-valence-electron atom with an effective core potential constructed to replace the boundary atom of the environment part and to form a pseudobond with the boundary atom of the active part. The main idea of the pseudobond approach is as follows: one considers that a large molecule is partitioned into two parts, an active part and an environment part, by cutting a covalent σ-bond Y-X. Y and X refer to boundary atoms of the environment part and the active part, respectively. Instead of using a hydrogen atom to cap the free valence of X atom as in the conventional link-atom approach, a pseudobond Yps-X is formed by replacing the Y atom with a one-free-valence boundary Y atom (Yps). The Yps atom is parametrized to make the Yps-X pseudobond mimic the original Y-X bond with similar bond length and strength, and also to have similar effects on the rest of the active part.34,35 In the pseudobond approach, the Yps atom and all atoms in the active part form a well-defined QM subsystem which can be treated by a QM method. Excluding Y atom, the remaining atoms in the environment part form the MM subsystem treated by a MM method. The pseudobond ab initio QM/MM approach has been demonstrated to be powerful in studies of enzyme reactions.36,37,38
In order to use the exactly same Amber force field in our QM/MM calculations as that used in our MD simulations, we developed and used a revised version of Gaussian03 and Amber7 programs, instead of the revised Gaussian03 and Tinker programs, to perform the QM/MM calculations in this study. In all of our QM/MM calculations, all atoms of (-)-cocaine and the side chains of S198, H438, and E325 were considered as the QM atoms, whereas the other atoms were regarded as MM atoms. Our QM/MM calculations were performed at the B3LYP/6-31*:Amber level, i.e. the QM calculations were carried out at the B3LYP/6-31G* level while the MM calculations were carried out by using the Amber force field implemented in the Amber7 program. The geometry optimizations were converged to the default criteria of the Gaussian03 (for QM part) and Amber7 (for MM part) programs. The QM/MM-optimized ES, TS1, and INT geometries were used to perform vibrational frequency calculations on the QM subsystem at the same QM/MM level. The vibrational frequency calculations confirmed that the optimized ES and INT1 geometries are indeed associated with local minima on the potential energy surface, whereas the optimized TS1 geometry is indeed associated with a first-order saddle point on the potential energy surface.
Most of the MD simulations and QM/MM calculations were performed in parallel on an HP Superdome (with 256 processors) and an IBM X-series Cluster (with 1,360 processors) at the Center for Computational Sciences, University of Kentucky. Some computations were carried out on a 34-processors IBM x335 Linux cluster and SGI Fuel workstations in our own lab.
Materials for in vitro studies
Cloned pfu DNA polymerase and Dpn I endonuclease were obtained from Stratagene (La Jolla, CA). [3H](-)-cocaine (50 Ci/mmol) was purchased from PerkinElmer Life Sciences (Boston, MA). The expression plasmid pRc/CMV was a gift from Dr. O. Lockridge, University of Nebraska Medical Center (Omaha, NE). All oligonucleotides were synthesized by the Integrated DNA Technologies, Inc (Coralville, IA). The QIAprep Spin Plasmid Miniprep Kit and Qiagen plasmid purification kit and QIAquick PCR purification kit were obtained from Qiagen (Santa Clarita, CA). Human embryonic kidney 293T/17 cells were from ATCC (Manassas, VA). Dulbecco’s modified Eagle’s medium (DMEM) was purchased from Fisher Scientific (Fairlawn, NJ). 3, 3′, 5, 5′-Tetramethylbenzidine (TMB) was obtained from Sigma (Saint Louis, Missouri). Anti-butyrylcholinesterase (mouse monoclonal antibody, Product # HAH002-01) was purchased from AntibodyShop (Gentofte, Denmark) and goat anti-mouse IgG HRP conjugate was from Zymed (San Francisco, CA).
