Abstract
The recently identified type VI secretion systems (T6SS) have a crucial function in the virulence of various proteobacteria, including the human pathogen Vibrio cholerae. T6SS are encoded by a conserved gene cluster comprising approximately 15 open reading frames, mediating the appearance of Hcp and VgrG proteins in cell culture supernatants. Here, we analysed the function of the V. cholerae T6SS member ClpV, a specialized AAA+ protein. ClpV is crucial for a functional T6SS and interacts through its N-terminal domain with the VipA/VipB complex that is composed of two conserved and essential members of T6SS. Transferring ClpV substrate specificity to a distinct AAA+ protein involved in proteolysis caused degradation of VipA but not Hcp or VgrG2, suggesting that VipA rather than Hcp/VgrG2 functions as a primary ClpV substrate. Strikingly, VipA/VipB form tubular, cogwheel-like structures that are converted by a threading activity of ClpV into small complexes. ClpV-mediated remodelling of VipA/VipB tubules represents a crucial step in T6S, illuminating an unexpected role of an ATPase component in protein secretion.
Keywords: AAA+ protein, chaperone, protein secretion
Introduction
Bacterial pathogenesis mainly relies on the activity of effector molecules that are either exported to the extracellular environment or directly transferred into host cells. In Gram-negative pathogens, the presence of outer and inner membranes, separated by the periplasmic space, adds an additional layer of complexity to the transport process. So far six distinct secretion systems (referred to as T1SS–T6SS (type VI secretion system)) have been described to export proteins through this multilayer cell envelope. The secretory machineries are distinguished by conserved structural components, the secreted effectors and the mechanism of substrate export. T2SS and T5SS mediate the secretion of proteins through the outer membrane in a contiguous step after substrate export into the periplasm through the Sec system. In contrast, T1SS, T3SS and T4SS transport effectors in a single step through the inner and outer membrane, with the exception of Bordetella pertussis T4SS that secretes pertussis toxin in two steps. Less is known about the organization and mechanism of T6SS that have been recently described by Mekalanos and co-workers (Pukatzki et al, 2006). T6SS components are usually encoded by a single gene cluster consisting of approximately 15 conserved open reading frames. This gene cluster is present in proteobacterial pathogens and symbionts, including Vibrio cholerae, Escherichia coli, Pseudomonas aeruginosa, Agrobacterium tumefaciens and Rhizobium leguminosarum (Das and Chaudhuri, 2003).
A hallmark of all T6SS is the presence of Hcp and VgrG proteins in culture supernatants (Dudley et al, 2006; Mougous et al, 2006; Pukatzki et al, 2006; Schell et al, 2007; Zheng and Leung, 2007; Wu et al, 2008). Species-specific secretory proteins of T6SS have been identified in a few cases (Dudley et al, 2006; Zheng and Leung, 2007). All exported proteins lack conventional N-terminal hydrophobic signal sequences and appear as unprocessed polypeptides.
The precise functions of T6SS secretory proteins are still unknown; however, implications in bacterial virulence have been reported for various species. In V. cholerae, secretion-deficient mutants strongly attenuate cytotoxicity towards the model host Dictyostelium discoideum (Pukatzki et al, 2006). T6SS mutants are associated with reduced virulence of Burkholderia mallei, Salmonella enterica and Edwardsiella tarda (Rao et al, 2004; Parsons and Heffron, 2005; Schell et al, 2007; Zheng and Leung, 2007). B. mallei Hcp is produced and secreted during infection, whereas P. aeruginosa Hcp seems to be preferentially exported during chronic infections in cystic fibrosis patients (Mougous et al, 2006; Schell et al, 2007). In R. leguminosarum, the T6SS controls specificity of host cell infection (Bladergroen et al, 2003). Interestingly, T6SS mutants of V. cholerae and V. parahaemolyticus also exhibit defects in biofilm formation (Enos-Berlage et al, 2005; Mueller et al, 2007).
Little is known about the mechanism of protein secretion by T6SS. How are T6SS secretory proteins targeted to the putative membrane-embedded translocon and which cellular components drive their export? T1SS to T5SS comprise at least one membrane-associated factor that mediates protein secretion by ATP hydrolysis (Remaut and Waksman, 2004). In case of T6SS, two proteins may function as energizing components: IcmF and ClpV. On the basis of sequence analysis, IcmF is predicted to be located in the inner membrane and to consist of a cytosolic and a large periplasmic domain. The cytosolic part harbours a conserved Walker A motif, implicating a function as an ATPase during type VI protein secretion. Indeed, icmF mutants prevent Hcp secretion (Pukatzki et al, 2006; Zheng and Leung, 2007; Wu et al, 2008). On the other hand, IcmF variants that are expected to be deficient in ATP binding and/or hydrolysis still support Hcp and VgrG export in E. tarda (Zheng and Leung, 2007), suggesting that ClpV might represent the central energy source for T6S.
ClpV is a member of the AAA+ (ATPases associated with various cellular activities) protein family (Schlieker et al, 2005). AAA+ proteins are oligomeric ring-like machines that bind ATP through the conserved AAA domain and convert the energy of ATP hydrolysis into a mechanical force to remodel a variety of substrates (Hanson and Whiteheart, 2005). Many cellular processes, including membrane fusion, protein repair and degradation, are controlled by AAA+ proteins; however, a function in protein secretion has not been described so far.
Here, we analysed the function of the AAA+ protein ClpV in V. cholerae T6S. We demonstrate that ClpV is strictly required for the secretion of Hcp and VgrG2. ClpV, however, does not directly exert an effect on the putative secretory proteins, but remarkably disintegrates a tubular, cogwheel-like protein complex, which is composed of VipA and VipB, conserved and essential members of T6SS. We propose that ClpV-mediated remodelling of VipA/VipB tubules is a crucial step in T6SS biogenesis.
Results
ClpV is an essential energizing component of T6SS
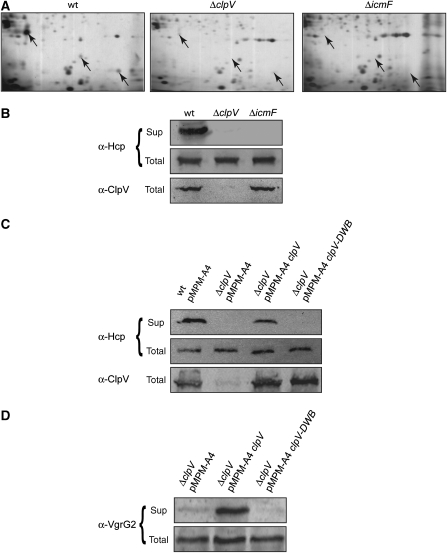
To investigate the role of the AAA+ protein ClpV in T6SS, we used V. cholerae V52 as a model organism as this strain constitutively expresses the components of the T6SS vas (virulence-associated secretion) gene cluster when grown in rich medium (Pukatzki et al, 2006). To test for a role of ClpV in T6S, we generated a chromosomal clpV in-frame deletion strain, termed V.c. ΔclpV. As a control, we constructed a V.c. ΔicmF mutant that had been previously demonstrated to be deficient in the secretion of Hcp, the major substrate of T6SS (Pukatzki et al, 2006). Comparison of the secretome of V.c. wild type, V.c. ΔclpV and V.c. ΔicmF cells after separation by 2D gel electrophoresis revealed that Hcp is lacking in both mutant strains (Figure 1A). Defects in Hcp secretion were confirmed by immunoblot analysis using Hcp-specific antibodies (Figure 1B). The deficiencies in Hcp secretion were neither caused by changes in Hcp synthesis nor by differences in Hcp stability, as both mutant strains harboured Hcp levels that were identical to those of V.c. wild-type cells (Figure 1B). Arabinose-controlled expression of plasmid-encoded clpV restored Hcp secretion, demonstrating that the observed secretion defect of V.c. ΔclpV is a direct effect. As AAA+ proteins remodel substrates in an ATP-fuelled process, we tested the ability of the ClpV E297A/E689A variant (referred to as ClpV-DWB), which harbours mutations in the Walker B motifs of both AAA domains, to complement V.c. ΔclpV. ClpV-DWB is deficient in ATP hydrolysis (data not shown) and did not restore Hcp secretion, demonstrating that an ATP-driven remodelling activity of ClpV is required for T6S (Figure 1C). Proteins of the VgrG family represent the second class of type VI secretory proteins. We therefore also tested for the secretion of V. cholerae VgrG2, which encompasses gp27- and gp5-like domains that are shared by all VgrG family members. Immunoblot analysis using VgrG2-specific antibodies confirmed the essential role of an active ClpV protein for T6SS (Figure 1D).
