Abstract
Human APOBEC3G exhibits anti-human immunodeficiency virus-1 (HIV-1) activity by deaminating cytidines of the minus strand of HIV-1. Here, we report a solution structure of the C-terminal deaminase domain of wild-type APOBEC3G. The interaction with DNA was examined. Many differences in the interaction were found between the wild type and recently studied mutant APOBEC3Gs. The position of the substrate cytidine, together with that of a DNA chain, in the complex, was deduced. Interestingly, the deamination reaction of APOBEC3G was successfully monitored using NMR signals in real time. Real-time monitoring has revealed that the third cytidine of the d(CCCA) segment is deaminated at an early stage and that then the second one is deaminated at a late stage, the first one not being deaminated at all. This indicates that the deamination is carried out in a strict 3′ → 5′ order. Virus infectivity factor (Vif) of HIV-1 counteracts the anti-HIV-1 activity of APOBEC3G. The structure of the N-terminal domain of APOBEC3G, with which Vif interacts, was constructed with homology modelling. The structure implies the mechanism of species-specific sensitivity of APOBEC3G to Vif action.
Keywords: APOBEC3G, cytidine deaminase, HIV, structure, Vif
Introduction
The apolipoprotein B messenger RNA-editing enzyme catalytic polypeptide (APOBEC)/activation-induced cytidine deaminase (AID) proteins are found in vertebrates and share the ability to mutate either DNA or RNA by deaminating cytidine to uridine (Rogozin et al, 2007). The APOPBEC1 deaminates and edits apolipoprotein B pre-mRNA (Navaratnam et al, 1993; Teng et al, 1993). AID deaminates immunoglobulin gene DNA, which is essential for the antigen-driven diversification of already rearranged immunoglobulin genes in the vertebrate adaptive immune system (Muramatsu et al, 1999). Human APOPBEC3G is expressed in spleen, testes, ovary and blood leukocytes, such as T lymphocytes and macrophages. APOBEC3G deaminates and modifies the newly synthesized cDNA minus strand of retroviruses such as the human immunodeficiency virus-1 (HIV-1) (Sheehy et al, 2002; Harris et al, 2003; Lecossier et al, 2003; Mangeat et al, 2003; Zhang et al, 2003; Beale et al, 2004; Greene, 2004; Harris and Liddament, 2004; Suspene et al, 2004; Yu et al, 2004; Esnault et al, 2005; Chelico et al, 2006). The modifications are supposed to induce degradation of the minus strand through action of uracil DNA glycosylase and apurinic–apyrimidinic endonuclease or the generation of G to A mutations upon synthesis of the plus strand, which may eliminate HIV-1 infectivity for virus infectivity factor (Vif)-deficient strains (Sheehy et al, 2002; Harris et al, 2003; Lecossier et al, 2003; Mangeat et al, 2003; Greene, 2004; Harris and Liddament, 2004; Esnault et al, 2005). Vif targets APOBEC3G for ubiquitination and proteasomal degradation, and thus abolishes the antiviral activity of APOBEC3G (Conticello et al, 2003; Marin et al, 2003; Yu et al, 2003; Mehle et al, 2004; Kobayashi et al, 2005).
In spite of the great attention to the APOBEC/AID protein family, the structural knowledge on this family is quite limited. The structure of APOBEC2 was reported last year (Prochnow et al, 2007), but APOBEC2 does not possess enzymatic activity. APOBEC3G is composed of 384 residues and possesses two consensus cytidine deaminase motifs of the zinc-finger type, His-X-Glu-X27-28-Pro-Cys-X2-Cys, in its N- and C-terminal domains (Harris and Liddament, 2004). It is known that the C-terminal deaminase domain is catalytically active, whereas the N-terminal one is not (Navarro et al, 2005). Recently, the solution structure of a mutant C-terminal deaminase domain, residues 198–384, was reported (Chen et al, 2008). To increase the solubility, mutation of five residues of the C-terminal domain was performed. Here, we present the solution structure of the wild-type C-terminal deaminase domain, residues 193–384, which possesses no mutation. Some difference in structure was found between the wild type and mutant deaminase domains of APOBEC3G. The mode of interaction with single-stranded DNA (ssDNA) was characterized by chemical shift perturbation analysis. Although the wild-type and mutant APOBEC3Gs share several features of the interactions, many differences in the interaction were also identified between them. The differences were supposed to be partly due to the introduction of the mutations themselves. The position of a substrate cytidine, and the position and polarity of an ssDNA chain in the complex were deduced from the results of analysis of the interaction. Very recently, the crystal structure of the C-terminal deaminase domain of APOBEC3G has been reported (Holden et al, 2008). The relation between our and their findings is also discussed.