Site-directed mutagenesis, protein expression, and in vitro activity assay
Site-directed mutagenesis of human BChE cDNA was performed by using the QuikChange method.39 Mutations were generated from wild-type human BChE in a pRc/CMV expression plasmid.40 Using plasmid DNA as template and primers with specific base-pair alterations, mutations were made by polymerase chain reaction with Pfu DNA polymerase, for replication fidelity. The PCR product was treated with Dpn I endonuclease to digest the parental DNA template. Modified plasmid DNA was transformed into Escherichia coli, amplified, and purified. The DNA sequences of the mutants were confirmed by DNA sequencing. BChE mutants were expressed in human embryonic kidney cell line 293T/17. Cells were grown to 80-90% confluence in 6-well dishes and then transfected by Lipofectamine 2000 complexes of 4 μg plasmid DNA per each well. Cells were incubated at 37 °C in a CO2 incubator for 24 hours and cells were moved to 60-mm culture vessel and cultured for four more days. The culture medium [10% fetal bovine serum in Dulbecco’s modified Eagle’s medium (DMEM)] was harvested for a BChE activity assay. To measure (-)-cocaine and benzoic acid, the product of (-)-cocaine hydrolysis catalyzed by BChE, we used sensitive radiometric assays based on toluene extraction of [3H]-(-)-cocaine labeled on its benzene ring.41 In brief, to initiate the enzymatic reaction, 100 nCi of [3H]-(-)-cocaine was mixed with 100 μl of culture medium. The enzymatic reactions proceeded at room temperature (25°C) with varying concentrations of (-)-cocaine. The reactions were stopped by adding 300 μl of 0.02 M HCl, which neutralized the liberated benzoic acid while ensuring a positive charge on the residual (-)-cocaine. [3H]benzoic acid was extracted by 1 ml of toluene and measured by scintillation counting. Finally, the measured (-)-cocaine concentration-dependent radiometric data were analyzed by using the standard Michaelis-Menten kinetics so that the catalytic parameters (kcat and KM) were determined along with the use of an enzyme-linked immunosorbent assay (ELISA)42 described below.
The ELISA buffers used in the present study are the same as those described in literature.42 The coating buffer was 0.1 M sodium carbonate/bicarbonate buffer, pH 9.5. The diluent buffer (EIA buffer) was potassium phosphate monobasic/potassium phosphate dibasic buffer, pH 7.5, containing 0.9% sodium chloride and 0.1% bovine serum albumin. The washing buffer (PBS-T) was 0.01 M potassium phosphate monobasic/potassium phosphate dibasic buffer, pH 7.5, containing 0.05% (v/v) Tween-20. All the assays were performed in triplicate. Each well of an ELISA microtiter plate was filled with 100 μl of the mixture buffer consisting of 20 μl culture medium and 80 μl coating buffer. The plate was covered and incubated overnight at 4°C to allow the antigen to bind to the plate. The solutions were then removed and the wells were washed four times with PBS-T. The washed wells were filled with 200 μl diluent buffer and kept shaking for 1.5 h at room temperature (25°C). After washing with PBS-T for four times, the wells were filled with 100 μl antibody (1:8,000) and were incubated for 1.5 h, followed by washing for four times. Then, the wells were filled with 100 μl goat anti-mouse IgG HRP conjugate complex diluted to a final 1:3,000 dilution, and were incubated at room temperature for 1.5 h, followed by washing for four times. The enzyme reactions were started by addition of 100 μl substrate (TMB) solution.42 The reactions were stopped after 15 min by the addition of 100 μl of 2 M sulfuric acid, and the absorbance was read at 460 nm using a Bio-Rad ELISA plate reader.
Protein purification
For the purpose of protein purification, we also constructed an expression plasmid pSecTag2A/N-His6 encoding the A199S/F227A/S287G/A328W/Y332G mutant of human BChE for high-level expression. The aforementioned serum expression system was replaced by Free Style 293 Expression System (Invitrogen). The system used 293F cells to replace the 293T cells in an orbit shaker bottle instead of using regular flask. HisPur™Cobalt Resin (Pierce) was used for the purification leading to highly purified enzyme (> 95% purity). The serum-free medium greatly facilitated rH-BChE purification processes.
In vivo studies
Subjects and drugs
Male NIH-Swiss mice (25-30 g) were obtained from Harlan Sprague-Dawley Inc. (Indianapolis, IN) and were housed in groups of 6 mice per cage. All mice were allowed ad libitum access to food and water, and were maintained on a 12-h light-dark cycle with lights on at 6:30 AM in a room kept at a temperature of 21-22°C. Experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals as adopted and promulgated by the National Institutes of Health. The experimental protocols were approved by the University Committee on the Use and Care of Animals at the University of Michigan.
(-)-Cocaine HCl (National Institute on Drug Abuse, Bethesda, MD) was dissolved in sterile water and was administered intraperitoneally at a volume of 0.01 mL/g. The BChE mutant (i.e. A199S/F227A/S287G/A328W/Y332G BChE) was diluted to difference concentrations in phosphate buffered saline and administered intravenously at a volume of 0.2 mL/mouse.
Behavioral assay
Cocaine-induced toxicity was characterized by the occurrence of convulsions and lethality. Cocaine-induced convulsions were defined as loss of righting posture for at least 5 s with the simultaneous presence of clonic limb movements.31 Lethality was defined as cessation of observed movement and respiration. Following cocaine administration, mice were immediately placed individually in Plexiglas containers (16 × 28 × 20 cm high) for observation. The presence or absence of convulsions and lethality were recorded for 60 min following cocaine administration.
