Abstract
Complement activation contributes to antibody-mediated allograft rejection, but increasing evidence also implicates complement proteins produced locally within the graft, in part by infiltrating mononuclear cells, as important mediators of tissue injury. To test this concept in transplant recipients we evaluated complement, complement regulator and Tcell/proinflammatory marker gene expression by quantitative real-time polymerase chain reaction in 71 archived heart transplant biopsies and correlated the results with the histologic grade of rejection. Significantly more transcripts encoding alternative pathway components factor B, C3 and properdin, and C3a receptor and C5a receptor, were detected in grade 3 vs. grade 0 or 1 biopsies. The grade 3 rejections also contained significantly higher amounts of CD3, interferon γ, perforin and granzyme B genes. In addition to providing supportive evidence for a pathogenic role of graft-derived complement in human heart transplant injury, these correlations suggest that molecular profiling of complement gene expression could be useful in the diagnosis of human allograft rejection.
Keywords: heart transplantation, human, complement, gene expression
Introduction
In the setting of transplantation, the complement cascade has an established pathogenic role as an effector arm of antibody mediated injury to the allograft. Recent studies from a number of laboratories have identified additional roles for complement in transplantation, including functioning as a key modulator of ischemia reperfusion injury and in regulating the strength and effector function of alloreactive T cells (1-8). In vitro and in vivo animal model studies have further shown that these effects involve activation of complement produced by the graft and/or by immune cells, and do not require serum/liver-derived complement components (3, 6-8). Supporting a role for graft-derived complement as a mediator of transplant injury is the report that kidneys expressing a particular polymorphic variant of C3 have worse outcomes than those without this polymorphism (9). Whether graft-derived (as opposed to systemic/liver derived) complement contributes to human heart transplant injury has not been evaluated. If graft-derived complement participates in allograft injury we reasoned that complement component gene expression should be detectable in biopsies of human heart transplants and that allograft rejection would be associated with upregulated expression of complement component genes.
Materials and Methods
Biopsy material
We obtained frozen archived tissue from cardiac allograft biopsies that had been collected as part of routine clinical care between 2003 and 2004 and stored in the Department of Anatomic Pathology at The Cleveland Clinic. Grade of rejection was determined by the clinical pathologist at the time of the biopsy using the 1990 ISHLT working formulation. The light microscopic histopathologic features of antibody mediated rejection as defined by the 2004 working formulation (10, 11) were absent in these biopsies. We identified 20 biopsies from 20 different patients with a histologic grade of 0, 13 biopsies from 13 individual patients with a histologic grade of 1A, and 38 biopsies from 38 separate patients with a histologic grade of 3A. Biopsies were obtained as part of a routine surveillance protocol, not in response to clinical symptoms. During this time period immunosuppression consisted of prednisone, mycophenolate mofetil and a calcineurin inhibitor (cyclosporine or tacrolimus). Induction therapy was only used selectively in sensitized patients. All patient materials were completely de-identified and this study was approved by our Institutional Review Board. Total RNA from a normal human heart was obtained from BioChain Institute, Inc. (Hayward, California).
Gene expression by quantitative real time PCR
Total RNA was isolated using the RNeasy Mini kit (QIAGEN Valencia, CA) and reverse-transcribed with the High Capacity cDNA Archive Kit (Applied Biosystems Foster City, CA). TaqMan Gene Expression Assays for factor B (fB), factor D (fD), properdin, C4, C3, C5, C3a receptor (C3aR), C5a receptor (C5aR), decay accelerating factor (DAF; CD55), membrane cofactor protein (CD46), protectin (CD59), CD3, granzyme B, perforin and interferon γ (IFNγ) were purchased from Applied Biosystems.
The cDNA was pre-amplified for 10 cycles with a pooled mix of gene expression assays, then diluted and analyzed by gene-specific real-time quantitative PCR on an Applied Biosystems 7500 Fast System. To test whether the multiplex preamplification step introduced bias into the results we compared results using preamplified and unamplified cDNA using the ΔΔCt method (Applied Biosystems). The 18S assay was not included in the preamplification primer pool. The ΔCt for the preamplified samples was calculated by subtracting the Ct for the 18S gene from the Ct for each gene of interest.
The ΔCt for the unamplified samples is calculated the same way.
The ΔΔCt is the difference between the preamplified ΔCt and the unamplified ΔCt.
A ΔΔCt value of zero means the expression pattern in the preamplified sample is the same as in unamplified cDNA. All of the genes tested in this study had ΔΔCts of less than 1.5 (data not shown). Relative gene expression (RQ) was determined by the 2-ΔΔCT method (12) using the 18S ribosomal gene as an endogenous control and cDNA from human PBMCs as calibrator. For each gene, the RQs for grades 1 and 3 samples were normalized to the grade 0 mean RQ. The means of the normalized grade 1 and 3 RQs were then calculated. A two-tailed Student's t test was used to determine the statistical significance of the difference in gene expression among groups.