Moreover, we succeeded for the first time in monitoring the deamination reaction in real-time using NMR signals. This method enabled us to address the dynamical aspects of the deamination reaction by APOBEC3G. Deamination in a very strict 3′ → 5′ order was detected for consecutive cytidine residues of ssDNA.
The N-terminal domain of APOBEC3G is required for encapsidation of APOBEC3G into virion (Cen et al, 2004; Harris and Liddament, 2004). It is also known that Vif of HIV-1 interacts with the N-terminal domain of APOBEC3G (Conticello et al, 2003; Harris and Liddament, 2004). The structure of the N-terminal domain has not been reported. The N- and C-terminal domains of APOBEC3G exhibit sequence homology. So, the structure of the N-terminal domain is constructed with homology modelling on the basis of the structure of the wild-type C-terminal domain. The constructed structure suggests the origin of species-specific sensitivity of APOBEC3G to Vif action.
Results
Structure of the wild-type deaminase domain of APOBEC3G
Figure 1A shows SDS–polyacrylamide gel electrophoresis of the purified deaminase domain of wild-type APOBEC3G. The purity was estimated to be over 99%. Figure 1B shows the 1H–15N HSQC spectrum of the deaminase domain of wild-type APOBEC3G. The HSQC peaks are sharp and well dispersed, which guarantees that the sample prepared with our procedure possesses the native structure, not being in an aggregated state, and is suitable for structure determination. Thus, it was found that wild-type APOBEC3G, instead of the mutant, could be used for the structure determination.
Figure 1.
SDS–polyacrylamide gel electrophoresis, HSQC spectrum and structure of the deaminase domain of the wild-type APOBEC3G. (A) SDS–polyacrylamide gel electrophoresis of the purified APOBEC3G. (B) 1H–15N HSQC spectrum. (C) A stereo view of superposition of the main chains of 10 final structures. N and C indicate L193 and N384, respectively. α-helices and β-strands are coloured red and blue, respectively. (D) The main chain of a representative structure with the lowest energy. α-helices and β-strands are coloured red/yellow and blue, respectively.
The structure of wild-type APOBEC3G was calculated on the basis of distance and dihedral angle constraints. The structure statistics are presented in Table I. The average pairwise r.m.s.d. between the 10 final structures as to the backbone atoms of the all secondary elements was 0.55±0.110 Å. Figure 1C and D shows the structures of wild-type APOBEC3G. Wild-type APOBEC3G is composed of five β-strands, β1–β5, and five α-helices, α1–α5, which are arranged in the order of β1-β2/β2′-α1-β3-α2-β4-α3-β5-α4-α5. The second β-strand is interrupted and divided into two short β-strands, β2 and β2′.
Table 1.