Figure 1.
ClpV is an essential energizing component of T6SS. (A) Culture supernatants of V. cholerae V52 wild-type (V.c. wt) and V. cholerae V52 ΔclpV and ΔicmF mutant cells were separated by 2D gel electrophoresis followed by silver staining. Protein spots that were specifically present in V.c. wt but missing in V.c. ΔclpV and V.c. ΔicmF mutants (arrows) were identified by mass spectrometry as full-length Hcp or Hcp degradation products. (B) The deficiencies of V.c. ΔclpV and V.c. ΔicmF mutants in Hcp secretion were verified by immunoblot analysis using Hcp-specific antibodies. Sup: culture supernatant; total: total cell extract. (C, D) Complementation of the V.c. ΔclpV secretion defect by plasmid-encoded clpV and clpV-DWB using Hcp- or VgrG2-specific antibodies. ClpV levels were monitored by immunoblot analysis using ClpV-specific antibodies.
The ClpV N-domain is essential for T6S and mediates binding to the VipA/VipB complex
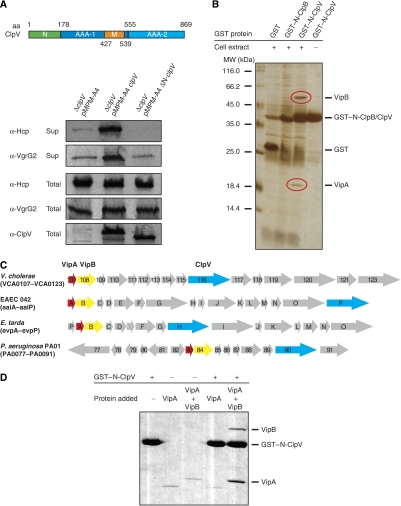
AAA+ proteins share the AAA domain as a common feature for ATP hydrolysis and oligomerization. Functional specificity is gained through the presence of additional domains that are missing in other family members. ClpV harbours two extra domains, an N-terminal domain (N-domain) and a middle domain (M-domain) that is inserted into the first AAA domain (Figure 2A). The ClpV N-domain exhibits weak sequence homology to the N-domains of ClpA, ClpB and ClpC. These N-domains are attached through flexible linkers to the corresponding AAA+ protein rings and mediate substrate specificity either directly or indirectly by serving as a binding platform for adaptor proteins that deliver their protein cargo upon binding to the AAA+ partner protein (Erbse et al, 2006; Kirstein et al, 2006). The role of M-domains in AAA+ protein function is less clear and their analysis is hampered by intimate contacts between M-domains and AAA domains and the contribution of M-domains to AAA+ oligomerization (Kedzierska et al, 2003; Mogk et al, 2003; Haslberger et al, 2007). We therefore focused on the function of the ClpV N-domain in T6S and generated a truncated ClpV variant lacking the N-domain (ΔN-ClpV). ΔN-ClpV was produced to ClpV wild-type level; however, it did not restore Hcp and VgrG2 secretion in V.c. ΔclpV mutant cells (Figure 2A). Purified ΔN-ClpV did neither exhibit defects in oligomerization nor in ATPase activity, excluding defects in its structural integrity as the reason for the observed secretion defect and demonstrating a direct, essential function of the ClpV N-domain in T6S (Supplementary Figure 1).
Figure 2.
The ClpV N-domain is essential for ClpV activity and mediates binding to the VipA/VipB complex. (A) Upper part: schematic domain organization of V. cholerae ClpV. N: N-terminal domain; M: middle domain. Domain boundaries are indicated (amino-acid (aa) numbering). Lower part: complementation analysis of the V.c. ΔclpV secretion defect by plasmid-encoded clpV and its deletion variant lacking the N-terminal domain (ΔN-clpV) using Hcp- and VgrG2-specific antibodies. ClpV levels were monitored by immunoblot analysis using ClpV-specific antibodies. Sup: culture supernatant; total: total cell extract. (B) Purified GST–N-ClpV was coupled to glutathione beads and incubated with soluble cell extracts of V. cholerae V52 ΔclpV. Cell extract incubations with either GST or GST–N-ClpB served as a control. Proteins that were specifically co-purified with GST–N-ClpV (red circles) were identified by mass spectrometry as VCA0107 and VCA0108 and were termed VipA (ClpV-interacting protein A) and VipB, respectively. The positioning of GST, GST–N-ClpV and GST–N-ClpB is indicated. A protein standard is given. (C) Schematic representation of the T6SS encoding gene clusters of V. cholerae, enteroaggregative Escherichia coli (EAEC 42), Edwardsiella tarda and Pseudomonas aeruginosa. VipA, VipB and ClpV encoding genes are coloured in red, yellow and blue, respectively. (D) Purified GST–N-ClpV was coupled to glutathione beads and incubated with purified VipA or VipA/VipB complex. Incubation of VipA or VipA/VipB with empty glutathione beads (−GST–N-ClpV) served as a control. The specific co-sedimentation of VipA/VipB with GST–N-ClpV demonstrates complex formation.
The essential function of the ClpV N-domain in T6S suggests that this extra domain is required for specific protein–protein interactions. To identify proteins that interact with the ClpV N-domain, we fused the extra domain to GST, yielding GST–N-ClpV. The fusion protein was purified and incubated with a soluble cell extract derived from V.c. ΔclpV mutant cells. Interacting proteins were isolated by co-purification with GST–N-ClpV using affinity chromatography. As controls, the same cell extract was either incubated with GST only or a GST–N-ClpB fusion variant harbouring the N-terminal domain of V. cholerae ClpB, a structurally closely related AAA+ protein. Two protein bands were specifically isolated upon GST–N-ClpV pull down and identified by mass spectrometry as VCA0107 and VCA0108 (Figure 2B). VCA0107 and VCA0108 are encoded within the vas gene cluster and represent conserved members of all T6SS. Both genes are always located adjacent to each other and are arranged in the same direction, suggesting a functional interaction (Figure 2C). VCA0107 and VCA0108 represent the first identified ClpV-interacting proteins and were therefore termed VipA and VipB (ClpV-interacting protein), respectively. Immunoblot analysis revealed that VipA and VipB are not present in V. cholerae wild-type supernatants (data not shown), indicating that they do not represent secretory substrates of T6SS and suggesting that they might function as adaptor proteins of ClpV, targeting secretory proteins to the AAA+ chaperone.