Drug administration
The mouse was placed in a small restraint chamber (Outer tube diameter: 30 mm, Inner tube diameter: 24 mm, Model #BS4-34-0012, Harvard Apparatus, Inc., Holliston, MA) that left the tail exposed. The tail was cleansed with an alcohol wipe and a 30G1/2 precision glide needle (Fisher Scientific, Pittsburgh, PA) was inserted into one of the side veins for infusion. The intravenous injection volume of BChE mutant (i.e. 0, 0.01, and 0.03 mg) was 0.2 mL per mouse and it was given 1 min before intraperitoneal administration of cocaine 180 mg/kg. To staunch the bleeding, sterile gauze and pressure were applied to the injection site.
Data analysis
Data from the behavioral toxicity studies (i.e. % of mice showing affected responses) were analyzed with Chi-square probability test with one tail. Protective effects of the BChE mutant were compared with those of phosphate buffered saline. The criterion for significance was set at p < 0.05.
Supplementary Material
Acknowledgements
This work was supported by NIH (grants DA013930 and DA021416). The authors also acknowledge the Center for Computational Sciences (CCS) at University of Kentucky for supercomputing time on an HP Superdome (with 4 nodes and 256 processors) and an IBM X-series Cluster with 1,360 processors.
References
- 1.Mendelson JH, Mello NK. New Engl. J. Med. 1996;334:965–972. doi: 10.1056/NEJM199604113341507. [DOI] [PubMed] [Google Scholar]
- 2.Singh S. Chem. Rev. 2000;100:925–1024. doi: 10.1021/cr9700538. [DOI] [PubMed] [Google Scholar]
- 3.Paula S, Tabet MR, Farr CD, Norman AB, Ball WJ., Jr. J. Med. Chem. 2004;47:133–142. doi: 10.1021/jm030351z. [DOI] [PubMed] [Google Scholar]
- 4.Gorelick DA. Drug Alcohol Depend. 1997;48:159–165. doi: 10.1016/s0376-8716(97)00119-1. [DOI] [PubMed] [Google Scholar]
- 5.Redish AD. Science. 2004;306:1944–1947. doi: 10.1126/science.1102384. [DOI] [PubMed] [Google Scholar]
- 6.Meijler MM, Kaufmann GF, Qi LW, Mee JM, Coyle AR, Moss JA. J. Am. Chem. Soc. 2005;127:2477–2484. doi: 10.1021/ja043935e. [DOI] [PubMed] [Google Scholar]
- 7.Carrera MRA, Kaufmann GF, Mee JM, Meijler MM, Koob GF, Janda KD. Proc. Natl. Acad. Sci. USA. 2004;101:10416–10421. doi: 10.1073/pnas.0403795101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landry DW, Zhao K, Yang GX-Q, Glickman M, Georgiadis TM. Science. 1993;259:1899–1901. doi: 10.1126/science.8456315. [DOI] [PubMed] [Google Scholar]
- 9.Zhan C-G, Deng S-X, Skiba JG, Hayes BA, Tschampel SM, Shields GC, Landry DW. J. Comput. Chem. 2005;26:980–986. doi: 10.1002/jcc.20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamendulis LM, Brzezinski MR, Pindel EV, Bosron WF, Dean RA. J. Pharmacol. Exp. Ther. 1996;279:713–717. [PubMed] [Google Scholar]
- 11.Sun H, Pang YP, Lockridge O, Brimijoin S. Mol. Pharmacol. 2002;62:220–224. doi: 10.1124/mol.62.2.220. [DOI] [PubMed] [Google Scholar]
- 12.Hamza A, Cho H, Tai H-H, Zhan C-G. J. Phys. Chem. B. 2005;109:4776–4782. doi: 10.1021/jp0447136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gateley SJ. Biochem. Pharmacol. 1991;41:1249–1254. doi: 10.1016/0006-2952(91)90665-r. [DOI] [PubMed] [Google Scholar]
- 14.Darvesh S, Hopkins DA, Geula C. Nature Rev. Neurosci. 2003;4:131–138. doi: 10.1038/nrn1035. [DOI] [PubMed] [Google Scholar]
- 15.Giacobini E, editor. Butyrylcholinesterase: Its Function and Inhibitors. Dunitz Martin Ltd; Great Britain: 2003. [Google Scholar]
- 16.Poet TS, McQueen CA, Halpert JR. Drug Metab. Dispos. 1996;24:74–80. [PubMed] [Google Scholar]
- 17.Pan W-J, Hedaya MA. J. Pharm. Sci. 1990;88:468–476. doi: 10.1021/js980283h. [DOI] [PubMed] [Google Scholar]