NMR and refinement statistics for protein structures
| Protein | |
|---|---|
| NMR distance and dihedral constraints | |
| Distance constraints | |
| Total NOE | 2380 |
| Intra-residue | 1117 |
| Inter-residue | 1263 |
| Sequential (∣i−j∣=1) | 581 |
| Medium range (∣i−j∣<4) | 311 |
| Long range (∣i−j∣>5) | 371 |
| Hydrogen bonds | 112 |
| Total dihedral angle restraints | |
| φ | 131 |
| ψ | 131 |
| Structure statistics | |
| Violations (mean and s.d.) | |
| Distance constraints (Å) | 0.0779±0.0008 |
| Dihedral angle constraints (deg) | 0.7545±0.0703 |
| Maximum dihedral angle violation (deg) | 7.718 |
| Maximum distance constraint violation (Å) | 0.478 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.0044±0.0001 |
| Bond angles (deg) | 0.6595±0.0055 |
| Impropers (deg) | 0.4181±0.0067 |
| Average pairwise r.m.s.d. between 10 structures (Å) | |
| Heavya | 1.38±0.22 |
| Backbonea | 0.71±0.14 |
| Heavyb | 1.26±0.20 |
| Backboneb | 0.55±0.11 |
| Ramachandran plot appearance | |
| Most favoured regions (%) | 71.5 |
| Additional allowed (%) | 25.9 |
| Generously allowed (%) | 2.3 |
| Disallowed regions (%) | 0.2 |
| aThese calculations included residues 221–242 and 258–379. | |
| bThese calculations included residues 221–228, 231–232, 240–242, 258–269, 276–283, 291–300, 305–311, 322–331, 333–338, 340–350 and 364–379. | |
In Figure 2, the structure of the wild-type APOBEC3G determined in solution by NMR (this study) is compared with the structure of the mutant carrying five substituted residues determined in solution by NMR (Chen et al, 2008), and also with the structure of the wild type determined for crystal by X-ray (Holden et al, 2008). The three structures are mostly similar to each other. An extra α-helical element, α0 (P199-F204), is formed for the wild type in solution (Figure 2A), but not for the mutant in solution (Figure 2B). The formation of α0 for the wild type in solution was supported by the secondary structure identification on the basis of chemical shifts of Cα, Cβ, C′, N and Hα with either CSI (Wishart and Sykes, 1994) or TALOS (Cornilescu et al, 1999). α0 (P199-F206) is also present for the wild type in crystal (Figure 2C). The wild type is composed of either L193-N384 (this study) or M197-Q380 (Holden et al, 2008), and the mutant of D198-N384. The lack of α0 in the mutant may be due to the lack of several N-terminal residues. Because of the lack of long-range NOEs involving α0, the relative position of α0 as to the rest of APOBEC3G was poorly determined (Figure 1C). Therefore, it is hard to discuss the difference in the relative position of α0 between the wild-type APOPBEC3Gs in solution and crystal.
Figure 2.
Comparison of the structures of the deaminase domain of APOBEC3G. (A) The wild type in solution by NMR (this study). (B) The mutant carrying five substitutions in solution by NMR (Chen et al, 2008). (C) The wild type in crystal by X-ray (Holden et al, 2008).
The second β-strand is interrupted and divided into two short β-strands for both the wild type and mutant in solution (Figure 2A and B). The second β-strand of the wild type in crystal is not divided but continuous, although a bulge exists in this β-strand (Figure 2C). Typical NOEs expected for a β-strand structure were not detected for the interrupted region. The secondary structure identification on the basis of chemical shifts of Cα, Cβ, C′, N and Hα also suggests the interruption at the central 3–5 residues of the second β-strand, although a β-strand-like structure is implied for the other region of the strand. Thus, it is not likely that the continuous β-strand is stably formed in solution.
The mutation of either F202 or T203 of α0 to Ala causes the decrease of the activity of APOBEC3G (Supplementary data of Chen et al, 2008). Similarly, the mutation of W232, V233, L235, F241 or L242 of β2 to Lys and that of either W232, V233, L235 or L242 of β2 to Ala cause the decrease of the activity of APOBEC3G (Supplementary data of Chen et al, 2008). Therefore, the structural differences detected for α0 and β2 may have some functional relevance.