To verify the interaction between the ClpV N-domain and VipA/VipB in vitro, we produced VipA and VipB as N-terminal SUMO fusions in E. coli allowing for the purification of authentic proteins after removal of the SUMO moiety by Ulp1. Although soluble VipA could be purified following this strategy, a SUMO–VipB fusion formed inclusion bodies upon overproduction. VipB aggregation was also observed when it was overproduced as an authentic protein (data not shown). To allow for purification of soluble VipB and to test for complex formation between VipA and VipB, N-terminally His6-tagged VipA and VipB were co-produced in E. coli. Co-purification of soluble VipB with His6-VipA after Ni-NTA affinity purification confirmed the interaction of both proteins and suggests a chaperone-like activity of VipA towards VipB (data not shown). Notably, the yield of purified VipA/VipB complex was rather minor, despite the presence of high levels of soluble VipA/VipB, implying limited accessibility of the His6 tag. Comparable results were obtained when the His6 tag was attached to the C terminus of VipB (data not shown). The subsequent use of VipA and His6–VipA/VipB in GST pull-down experiments revealed that VipA interacts only with the ClpV N-domain when complexed to VipB but not alone, suggesting that the primary contact between the VipA/VipB complex and the N-domain might be mediated by VipB (Figure 2D). Furthermore, these results demonstrate that no additional factors are required for the binding of VipA/VipB to the ClpV N-domain, and accordingly VipA/VipB were also isolated in complex with GST–N-ClpV when soluble cell extracts from V.c. ΔclpV Δhcp1 Δhcp2 mutant cells, lacking the main secretory protein of T6SS, were used in the pull-down experiments (data not shown).
VipA and VipB are crucial for type VI protein secretion
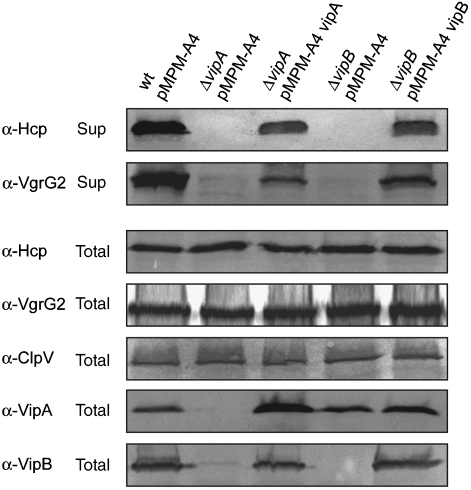
To analyse the function of VipA and VipB in T6S, we constructed in-frame deletion mutants of vipA and vipB. Although not affecting total Hcp or VgrG2 levels, V.c. ΔvipA and V.c. ΔvipB mutants did no longer secrete Hcp and VgrG2, demonstrating their essential function for T6SS (Figure 3). Both mutant strains could be complemented by plasmid-encoded vipA and vipB, respectively, demonstrating that the secretion defect is directly caused by the absence of either VipA or VipB but not by polar effects on downstream-located gene cluster members. Notably, the levels of VipB were strongly reduced in V.c. ΔvipA mutant cells. Normal VipB levels were restored when plasmid-encoded vipA was expressed in trans, indicating that VipB is unstable and prone to degradation in the absence of its interaction partner VipA (Figure 3).
Figure 3.
VipA and VipB are crucial for T6SS. Analysis of Hcp and VgrG2 secretion in V. cholerae V52 wild-type (wt) and V. cholerae V52 ΔvipA or ΔvipB mutant cells harbouring either a vector control (pMPM-A4) or plasmid-encoded vipA or vipB. Hcp and VgrG2 secretion was monitored by immunoblot analysis using specific antibodies. VipA and VipB levels were determined using VipA- and VipB-specific antibodies. Sup: culture supernatant; total: total cell extract.
VipA becomes substrate of a hybrid AAA+ protein harbouring the ClpV N-domain
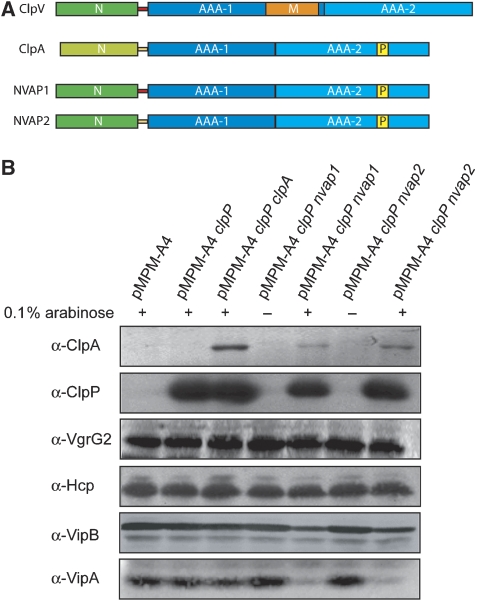
On the basis of the presented findings, we speculated that the VipA/VipB complex might function as an adaptor protein by delivering bound cargo to ClpV after docking to the ClpV N-domain. Transferred substrates could be threaded through the central channel of oligomeric ClpV in a subsequent step. Such a threading activity has been demonstrated for various AAA+ proteins involved in protein quality control and could allow for ClpV-mediated protein export through T6SS. To identify translocated ClpV substrates, we first attempted to convert ClpV into an AAA+ chaperone that cooperates with the bacterial peptidase ClpP. Such a ClpV variant would allow for the identification of threaded substrates by monitoring their degradation in the additional presence of ClpP. The approach is based on the finding that the incorporation of the ‘ClpP interaction loop', a conserved structural motif of ClpP-cooperating AAA+ proteins, into ClpB converts ClpB from a refolding into a degrading disaggregase upon addition of ClpP (Weibezahn et al, 2004). A similar strategy, however, failed in case of ClpV and resulted in an inactive ClpV variant exhibiting severe oligomerization defects and no ATPase activity (data not shown). We therefore followed an alternative approach that aimed to transfer the substrate specificity of ClpV to the ClpP-cooperating AAA+ protein ClpA by exchanging the homologous N-domains. This strategy is based on the finding that the substrate specificity of AAA+ proteins can be exchanged by replacing extra domains involved in adaptor protein and/or substrate interaction (Kirstein et al, 2006). The generated fusion proteins, referred to as NVAP1 and NVAP2, consisted of the N-domain of V. cholerae ClpV and the AAA domains of E. coli ClpA (Figure 4A). The protein fusions differed slightly with respect to the linker sequences that connect the ClpV N-domain and the first AAA domain of E. coli ClpA (Supplementary Figure 2). Both fusion proteins (NVAP1 and NVAP2) were expressed together with E. coli ClpP in V.c. ΔclpV mutant cells, and changes in the total levels of specific proteins were monitored by immunoblot analysis. As a control, E. coli ClpP was expressed either alone or in combination with full-length E. coli ClpA. Although changes in the total amount of the secretory proteins, Hcp and VgrG2, were not observed upon arabinose-induced co-expression of NVAP1/2 and ClpP, VipB levels were slightly reduced and VipA levels were strongly reduced (Figure 4B). Reduced VipA levels could be attributed to NVAP/ClpP-mediated degradation as neither the expression of ClpP alone nor the co-expression of ClpA/ClpP affected VipA or VipB levels (Figure 4B). The slight reduction of VipB levels might be caused by the absence of VipA, which is a prerequisite for VipB stability. These findings confirm that the specificity of ClpV towards VipA/VipB can be transferred to a different AAA+ protein by exchanging N-domains and suggest that VipA/VipB are located in close proximity to the substrate-processing pore site of the interacting AAA+ protein. The observed degradation of VipA by the NVAP chimaeras also raises the possibility that the VipA/VipB complex, but not the putative secretory proteins Hcp and VgrG, represents the primary substrates of ClpV.