Results and discussion
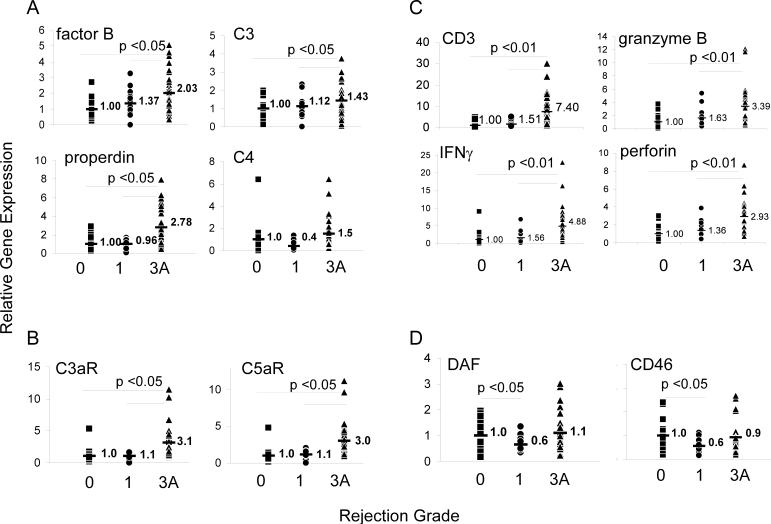
The results of our analysis are summarized in Figure 1 and are expressed as fold increase versus values in grade 0 biopsies (normalized to a mean of 1). Notably, gene products for all components were detectable in 89% of the tissue samples regardless of histologic grade. As a comparison, we also tested for complement/regulator gene expression in RNA obtained from a normal heart. Normal heart tissue contained 100−200-fold less RNA for C3, fB, C3aR and C5aR, and 5-fold less RNA for CD46 and CD55 than any of the biopsies (data not shown) revealing that transplantation with or without detectable intragraft infiltrates induces significant upregulation of these genes. Because our study was designed to assess gene product by PCR (as opposed to protein), the detected complement components must have derived from the graft tissue and could not reflect deposition/activation of serum complement as a consequence of antibody mediated allograft injury.
Figure 1. Gene expression profiles in heart biopsies measured by qPCR.
Relative gene expression values for A) complement components Factor B, C3, properdin and C4, B) complement receptors C5aR and C3aR, C) T cell/proinflammatory genes CD3, perforin, granzyme B and IFNγ, and D), complement regulatory proteins DAF/CD55 and CD46 are shown. The results for Grade 1 and 3 biopsies were normalized to the grade 0 mean. (■ Grade 0;  Grade 1; ▲ Grade 3). Numbers in each panel represent the mean fold increase for each group. All samples were also tested for the expression of factor D, C5 and CD59 and there were no significant differences among the groups for each of these genes (data not shown).
Grade 1; ▲ Grade 3). Numbers in each panel represent the mean fold increase for each group. All samples were also tested for the expression of factor D, C5 and CD59 and there were no significant differences among the groups for each of these genes (data not shown).
ISHLT Grade 3 biopsies (moderate rejection or 2R) contained significantly more transcripts than grade 0 or 1 biopsies for fB and properdin (alternative pathway components) and C3 (Fig 1A) and C5aR/C3aR genes (Fig 1B) consistent with experimental data revealing roles for these molecules as mediators of graft injury. In further accord with animal model studies revealing alternative but not classical pathway complement as mediating transplant injury (3, 13), C4 expression (Fig 1A) was not different between grade 0−1 and grade 3 biopsies. Factor D and C5 expression were also not different among groups (not shown). We did not detect significant differences in expression of any of these tested genes between grade 1 and grade 0 biopsies. Consistent with the histological diagnosis of cellular rejection, grade 3 biopsies also contained significantly higher expression of the T cell/proinflammatory genes, CD3, perforin, granzyme B and IFNγ than grade 1 or grade 0 biopsies (Figure 1C).
Our technical approach did not permit us to define the specific cellular source of each complement component. Studies of animal models and human cells lines have shown that epithelial and endothelial cells upregulate complement production in response to various proinflammatory stimuli including ischemia/reperfusion (14-18), but the detected correlation with histologic grade of injury and with increased expression of T cell gene products strongly suggests the possibility that immune cells infiltrating the allograft are a dominant source of complement within the graft.
In contrast to the upregulated expression of alternative pathway complement components, expression of complement regulator genes, DAF and CD46 (Figure 1D) but not CD59 (not shown) were significantly lower in grade 1 biopsies vs. either grade 0 or grade 3 biopsies. A relative diminution of complement regulator expression could permit enhanced complement activation, thereby potentially amplifying local injury to the graft. Such an effect would be consistent with our previously published work (3) showing that DAF transiently down regulates during T cell/APC interactions which in turn permits local complement activation that enhances T cell responses.
It is worth emphasizing that the light microscopic histopathologic features of antibody-mediated rejection were absent in these biopsies. Insufficient tissue was available for C4d staining and we do not have the ability to assess serum alloantibody in the patients from which these biopsies were obtained. Despite these technical limitations, this report represents the first documentation (to our knowledge) that complement gene products are detectable in human allograft biopsies and that they are upregulated in association with graft injury. In addition to supporting the hypothesis that graft-derived complement participates in heart transplant injury, these correlative data raise the possibility that molecular profiling of complement gene expression could be useful diagnostically and suggest that complement components are potential targets for adjunctive therapies aimed at preventing human heart transplant injury.