- 18.Larsen NA, Turner JM, Stevens J, Rosser SJ, Basran A, Lerner RA, Bruce NC, Wilson IA. Nature Struct. Biol. 2002;9:17–21. doi: 10.1038/nsb742. [DOI] [PubMed] [Google Scholar]
- 19.Turner JM, Larsen NA, Basran A, Barbas CF, Bruce NC, Wilson IA, Lerner RA. Biochemistry. 2002;41:12297–12307. doi: 10.1021/bi026131p. [DOI] [PubMed] [Google Scholar]
- 20.Sun H, Yazal JE, Lockridge O, Schopfer LM, Brimijoin S, Pang YP. J. Biol. Chem. 2001;276:9330–9336. doi: 10.1074/jbc.M006676200. [DOI] [PubMed] [Google Scholar]
- 21.Zhan C-G, Zheng F, Landry DW. J. Am. Chem. Soc. 2003;125:2462–2474. doi: 10.1021/ja020850+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao Y, Atanasova E, Sui N, Pancook JD, Watkins JD, Brimijoin S. Mol. Pharmacol. 2005;67:204–211. doi: 10.1124/mol.104.006924. [DOI] [PubMed] [Google Scholar]
- 23.Zhan C-G, Gao D. Biophysical J. 2005;89:3863–3872. doi: 10.1529/biophysj.105.070276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao D, Zhan C-G. J. Phys. Chem. B. 2005;109:23070–23076. doi: 10.1021/jp053736x. [DOI] [PubMed] [Google Scholar]
- 25.Pan Y, Gao D, Yang W, Cho H, Yang G-F, Tai H-H, Zhan C-G. Proc. Natl. Acad. Sci. USA. 2005;102:16656–16661. doi: 10.1073/pnas.0507332102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao D, Zhan C-G. Proteins. 2006;62:99–110. doi: 10.1002/prot.20713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao D, Cho H, Yang W, Pan Y, Yang G-F, Tai H-H, Zhan C-G. Angew. Chem. Int. Ed. 2006;45:653–657. doi: 10.1002/anie.200503025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan Y, Gao D, Yang W, Cho H, Zhan C-G. J. Am. Chem. Soc. 2007;129:13537–13543. doi: 10.1021/ja073724k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brimijoin S, Gao Y, Anker JJ, Gliddon LA, LaFleur D, Shah R, Zhao Q, Singh M, Carroll MEA. Neuropsychopharmacology. 2008 doi: 10.1038/sj.npp.1301666. advance online publication, 16 January 2008; doi:10.1038/sj.npp.1301666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Case DA, et al. Amber7. University of California; San Francisco: 2002. [Google Scholar]
- 31.Ko M-C, Bowen LD, Narasimhan D, Berlin AA, Lukacs NW, Sunahara RK, Cooper ZD, Woods JH. J. Pharmacol. Exp. Therap. 2007;320:926–933. doi: 10.1124/jpet.106.114223. [DOI] [PubMed] [Google Scholar]
- 32.Nicolet Y, Lockridge O, Masson P, Fontecilla-Camps JC, Nachon F. J. Biol. Chem. 2003;278:41141–41147. doi: 10.1074/jbc.M210241200. [DOI] [PubMed] [Google Scholar]
- 33.Frisch MJ, et al. Gaussian 03, Revision A.1. Gaussian; Pittsburgh, PA: 2003. [Google Scholar]
- 34.Zhang Y, Lee T, Yang W. J. Chem. Phys. 1999;110:46–54. [Google Scholar]
- 35.Zhang Y. J. Chem. Phys. 2005;122:024114. doi: 10.1063/1.1834899. [DOI] [PubMed] [Google Scholar]
- 36.Corminboeuf C, Hu P, Tuckerman ME, Zhang Y. J. Am. Chem. Soc. 2006;128:4530–4531. doi: 10.1021/ja0600882. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Liu H, Yang W. J. Chem. Phys. 2000;112:3483–3492. [Google Scholar]
- 38.Hu P, Zhang Y. J. Am. Chem. Soc. 2006;128:1272–1278. doi: 10.1021/ja056153+. [DOI] [PubMed] [Google Scholar]
- 39.Braman J, Papworth C, Greener A. Methods Mol. Biol. 1996;57:31–44. doi: 10.1385/0-89603-332-5:31. [DOI] [PubMed] [Google Scholar]
- 40.Masson P, Xie W, Froment M-T, Levitsky V, Fortier P-L, Albaret C, Lockridge O. Biochim. Biophys. Acta. 1999;1433:281–293. doi: 10.1016/s0167-4838(99)00115-6. [DOI] [PubMed] [Google Scholar]
- 41.Sun H, Shen ML, Pang YP, Lockridge O, Brimijoin S. J. Pharmacol. Exp. Ther. 2002;302:710–716. doi: 10.1124/jpet.302.2.710. [DOI] [PubMed] [Google Scholar]
- 42.Brock A, Mortensen V, Loft AGR, Nørgaard-Pedersen B. J. Clin. Chem. Clin. Biochem. 1990;28:221–224. doi: 10.1515/cclm.1990.28.4.221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.