Figure 4.
Transplanting the ClpV N-domain to the ClpA/ClpP proteolytic machinery results in VipA degradation. (A) Schematic domain organization of V. cholerae ClpV, E. coli ClpA and the hybrid NVAP1/2 (ClpV/ClpA) fusion proteins. The flexible linker sequences, connecting the N-terminal domain and the AAA-1 domain of ClpV or ClpA are coloured in red and yellow, respectively. The P-loop motif (P), which is crucial for the interaction of ClpA with the peptidase ClpP, is indicated. N: N-terminal domain; M: middle domain. (B) V. cholerae V52 ΔclpV mutant cells harbouring the indicated plasmids were grown in the absence or presence of 0.1% (w/v) arabinose as indicated. Total levels of ClpA, ClpP, VgrG, Hcp, VipA and VipB were determined by immunoblot analysis using specific antibodies. A full-colour version of this figure is available at The EMBO Journal Online.
Cogwheel-like VipA/VipB tubules are remodelled by ClpV
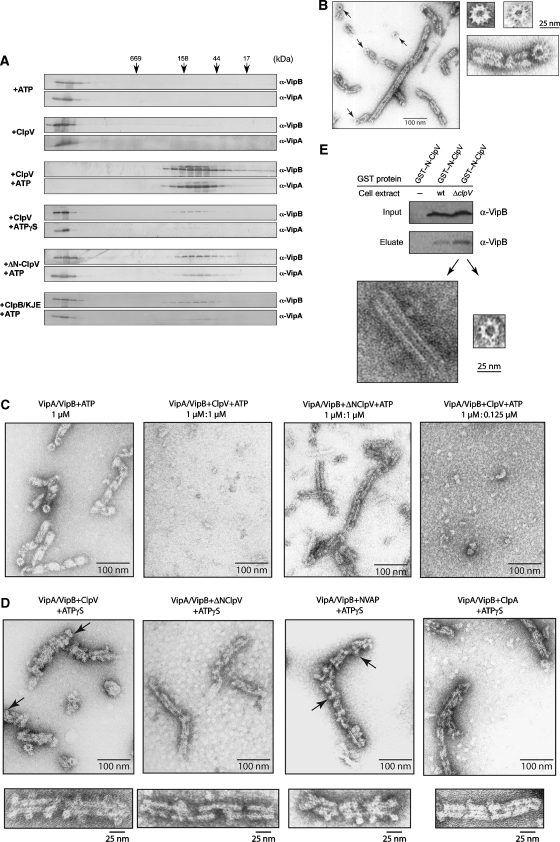
To analyse whether VipA/VipB functions as a substrate for ClpV, we tested in vitro whether ClpV exhibits a remodelling activity towards the interacting proteins. Characterization of the VipA/VipB complex by size-exclusion chromatography revealed that VipA/VipB eluted in the void volume of the Superose 6 column, indicating a complex size larger than 2000 kDa (Figure 5A). In contrast, VipA alone eluted as a much smaller species (approximately 80 kDa), suggesting that the formation of larger complexes is driven by its association with VipB (data not shown). Strikingly, upon incubation with ClpV and an ATP regenerating system, the large VipA/VipB complex became dissociated (Figure 5A). VipA and VipB remained associated, however, in a much smaller complex of approximately 80 kDa. The remodelling activity of ClpV was dependent on ATP hydrolysis, as no or inefficient VipA/VipB complex dissociation was observed in the absence of nucleotide or in the presence of non-hydrolysable ATPγS, respectively (Figure 5A). In the case of ClpV+ATPγS, only a minor fraction of VipA/VipB was eluting at the position of the smaller complexes, whereas the majority still eluted in the void volume. The occurrence of small-sized VipA/VipB complexes under such conditions might represent a rather unspecific chaperone activity towards a subfraction of VipA/VipB, as it is observed in all control experiments using various chaperones (see below). ATP-dependent remodelling of the VipA/VipB complex mirrors the necessity of T6S for active, ATP-hydrolysing ClpV. Along the same line, ΔN-ClpV, which does not support type VI protein secretion and does not interact with VipA/VipB, did not exhibit an efficient remodelling activity towards VipA/VipB (Figure 5A). To exclude the scenario that the observed ClpV-mediated disintegration of the VipA/VipB complex simply reflects a general disaggregation activity towards misfolded, aggregated VipA/VipB, we monitored the activity of ClpB and the DnaK chaperone machinery (KJE), which together form a bi-chaperone system to solubilize aggregated proteins, towards VipA/VipB. ClpB/KJE did not dissociate the VipA/VipB complex, underlining the highly specific activity of ClpV (Figure 5A). Along the same line, treatment of VipA/VipB with structurally related AAA+ proteins (ClpB, Hsp104) or the GroEL chaperone did not result in complex disintegration (data not shown).
Figure 5.
ClpV converts cogwheel-like VipA/VipB tubules to small complexes. (A) VipA/VipB complexes were incubated in the presence of the indicated components for 30 min at 30°C. Complex integrity was monitored by size-exclusion chromatography and eluted fractions were analysed by immunoblotting using VipA- and VipB-specific antibodies. Elution positions of protein standards are indicated by arrows. (B) Morphology of VipA/VipB complexes monitored by electron microscopy. Arrows indicate cogwheel-like structures. Respective scale bars are given. (C) VipA/VipB complexes were incubated without or with ClpV or ΔN-ClpV (+ATP) for 30 min at 30°C and analysed by electron microscopy. Respective scale bars are given. (D) VipA/VipB complexes were incubated with ClpV, ΔN-ClpV, NVAP1 and ClpA (each+ATPγS) and analysed by electron microscopy. Arrows indicate dot-like structures that are associated with VipA/VipB tubules and exhibiting a central hole. Respective scale bars are given. (E) Purified GST–N-ClpV was coupled to glutathione beads and incubated with soluble cell extracts of V. cholerae V52 wild type and ΔclpV. Bound proteins were eluted by the addition of glutathione and analysed by immunoblot analysis using VipB-specific antibodies and by electron microscopy. Equal amounts of VipB present in the used cell extracts were confirmed by western blot (input control).
Next, we analysed the structure of the large VipA/VipB complexes by electron microscopy (Figure 5B). Interestingly, VipA/VipB formed regular, tubular structures with a diverse length of 25–500 nm, implying possible breakage of the tubules upon cell lysis and purification. Accordingly, longer tubules appear to be built of individual shorter segments. In several instances, top views of the tubular complexes were detected, revealing a cogwheel-like structure harbouring 12 prominent cogs (Figure 5B). Such cogwheels can also be observed at the end of a VipA/VipB tubule and explain the longitudinal stripes that are visible on side views of the tubules (Figure 5B). The overall diameter of the cogwheel structure measured approximately 300 Å with a central pore measuring 100 Å. The formation of such tubular complexes might also explain the reduced accessibility of affinity tags attached to either VipA or VipB that was observed upon protein purification.