Abbreviations
- C3aR
C3a receptor
- C5aR
C5a receptor
- DAF
decay accelerating factor
- fB
factor B
- qPCR
quantitative real-time polymerase chain reaction
- IFNγ
interferon gamma
References
- 1.Zhou W, Peng Q, Li K, Sacks SH. Role of dendritic cell synthesis of complement in the allospecific T cell response. Mol Immunol. 2006 doi: 10.1016/j.molimm.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Peng Q, Li K, Patel H, Sacks SH, Zhou W. Dendritic cell synthesis of C3 is required for full T cell activation and development of a Th1 phenotype. J Immunol. 2006;176(6):3330. doi: 10.4049/jimmunol.176.6.3330. [DOI] [PubMed] [Google Scholar]
- 3.Heeger PS, Lalli PN, Lin F, et al. Decay-accelerating factor modulates induction of T cell immunity. J Exp Med. 2005;201(10):1523. doi: 10.1084/jem.20041967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marsh JE, Farmer CK, Jurcevic S, Wang Y, Carroll MC, Sacks SH. The allogeneic T and B cell response is strongly dependent on complement components C3 and C4. Transplantation. 2001;72(7):1310. doi: 10.1097/00007890-200110150-00022. [DOI] [PubMed] [Google Scholar]
- 5.Pratt JR, Basheer SA, Sacks SH. Local synthesis of complement component C3 regulates acute renal transplant rejection. Nat Med. 2002;8(6):582. doi: 10.1038/nm0602-582. [DOI] [PubMed] [Google Scholar]
- 6.Lalli PN, Strainic MG, Lin F, Medof ME, Heeger PS. Decay accelerating factor can control T cell differentiation into IFN-gamma-producing effector cells via regulating local C5a-induced IL-12 production. J Immunol. 2007;179(9):5793. doi: 10.4049/jimmunol.179.9.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farrar CA, Zhou W, Lin T, Sacks SH. Local extravascular pool of C3 is a determinant of postischemic acute renal failure. Faseb J. 2006;20(2):217. doi: 10.1096/fj.05-4747com. [DOI] [PubMed] [Google Scholar]
- 8.Zhou W, Medof ME, Heeger PS, Sacks S. Graft-derived complement as a mediator of transplant injury. Curr Opin Immunol. 2007;19(5):569. doi: 10.1016/j.coi.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Brown KM, Kondeatis E, Vaughan RW, et al. Influence of donor C3 allotype on late renal-transplantation outcome. N Engl J Med. 2006;354(19):2014. doi: 10.1056/NEJMoa052825. [DOI] [PubMed] [Google Scholar]
- 10.Reed EF, Demetris AJ, Hammond E, et al. Acute antibody-mediated rejection of cardiac transplants. J Heart Lung Transplant. 2006;25(2):153. doi: 10.1016/j.healun.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Stewart S, Winters GL, Fishbein MC, et al. Revision of the 1990 working formulation for the standardization of nomenclature in the diagnosis of heart rejection. J Heart Lung Transplant. 2005;24(11):1710. doi: 10.1016/j.healun.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Lin T, Zhou W, Farrar CA, Hargreaves RE, Sheerin NS, Sacks SH. Deficiency of C4 from donor or recipient mouse fails to prevent renal allograft rejection. Am J Pathol. 2006;168(4):1241. doi: 10.2353/ajpath.2006.050360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laudes IJ, Chu JC, Huber-Lang M, et al. Expression and function of C5a receptor in mouse microvascular endothelial cells. J Immunol. 2002;169(10):5962. doi: 10.4049/jimmunol.169.10.5962. [DOI] [PubMed] [Google Scholar]
- 15.Selvan RS, Kapadia HB, Platt JL. Complement-induced expression of chemokine genes in endothelium: regulation by IL-1-dependent and -independent mechanisms. J Immunol. 1998;161(8):4388. [PubMed] [Google Scholar]
- 16.Sheerin NS, Zhou W, Adler S, Sacks SH. TNF-alpha regulation of C3 gene expression and protein biosynthesis in rat glomerular endothelial cells. Kidney Int. 1997;51(3):703. doi: 10.1038/ki.1997.101. [DOI] [PubMed] [Google Scholar]
- 17.Kitano E, Kitamura H. Synthesis of factor D by gastric cancer-derived cell lines. Int Immunopharmacol. 2002;2(6):843. doi: 10.1016/s1567-5769(02)00028-0. [DOI] [PubMed] [Google Scholar]
- 18.Pratt JR, Abe K, Miyazaki M, Zhou W, Sacks SH. In situ localization of C3 synthesis in experimental acute renal allograft rejection. Am J Pathol. 2000;157(3):825. doi: 10.1016/S0002-9440(10)64596-8. [DOI] [PMC free article] [PubMed] [Google Scholar]