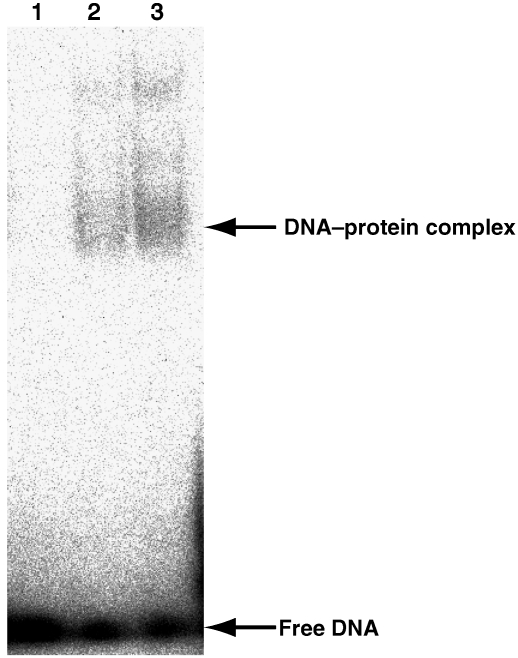
Binding of the wild-type deaminase domain to a ssDNA, as revealed by a gel retardation experiment
It was reported that minus strand deamination by APOBEC3G occurs preferentially at a CCCA sequence (Yu et al, 2004). 10-mer DNA, d(ATTCCCAATT), contains the CCCA sequence in the middle. Binding of the deaminase domain of the wild-type APOBEC3G to a short single-stranded 10-mer DNA was revealed by gel retardation experiments (Figure 3). To our best knowledge, this is the first demonstration by a gel retardation experiment that the deaminase domain can bind to a short ssDNA of only 10 residues, although the binding of full-length APOPBEC3G to another 10-mer DNA was demonstrated on the basis of the change in steady-state fluorescence depolarization of fluorescein-labelled DNA (Chelico et al, 2006). The apparent dissociation constant, Kd, was estimated to be ca. 130–190 μM, on the basis of the relative intensities of bands corresponding to either free DNA or the DNA–protein complex. Kd for the complex between the mutant deaminase domain and a 21-mer ssDNA was reported to be minimally 450 μM, on the basis of the results of a chemical shift perturbation experiment (Chen et al, 2008). Thus, the affinity of the wild type to 10-mer DNA is comparable or slightly higher than that of the mutant to 21-mer DNA. Therefore, 10-mer DNA was used for further analysis of the interaction using the chemical shift perturbation method.
Figure 3.
Gel retardation experiments indicating binding of the wild-type deaminase domain to a short ssDNA comprising 10 residues. 32P-labelled 10-mer DNA (200 nM), d(ATTCCCAATT), was incubated with 0, 40 and 80 μM of the deaminase domain of the wild-type APOBEC3G (lanes 1–3). The mixtures were run on a polyacrylamide gel and then exposed.
The differences in the interaction of the deaminase domain with ssDNA between the wild-type and mutant APOBEC3Gs, as revealed by chemical shift perturbation
Figure 4A and C shows the chemical shift perturbations for the deaminase domain of the wild-type APOBEC3G on binding of ssDNA, d(ATTCCCAATT). For comparison, Figure 4B shows the chemical shift perturbations reported for the mutant deaminase domain on binding of 21-mer ssDNA (Chen et al, 2008). In general, perturbations detected for the wild-type APOBEC3G:10-mer DNA complex were larger than those for the mutant APOBEC3G:21-mer DNA complex: the maximum perturbation for the mutant was 0.03 p.p.m., whereas six residues exhibited perturbation of greater than 0.03 p.p.m. for the wild type. The larger perturbations could be partly due to the higher oligonucleotide ratio to APOBEC3G (an APOBEC3G:DNA ratio is 1:10 for our case, whereas the ratio is 1:4 for Chen et al) and/or the use of the wild-type APOBEC3G. It is expected that the mode of the interaction may be deduced more precisely with the pronounced perturbation data obtained here.
Figure 4.
Mapping of chemical shift perturbations upon binding of DNA, surface electrostatic potential and proposed position of a substrate cytidine. (A) Chemical shift perturbations observed for the wild-type deaminase domain upon binding of 10-mer DNA. The residues exhibiting combined chemical shift perturbations as to HN and N of >0.03 p.p.m. and 0.02–0.03 p.p.m. are coloured red and yellow, respectively. The residues whose 1H–15N correlation peaks either disappeared or became notably weak, the relative intensity in a free state to that in a complex state being greater than 1.2, are coloured blue. (B) Chemical shift perturbations observed for the mutant deaminase domain upon binding of 21-mer DNA. The residues exhibiting combined chemical shift perturbations of >0.03 p.p.m. and 0.02–0.03 p.p.m. are coloured red and yellow, respectively. The structure and perturbation data reported for the mutant deaminase domain (Chen et al, 2008) were used to make this figure. (C) Left, the perturbations for the wild-type deaminase domain mapped on the surface representation; right, positive and negative surface potentials of the wild-type deaminase domain represented in blue and red, respectively. (D) A close-up of the deduced key interactive region of the wild-type APOBEC3G deaminase domain, with the proposed position of a substrate cytidine indicated by a dashed circle, viewed from two different angles. (E) Three possible positions of ssDNA relative to the APOBEC3G deaminase domain. Right, the position proposed by Chen et al, 2008 (dashed vertical line in blue) and that by Holden et al, 2008 (dashed kinked horizontal line in green); left, the third possible position.