Notably, VipA/VipB tubules were completely dissolved upon incubation with ClpV and ATP, whereas ΔN-ClpV did not exhibit such activity (Figure 5C). To exclude the possibility that the severing activity of ClpV can only be observed in the presence of high ClpV levels, which might not reflect the in vivo situation, we determined the cellular levels of VipA, VipB and ClpV by quantitative immunoblotting. VipA and VipB are approximately eight-fold more abundant than ClpV as monomeric species (VipA, VipB: 26 000 molecules per cell each, ClpV: 3400 molecules per cell; Supplementary Figure 3). Next, we used the determined in vivo ratio in the in vitro disassembly reaction. Sub-stoichiometric ClpV levels still mediated the disintegration of VipA/VipB tubules, indicating that ClpV acts catalytically and is functional under conditions that reflect the cellular chaperone versus substrate ratio (Figure 5C).
When VipA/VipB complexes were incubated with ClpV and the non-hydrolysable ATP analog ATPγS, a different picture was observed. In detail, the tubular structures remained, but now exhibited multiple dot-like structures at their surfaces. The round-shaped dots have a diameter of approximately 130 Å and in some cases exhibit a central pore, suggesting that they represent ClpV rings that decorate the VipA/VipB tubules (Figure 5D). In agreement with such assumption, dot-like structures that are attached to VipA/VipB tubules were not observed when ΔN-ClpV+ATPγS was used (Figure 5D). To provide direct evidence that the appearance of dot-like structures at the VipA/VipB tubules is caused by N-domain-mediated binding of the ClpV ring, we recorded EM pictures of VipA/VipB tubules incubated with either ClpA/ATPγS or NVAP1/ATPγS. VipA/VipB-associated dots were visible only in the presence of NVAP1/ATPγS, which harbours the substrate specificity determining extra domain of ClpV (Figure 5D).
To provide evidence that VipA/VipB tubules also exist in V. cholerae cells and do not form artificially upon production in E. coli, we purified VipA/VipB from V. cholerae wild-type and ΔclpV mutant cells by affinity chromatography using GST–N-ClpV coupled to glutathione sepharose (see also Figure 2B). Bound VipA/VipB complexes were released from the resin by the addition of glutathione and analysed by negative staining EM. Tubular, cogwheel-like structures were detectable in the eluate from GST–N-ClpV columns but were absent in control experiments using GST–N-ClpB affinity columns, demonstrating that VipA/VipB tubules exist in V. cholerae cells (Figure 5E, data not shown). Notably, the amount of bound VipA/VipB was higher when isolated from V. cholerae ΔclpV cells and, accordingly, more tubular structures were visible in EM (Figure 5E, data not shown). This finding suggests that the ClpV severing activity is also operative in vivo, leading to reduced numbers of VipA/VipB tubules in V. cholerae wild-type cells.
A conserved, pore-located ClpV residue is essential for VipA/VipB complex disintegration
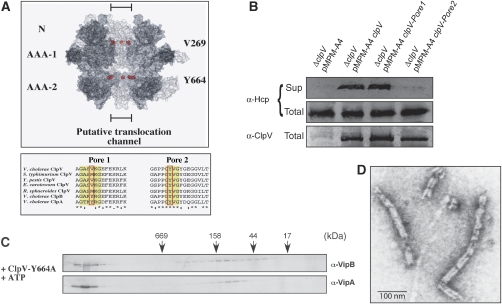
What is the basic mechanism of ClpV-mediated VipA/VipB complex dissociation? Many AAA+ family members share a common threading activity for substrate remodelling that is crucial for their functions in protein degradation, protein disaggregation and the severing of regular protein structures, including prion fibres and microtubules (Siddiqui et al, 2004; Weibezahn et al, 2004; Hinnerwisch et al, 2005; Roll-Mecak and Vale, 2008; Tessarz et al, 2008). Conserved aromatic residues that are positioned at the central pores of AAA+ proteins have been demonstrated to be crucial for substrate threading in various AAA+ members by directly contacting translocating substrates (Yamada-Inagawa et al, 2003; Schlieker et al, 2004; Siddiqui et al, 2004; Weibezahn et al, 2004). ClpV also harbours a conserved aromatic residue (Y664) at the central pore of the second AAA domain (pore2), whereas a methionine or valine residue is positioned at the corresponding pore site of AAA-1 (pore1) (Figure 6A). A hexameric model of ClpV confirmed the localization of the selected residues at the inner channel of the ClpV ring (Figure 6A). To test whether a ClpV threading activity is crucial for T6SS, we mutated the ClpV pore sites yielding ClpV-V269A (pore1) and ClpV-Y664A (pore2) and tested for their ability to restore Hcp and VgrG2 secretion in V.c. ΔclpV mutant cells. Although Hcp and VgrG2 were detectable in the culture supernatant upon expression of ClpV-V269A, ClpV-Y664A did not allow for type VI protein export (Figure 6B). Purified ClpV-Y664A did not reveal defects in hexamerization and exhibited significant ATPase activity (1.07/s compared with 1.7/s in the case of ClpV wild type) (Supplementary Figure 4); however, the ClpV pore mutant did not exhibit a remodelling activity towards the VipA/VipB complex (Figure 6C and D). Taken together, these findings indicate that the dissociation of VipA/VipB tubules relies on ClpV-mediated substrate threading and represents an integral part of T6SS biogenesis.
Figure 6.
A conserved pore-located residue of ClpV is crucial for T6S and remodelling of the VipA/VipB complex. (A) Upper part: surface representation of a ClpV hexamer model. Two monomers were removed from the hexamer to show the inside of the putative translocation channel. The positioning of N-terminal domains (N) and ATPase domains (AAA-1 and AAA-2) are indicated. Pore-located Val269 and Tyr664 are shown as CPK models and are coloured in red. Lower part: multiple sequence alignment of polypeptide segments located at the central pore entrance of the first or second AAA domain (pore 1 and pore 2, respectively) of various ClpV proteins. The protein sequence of the pore regions of V. cholerae ClpV was aligned with numerous other ClpV proteins from Salmonella typhimurium, Yersinia pestis, Erwinia carotowora and Rhodobacter sphaeroides. V. cholerae ClpA and ClpB are included as a reference. Conserved, pore-located residues that have been implicated in ClpA- or ClpB-mediated substrate threading are highlighted (red rectangle). (B) Complementation analysis of the V.c. ΔclpV secretion defect by plasmid-encoded clpV and pore mutant derivatives using Hcp-specific antibodies. ClpV levels were monitored by immunoblot analysis using ClpV-specific antibodies. Sup: culture supernatant, total: total cell extract. The ClpV-Y664A pore mutant does not efficiently dissociate VipA/VipB complexes as analysed by size-exclusion chromatography (C) and electron microscopy (D). A scale bar is indicated.
Discussion
Most bacterial secretion systems characterized so far require an ATPase component to drive the transport of effector molecules. Here, we report on the first detailed molecular dissection of the AAA+ protein ClpV, an essential energizing component of the recently identified T6SS. Using V. cholerae V52 as a model organism, we demonstrate that ATP hydrolysis by ClpV is crucial for the export of the type VI proteins Hcp and VgrG2. Our results are consistent with recent studies showing a central role of ClpV in T6SS of P. aeruginosa, E. tarda and EAEC (Dudley et al, 2006; Mougous et al, 2006; Zheng and Leung, 2007). Besides ClpV, T6SS comprises a second putative ATPase component, IcmF. As IcmF mutants that are expected to be deficient in their ATPase cycles still supported Hcp and VgrG export in E. tarda (Zheng and Leung, 2007), ClpV seems to represent the primary energizing component of T6SS.
ATPase components of T2SS, T3SS and T4SS also form hexameric assemblies and exhibit in parts structural similarity with AAA+ proteins, implying conserved mechanistic features and functions (Yeo and Waksman, 2004; Galan and Wolf-Watz, 2006). InvC, the central ATPase component of S. enterica T3SS, represents the best-characterized protein. InvC dissociates secretory proteins from escorting chaperones by unfolding and potentially pumps the unfolded substrate into the T3SS needle, which is too small to allow for the passage of folded proteins (Akeda and Galan, 2005). ClpV belongs to the Hsp100/Clp subgroup of AAA+ proteins and all family members characterized to date exhibit a threading/unfolding activity. Therefore, a similar mode of ClpV action has been suggested by unfolding the secretory proteins of T6SS, enabling their transport across the inner membrane through a putative type VI translocon (Kulasekara and Miller, 2007; Cascales, 2008; Filloux et al, 2008). Such a model implies physical interactions between ClpV and the secretory proteins; however, various pull-down experiments using tagged variants of the secretory proteins failed to provide evidence for an interaction of either Hcp or VgrG2 with ClpV (data not shown). Furthermore, using purified components, we did not observe a ClpV remodelling activity towards Hcp/VgrG2 (data not shown), suggesting that Hcp and VgrG2 might not function as ClpV substrates. Accordingly, several findings suggest that Hcp and VgrG do not represent classical secretory proteins but rather are integral, structural parts of T6SS. Purified Hcp forms hexameric rings with a large central pore of 40 Å, suggesting that Hcp may form a transmembrane-spanning channel through which other secretory substrates might be secreted (Mougous et al, 2006). In support of this hypothesis, EvpP, a species-specific type VI secretory protein of E. tarda, interacts with Hcp and requires Hcp for export (Zheng and Leung, 2007). Furthermore, the presence of Hcp and VgrG in culture supernatants mutually depends on each other, a finding that is not expected from classical secretory proteins (Pukatzki et al, 2006, 2007; Zheng and Leung, 2007). VgrG proteins have been suggested to represent surface-localized structures that mediate the penetration of host membranes allowing for the exposure of effector domains within the cytosol of target cells (Pukatzki et al, 2007). In agreement with such activity, direct cell-to-cell contact is required for the T6SS-mediated toxicity of V. cholerae V52 towards D. discoideum (Pukatzki et al, 2006). In conclusion, Hcp and VgrG might not represent classical effector proteins, but may rather be appended at the bacterial surface and potentially be released into the culture supernatant by shearing.
The crucial role of ClpV in T6S became apparent when VipA (VCA0107) and VipB (VCA0108) were identified as ClpV-binding partners. VipA and VipB represent conserved members of T6SS encoding gene clusters and their essential function in T6S was verified by nonpolar V. cholerae ΔvipA and ΔvipB deletion mutants. Complex formation between VipA and VipB was demonstrated by co-purification of VipB with His-tagged VipA and is mirrored by their stringent physical proximity in T6SS encoding gene clusters. An interaction between VipA (IglA) and VipB (IglB) has also been recently reported for their orthologues in Francisella novicida (de Bruin et al, 2007).
The association of VipA/VipB with ClpV is mediated by the N-terminal extra domain of ClpV. N-domains of AAA+ proteins are connected through flexible linkers with the ATPase rings and have crucial functions in substrate selection. Several findings demonstrate that the VipA/VipB complex represents the primary substrate of ClpV but does not function as an adaptor protein. First, ΔN-ClpV is deficient in restoring Hcp and VgrG secretion in V.c. ΔclpV mutant cells, demonstrating an essential function of the ClpV N-domain in T6S. Second, transferring the ClpV N-domain to ClpP-cooperating ClpA (NVAP1/2) resulted in VipA degradation, whereas Hcp and VgrG2 levels remained unaffected. Third, in vitro ClpV specifically remodelled the VipA/VipB complex in a process requiring ATP hydrolysis, thereby mirroring its essential function as the primary energy source of T6SS. Fourth, VipA/VipB complex dissociation was no longer observed in the case of the ClpV-Y664A pore mutant, indicating that ClpV-mediated substrate threading is an integral step during complex disintegration.
Interestingly, purified VipA/VipB complexes formed tubular, cogwheel-like structures. The formation of such large, regular complexes argues against the possibility that VipA/VipB misfold upon their heterologous production in E. coli leading to unspecific aggregation. In fact, VipA/VipB complexes purified from V. cholerae exhibited the same structural organization (Figure 5D; Supplementary Figure 5). The formation of tubules is potentially driven by VipB, as VipA alone does not form tubules. Furthermore, VipA alone does not interact with the ClpV N-domain, indicating that the formation of tubular structures upon VipA/VipB association is a prerequisite for ClpV interaction. ClpV converts the large tubular VipA/VipB structures into smaller complexes, an activity that is reminiscent of the activity of the AAA+ proteins Spastin and Vps4 in the severing of microtubules and ESCRTIII complexes (Ghazi-Tabatabai et al, 2008; Lata et al, 2008; Roll-Mecak and Vale, 2008). In the context of protein export, such an activity of an AAA+ protein is entirely unexpected and adds a remarkable new layer of complexity to bacterial protein secretion.
What might be the function of the VipA/VipB tubules and the severing activity of ClpV? Both VipA and VipB are cytosolic proteins and could not be detected in cell culture supernatants, largely excluding a ClpV-driven export of VipA or VipB (data not shown). Furthermore, we did not obtain evidence for an interaction between VipA/VipB and Hcp or VgrG2 in pull-down experiments, largely excluding a function of VipA/VipB as adaptor proteins directing the secretory proteins for export (data not shown). Instead, we suggest that the ClpV-mediated remodelling of VipA/VipB tubules directly represents an essential step in T6S. The severing activity of ClpV might allow to control the dynamics of VipA/VipB tubules by regulating the number and size of the complexes. Protein export through T6SS might also require the complete disassembly of VipA/VipB tubules at a certain stage of T6S biogenesis or transport, resembling the function of Vps4 in the formation of multivesicular bodies (Ghazi-Tabatabai et al, 2008; Lata et al, 2008). Consistent with both hypotheses, VipA/VipB tubules appear to be more abundant in V. cholerae ΔclpV mutant cells. How exactly the remodelling of VipA/VipB tubules contribute to T6S needs to be addressed in future studies. The presented findings set the stage to unravel a novel twist in the versatile secretion systems of bacteria.
Materials and methods
Strains and plasmids
Supplementary Table 1 lists all strains and plasmids used in this study. All bacterial strains were grown in LB media at 37°C if not stated otherwise. Antibiotics were used at the following concentrations: 2 μg/ml (V. cholerae) and 20 μg/ml (E. coli) chloramphenicol, 100 μg/ml (V. cholerae) and 50 μg/ml (E. coli) streptomycin, 200 μg/ml (V. cholerae) and 100 μg/ml (E. coli) ampicillin. PCR reactions and cloning procedures followed standard protocols.
V. cholerae in-frame gene deletions were performed as described before by using the pDS132 suicide plasmid for allelic replacement of the gene of interest (Philippe et al, 2004). Chromosomal deletion mutants were identified by colony PCR and additionally confirmed by immunoblot analysis using specific antibodies. V. cholerae deletion mutants were complemented by pMPM-A4 harbouring the gene of interest, allowing for arabinose-controlled gene expression. Total protein levels were adjusted to wild-type levels by titration of arabinose levels and comparing the signal intensities upon immunoblot analysis.