It is known that E259 is a catalytic residue for the deamination reaction (Mangeat et al, 2003; Shindo et al, 2003; Navarro et al, 2005). Chemical shift perturbation, including a decrease in the intensity of a correlation peak, was detected for residues surrounding E259. Perturbations were seen for the residues of the loop between α0 and β1 (an α0–β1 loop) (N208 and R215), those of a β2′–α1 loop (C243, A246, E254, R256 and H257), those of α1 (L260, C261, F262, L263, V265 and I266), those of β4 (V305 and S306), those of a β4–α3 loop (D316 and G319), that of β5 (I337), that of α4 (D348) and those of an α4–α5 loop (V351, D352 and D362) (Figure 4A). Similar perturbations were observed for the mutant APOBEC3G for the same segments (Figure 4B). On the other hand, the perturbations observed for the following segments are specific to the wild type, not being observed for the mutant: a β2–β2′ loop (R238), β2′ (G240 and L242), an α1–β3 loop (L271, D272, D274 and Q275), β3 (R278), a β3–α2 loop (T283 and S284) and the N-terminal region including α0 (S196, D198 and F204) (Figure 4A).
Differences in chemical shift perturbation between the wild-type and mutant APOBEC3Gs may partly be attributed to the mutations themselves. Perturbations for R238, G240, L242 and C243 were observed for the wild type, but not for the mutant (Figure 4A and B). These residues are close to C243, so the mutation of Cys243 to Ala may be responsible for the decrease in the perturbations for the mutant. Similarly, perturbations for G319 and Q322 were exclusively observed for the wild type (Figure 4A and B). The mutation of close Cys321 to Ala may be responsible for the decrease in the perturbations for the mutant. Moreover, perturbations were observed for the residues of α0 and those close to α0 for the wild type, whereas perturbations were not observed for the corresponding N-terminal region of the mutant (Figure 4A and B). As described previously, the mutation of either F202 or T203 of α0 causes the decrease of the activity of APOBEC3G. So, the interaction involving α0 may be meaningful. From these points of view, it is critical to have perturbation data on the wild-type APOBEC3G to deduce the interaction correctly.
Occurrence of the deamination reaction in an NMR tube
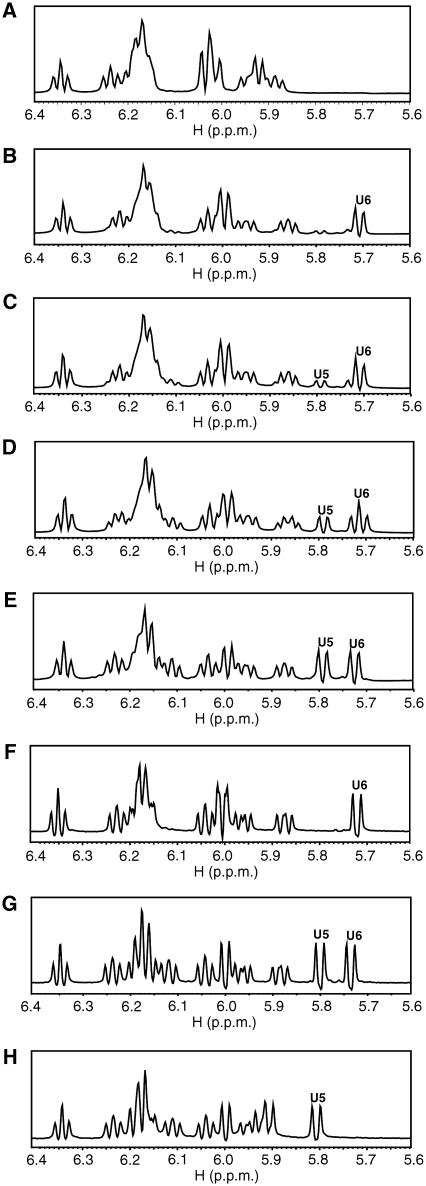
Figure 5A shows the pyrimidine (either cytidine or uridine residues) H5–C5 region of the 1H–13C HSQC spectrum of 10-mer DNA, d(A1T2T3C4C5C6A7A8T9T10). Three peaks corresponding to the C4, C5 and C6 residues of 10-mer DNA can be seen. It should be noted that the intensity of the left peak is two times greater than that of the right peak, and that the two signals overlap on the left peak. Figure 5B shows the same region of the 10-mer DNA obtained 24 h after the addition of the wild-type deaminase domain to the NMR tube, the molar ratio of DNA 10-mer:deaminase domain being 10:1. Two new peaks have appeared in Figure 5B, whereas one peak remains at the original position. The 13C chemical shift values of the two new peaks imply that these peaks originate from uridine residues. This suggests that the deamination reaction occurred in the NMR tube.
Figure 5.