For transformation of plasmids into V. cholerae, electrocompetent cells were prepared as described (Hamashima et al, 1995). Plasmid DNA (1 μl) was added to the electrocompetent cells and incubated on ice for 5 min. Cells were rapidly transferred to pre-cooled electrocuvettes (2 mm gap) and pulsed (1000 Ω, 1.8 kV and 25 μF). Immediately, 1 ml LB medium was added and cells were transferred into an Eppendorf tube, incubated for 45 min at 37°C and spread on LB plates with appropriate antibiotics.
Proteins
C-terminal His6-tagged wild-type and mutant ClpV were purified after overproduction from E. coli ΔclpB mutant cells following the instructions of the manufacturer (Macherey-Nagel). N-terminal His6-tagged ClpV N-domain, His6–VipA and His6–SUMO-fused VipA or VipB were purified accordingly after overproduction from E. coli XL1 Blue cells. SUMO fusion proteins were cleaved by the addition of purified His6–Ulp1, allowing for the isolation of VipA and VipB. The N-terminal GST-tagged N-domain of ClpV was overproduced in E. coli XL1 Blue cells and purified following the instructions of the manufacturer (GE Healthcare). Protein concentrations were determined with the Bio-Rad Bradford assay using bovine serum albumin as a standard.
Biochemical assays
The analysis of the V. cholerae secretome was performed as described (Pukatzki et al, 2006). For 2D gel electrophoresis, TCA-precipitated secretory proteins were dissolved in 2D buffer (6 M urea, 2 M thiourea, 2% (w/v) CHAPS, 0.3% (w/v) DTT, 0.5% (v/v) ampholytes (pH 3–10)) and applied to discontinuous 2D gels (9–16%). Finally, the gels were developed by silver staining. Protein spots that were present in V. cholerae wild type, but missing in V. cholerae deletion mutants were excised and analysed by mass spectrometry.
Size-exclusion chromatography of ClpV was performed at 4°C in MDH/2 buffer (25 mM Tris, pH 7.5, 75 mM KCl, 10 mM MgCl2 and 2 mM DTT) using a Superose 6 HR 10/30 column (Amersham Biosciences). Nucleotide-dependent oligomerization was followed in the presence or absence of 2 mM ATP in the running buffer. ClpV (6 μM) or derivatives were incubated in MDH/2 buffer in the absence or presence of nucleotides (2 mM ATP) for 5 min at room temperature prior to injection.
ATPase hydrolysis rates under steady-state conditions were determined as described earlier (Mogk et al, 2003). Briefly, 0.5 μM ClpV or derivatives were incubated in MDH/2 buffer for 5 min at 30°C before the reaction was started by the addition of 2 mM ATP (including [α-32P]ATP (0.1 μCi; Amersham Biosciences)). ATP hydrolysis was quantified by using the program MACBAS version 2.5 (Fuji), and rates of ATP hydrolysis were determined by using the program GRAFIT version 3.0 (Erithacus Software).
VipA/VipB complex disintegration was monitored by size-exclusion chromatography using a Superose 6 column (Amersham Biosciences) performed at room temperature in MDH buffer (50 mM Tris, pH 7.5, 150 mM KCl, 20 mM MgCl2 and 2 mM DTT). Prior to injection, 2 μM VipA/VipB was incubated for 30 min at 30°C in the presence of an ATP-regenerating system (2 mM ATP, 3 mM phosphoenolpyruvate and 20 ng/μl pyruvate kinase) and the following components: 3 μM ClpV and derivatives; 3 μM ClpB plus 1 μM DnaK, 0.2 μM DnaJ, 0.1 μM GrpE; 3 μM Hsp104/ClpB; 7 μM GroEL-D87K; 3 μM BSA. Eluted fractions were collected and analysed by immunoblotting.
For ex vivo GST pull-down experiments, 200 μl of glutathione sepharose beads was prepared in binding buffer (50 mM Tris pH 7.5, 100 mM NaCl, 5 mM Mg(Ac)2, 1 mM EDTA, 5% glycerol and 2 mM DTT). Next, 200 μg of GST-tagged protein was added to the beads and incubated for 2 h in an overhead shaker at 4°C. The beads were washed and transferred to a 15 ml Falcon tube. The prepared beads were incubated with soluble V. cholerae cell extracts (300 mg total protein content) for 2 h in an overhead shaker at room temperature. Centrifugation at 400 g for 8 min separated the beads from the supernatant, which was discarded. The beads were washed with washing buffer (binding buffer supplemented with 0.05% Triton X-100) and finally boiled in 60 μl of SDS sample buffer. For further analysis, 30 μl of the sample was separated on SDS–PAGE and analysed by silver staining.
In the case of in vitro pull-down experiments, 40 μl sepharose beads were prepared in binding buffer (50 mM HEPES pH 7.5, 100 mM NaCl, 10 mM Mg(Ac)2, 0.1 mM EDTA and 5 mM β-mercaptoethanol). GST–N-ClpV (10 μg) was added to the beads and incubated for 1 h in an overhead shaker at 4°C. Next, the putative interaction partners were added in 2.5-fold molar excess. The sample was incubated for 1 h in an overhead shaker at 4°C. The beads were spinned down for 2 min with 400 g at 4°C and the supernatant was discarded. The beads were washed and finally boiled with 20 μl SDS sample buffer and analysed by SDS–PAGE.
Generation of a hexameric ClpV model
The monomeric structure of ClpV has been modelled based on the homologous template ClpB (1qvr; Lee et al, 2003) using MODELLER (Sali and Blundell, 1993). The structural model was manually refined and energy minimized using SPDBV (Guex and Peitsch, 1997). In the M-domain, an insertion and a deletion were found at the tips of the helical region and remodelled using SPDBV. Insertions in the pore-facing regions of the AAA-1 domain were modelled using templates ClpA (1r6b; Xia et al, 2004) and ClpB1 (2p65; Wernimont et al, 2007). In AAA-2, missing residues in the template were remodelled based on ClpA (1ksf; Guo et al, 2002).
The hexameric complex of six ClpV monomers was built following a previous method (Diemand and Lupas, 2006). The model was analysed using the iMolTalk server (Diemand and Scheib, 2004) and interface contacts in the complex were detected with a distance threshold of 3.4 Å.
Electron microscopy
For negative staining, VipA/VipB complexes were treated as described above and applied to glow-discharged, carbon-coated 300 mesh copper grids. After washing with water, the specimens were stained with 2% uranyl acetate and finally dried. Micrographs were taken with a Zeiss EM10A electron microscope.
Supplementary Material
Supplementary Figures 1–5
Supplementary Table 1
Acknowledgments
We thank Fidelma Boyd for providing bacterial strains and reagents, Dominique Schneider for sending pDS132 and Joachim Reidl for experimental advice. We are indebted to Erich Sawruk for helping to establish the S2 laboratory. We thank Ulrike Ackermann for photographic work. We are grateful to Peter Tessarz for a critical reading of the paper and to Bernd Bukau for continuous support. This study was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG) to AM (MO970-2) and a Heisenberg Fellowship of the DFG to AM.