1H–13C HSQC spectra indicating cytidine to uridine conversion for 10-mer DNA through the deamination reaction in an NMR tube. The pyrimidine (either cytidine or uridine residues) H5–C5 region of the 1H–13C HSQC spectrum of 10-mer DNA, d(ATTCCCAATT) (A), and that recorded 24 h after the addition of the wild-type deaminase domain to the NMR tube (B). The same regions of mutant 10-mer DNAs with the C4C5U6 (C), C4U5U6 (D) and C4U5C6 (E) segments, respectively, for reference.
It was reported that 5′-CC-3′ is the consensus sequence for deamination by APOBEC3G, where C is the site of the deamination (Beale et al, 2004). Therefore, there are three possible patterns of deamination for the C4C5C6 segment of 10-mer DNA, C4C5U6, C4U5C6 and C4U5U6. It was also reported that the third cytidine of the CCCA sequence, underlined, is preferably deaminated by APOBEC3G (Yu et al, 2004; Chelico et al, 2006) and that the second cytidine is also deaminated to some extent (Chelico et al, 2006). To confirm the occurrence of the deamination, HSQC spectra of three mutant 10-mer DNAs in which the C4C5C6 segment was mutated to either C4C5U6, C4U5U6 or C4U5C6 were recorded for reference (Figure 5C–E). Then, it was found that the HSQC spectrum obtained 24 h after the addition of the wild-type deaminase domain (Figure 5B) was the same as that of the mutant 10-mer DNA with the C4U5U6 segment (Figure 5D). This clearly demonstrates that the deamination reaction by the wild-type deaminase domain actually occurred for C5 and C6 residues in the NMR tube.
Assignment of the H5–C5 correlation peaks for the U6 and U5 residues was performed in a straightforward way from the spectra of the mutant 10-mer DNAs with the C4C5U6 and C4U5C6 segments, respectively (Figure 5C and E), together with those for the C4, C5 and C6 residues. The assignments made are indicated in Figure 5.
Strictly regulated deamination of consecutive cytidine residues in a 3′ → 5′ order, as revealed by real-time monitoring of the reaction using NMR signals
Figure 6A–E shows the time chase of the 1H NMR spectra of 10-mer DNA after the addition of the wild-type deaminase domain. For reference, 1H NMR spectra of the three mutant 10-mer DNAs are also shown with the H5 assignments of the U5 and U6 residues. The H5 peak of the U6 residue is present in the spectrum recorded 30 min after the addition of the deaminase domain (Figure 6B). Generally, the H5 peak of a uridine residue is a doublet due to a 3JH5H6 coupling. The intensity is comparable to that seen in the spectrum recorded 24 h after the addition. This indicates that the conversion of a C6 residue to a U6 residue through deamination by the wild-type deaminase domain has been almost completely accomplished within 30 min. The intensity of the H5 peak of the U5 residue is very weak in Figure 6B, but the intensity gradually increases (Figure 6C), becomes roughly half of the final intensity level in 4.5 h (Figure 6D), and finally reaches the same level as that of the U6 residue (Figure 6E). These results indicate that deamination occurs almost exclusively for the C6 residue at an early stage and that then deamination occurs for the C5 residue with a much slower reaction rate. Not only the C6 residue but also the C5 one is fully converted to a uridine residue at the end of the reaction. This is the first report that a strictly regulated deaminase reaction of consecutive cytidine residues in a 3′ → 5′ order can be directly monitored in real-time using NMR signals.