References
- Akeda Y, Galan JE (2005) Chaperone release and unfolding of substrates in type III secretion. Nature 437: 911–915 [DOI] [PubMed] [Google Scholar]
- Bladergroen MR, Badelt K, Spaink HP (2003) Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol Plant Microbe Interact 16: 53–64 [DOI] [PubMed] [Google Scholar]
- Cascales E (2008) The type VI secretion toolkit. EMBO Rep 9: 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Chaudhuri K (2003) Identification of a unique IAHP (IcmF associated homologous proteins) cluster in Vibrio cholerae and other proteobacteria through in silico analysis. In Silico Biol 3: 287–300 [PubMed] [Google Scholar]
- de Bruin OM, Ludu JS, Nano FE (2007) The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol 7: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diemand AV, Lupas AN (2006) Modeling AAA+ ring complexes from monomeric structures. J Struct Biol 156: 230–243 [DOI] [PubMed] [Google Scholar]
- Diemand AV, Scheib H (2004) iMolTalk: an interactive, internet-based protein structure analysis server. Nucleic Acids Res 32: W512–W516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP (2006) Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol Microbiol 61: 1267–1282 [DOI] [PubMed] [Google Scholar]
- Enos-Berlage JL, Guvener ZT, Keenan CE, McCarter LL (2005) Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol Microbiol 55: 1160–1182 [DOI] [PubMed] [Google Scholar]
- Erbse A, Schmidt R, Bornemann T, Schneider-Mergener J, Mogk A, Zahn R, Dougan DA, Bukau B (2006) ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature 439: 753–756 [DOI] [PubMed] [Google Scholar]
- Filloux A, Hachani A, Bleves S (2008) The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154: 1570–1583 [DOI] [PubMed] [Google Scholar]
- Galan JE, Wolf-Watz H (2006) Protein delivery into eukaryotic cells by type III secretion machines. Nature 444: 567–573 [DOI] [PubMed] [Google Scholar]
- Ghazi-Tabatabai S, Saksena S, Short JM, Pobbati AV, Veprintsev DB, Crowther RA, Emr SD, Egelman EH, Williams RL (2008) Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure 16: 1345–1356 [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- Guo F, Maurizi MR, Esser L, Xia D (2002) Crystal structure of ClpA, an Hsp100 chaperone and regulator of ClpAP protease. J Biol Chem 277: 46743–46752 [DOI] [PubMed] [Google Scholar]
- Hamashima H, Iwasaki M, Arai T (1995) A simple and rapid method for transformation of Vibrio species by electroporation. Methods Mol Biol 47: 155–160 [DOI] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW (2005) AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol 6: 519–529 [DOI] [PubMed] [Google Scholar]
- Haslberger T, Weibezahn J, Zahn R, Lee S, Tsai FT, Bukau B, Mogk A (2007) M domains couple the ClpB threading motor with the DnaK chaperone activity. Mol Cell 25: 247–260 [DOI] [PubMed] [Google Scholar]
- Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL (2005) Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell 121: 1029–1041 [DOI] [PubMed] [Google Scholar]
- Kedzierska S, Akoev V, Barnett ME, Zolkiewski M (2003) Structure and function of the middle domain of ClpB from Escherichia coli. Biochemistry 42: 14242–14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein J, Schlothauer T, Dougan DA, Lilie H, Tischendorf G, Mogk A, Bukau B, Turgay K (2006) Adaptor protein controlled oligomerization activates the AAA+ protein ClpC. EMBO J 25: 1481–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulasekara HD, Miller SI (2007) Threonine phosphorylation times bacterial secretion. Nat Cell Biol 9: 734–736 [DOI] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W (2008) Helical structures of ESCRT-III are disassembled by VPS4. Science 321: 1354–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Sowa ME, Watanabe Y, Sigler PB, Chiu W, Yoshida M, Tsai FT (2003) The structure of ClpB. A molecular chaperone that rescues proteins from an aggregated state. Cell 115: 229–240 [DOI] [PubMed] [Google Scholar]
- Mogk A, Schlieker C, Strub C, Rist W, Weibezahn J, Bukau B (2003) Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP-hydrolysis and chaperone activity. J Biol Chem 278: 15–24 [DOI] [PubMed] [Google Scholar]
- Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ (2006) A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312: 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller RS, McDougald D, Cusumano D, Sodhi N, Kjelleberg S, Azam F, Bartlett DH (2007) Vibrio cholerae strains possess multiple strategies for abiotic and biotic surface colonization. J Bacteriol 189: 5348–5360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DA, Heffron F (2005) sciS, an icmF homolog in Salmonella enterica serovar typhimurium, limits intracellular replication and decreases virulence. Infect Immun 73: 4338–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe N, Alcaraz JP, Coursange E, Geiselmann J, Schneider D (2004) Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51: 246–255 [DOI] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ (2007) Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci USA 104: 15508–15513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ (2006) Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA 103: 1528–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao PS, Yamada Y, Tan YP, Leung KY (2004) Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol Microbiol 53: 573–586 [DOI] [PubMed] [Google Scholar]
- Remaut H, Waksman G (2004) Structural biology of bacterial pathogenesis. Curr Opin Struct Biol 14: 161–170 [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A, Vale RD (2008) Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature 451: 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234: 779–815 [DOI] [PubMed] [Google Scholar]
- Schell MA, Ulrich RL, Ribot WJ, Brueggemann EE, Hines HB, Chen D, Lipscomb L, Kim HS, Mrazek J, Nierman WC, Deshazer D (2007) Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol Microbiol 64: 1466–1485 [DOI] [PubMed] [Google Scholar]
- Schlieker C, Weibezahn J, Patzelt H, Tessarz P, Strub C, Zeth K, Erbse A, Schneider-Mergener J, Chin JW, Schultz PG, Bukau B, Mogk A (2004) Substrate recognition by the AAA+ chaperone ClpB. Nat Struct Mol Biol 11: 607–615 [DOI] [PubMed] [Google Scholar]
- Schlieker C, Zentgraf H, Dersch P, Mogk A (2005) ClpV, a unique Hsp100/Clp member of pathogenic proteobacteria. Biol Chem 386: 1115–1127 [DOI] [PubMed] [Google Scholar]
- Siddiqui SM, Sauer RT, Baker TA (2004) Role of the processing pore of the ClpX AAA+ ATPase in the recognition and engagement of specific protein substrates. Genes Dev 18: 369–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessarz P, Mogk A, Bukau B (2008) Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol Microbiol 68: 87–97 [DOI] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FT, Mogk A, Bukau B (2004) Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119: 653–665 [DOI] [PubMed] [Google Scholar]
- Wernimont A, Lew J, Kozieradzki I, Lin Y, Hassanali A, Zhao Y, Arrowsmith C, Edwards A, Weigelt J, Sundstrom M, Bochkarev A, Hui R, Artz J (2007) Crystal structure of the first nucleotide binding domain of chaperone ClpB1, putative from Plasmodium vivax, Structural Genomics Consortium
- Wu HY, Chung PC, Shih HW, Wen SR, Lai EM (2008) Secretome analysis uncovers an Hcp-family protein secreted via a type VI secretion system in Agrobacterium tumefaciens. J Bacteriol 190: 2841–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D, Esser L, Singh SK, Guo F, Maurizi MR (2004) Crystallographic investigation of peptide binding sites in the N-domain of the ClpA chaperone. J Struct Biol 146: 166–179 [DOI] [PubMed] [Google Scholar]
- Yamada-Inagawa T, Okuno T, Karata K, Yamanaka K, Ogura T (2003) Conserved pore residues in the AAA protease FtsH are important for proteolysis and its coupling to ATP hydrolysis. J Biol Chem 278: 50182–50187 [DOI] [PubMed] [Google Scholar]
- Yeo HJ, Waksman G (2004) Unveiling molecular scaffolds of the type IV secretion system. J Bacteriol 186: 1919–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Leung KY (2007) Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol 66: 1192–1206 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–5
Supplementary Table 1