Figure 6.
Time chase of 1H NMR spectra indicating deamination of two cytidine residues in a strict 3′ → 5′ order. 1H NMR spectrum of 10-mer DNA (A), and ones recorded 0.5 h (B), 1.5 h (C), 4.5 h (D) and 24 h (E) after the addition of the wild-type deaminase domain to the NMR tube. 1H NMR spectra of mutant 10-mer DNAs with the C4C5U6 (F), C4U5U6 (G) and C4U5C6 (H) segments, respectively, for reference.
It was noted that the H5 peak of the U6 residue is a doublet at an early stage (Figure 6B), next becomes a triplet at a middle stage (Figure 6D) and finally becomes a doublet again (Figure 6E). It was found that the H5 doublet of the U6 residue of the C4U5U6 segment appears slightly in a down-field region compared with that of the C4C5U6 segment: the position of the right half of the former doublet almost coincides with that of the left half of the latter doublet. Therefore, the H5 peak of the U6 residue becomes a triplet when roughly half of the C5 residue has been converted to U5 in the course of the reaction (Figure 6D).
The structure of the N-terminal domain of APOBEC3G constructed on the basis of the structure of the C-terminal domain
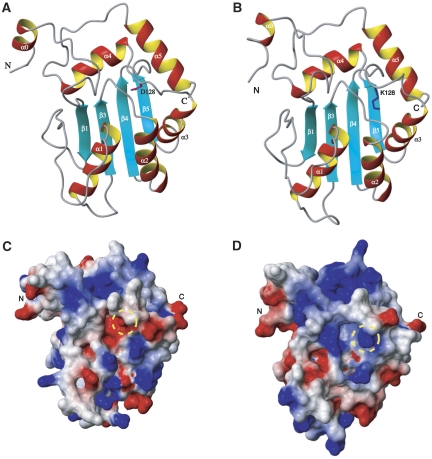
The N- and C-terminal domains of human APOBEC3G exhibit the sequence homology, the sequence identity and similarity being 36 and 54%, respectively. So, the structure of the N-terminal domain of human APOBEC3G (residues 10–192) was constructed with homology modelling on the basis of the structure of the C-terminal wild-type deaminase domain of human APOBEC3G (Figure 7A), using the program MODELLER 9v3 (Marti-Renom et al, 2000). To address the origin of species-specific sensitivity of APOBEC3G to Vif action, which is discussed below, the structure of the N-terminal domain of APOBEC3G of African green monkey (AGM) (residues 10–192) is also constructed in the same way (Figure 7B). The sequence identity and similarity between the N-terminal domain of AGM APOBEC3G and the C-terminal domain of human APOBEC3G are 38 and 53%, respectively. As discussed below, the residue at the position 128 governs the species-specific sensitivity of APOBEC3G to Vif action, and is indicated in Figure 7A and B. Surface potentials of the N-terminal domains of human and AGM APOBEC3Gs are shown in Figure 7C and D, respectively, the residue at the position 128 being indicated with a circle. D128 of human APOBEC3G and K128 of AGM APOBEC3G locate at the surface, which is suitable for these residues to interact with Vif.
Figure 7.
The structures of the N-terminal domains (residues 10–192) that interact with Vif. The structures of the N-terminal domains of human (A) and African green monkey (B) APOBEC3Gs, constructed with homology modelling on the basis of the structure of the C-terminal deaminase domain of human APOBEC3G. The surface potentials of the N-terminal domains of human (C) and African green monkey (D) APOBEC3Gs. The residue at the position 128 is indicated with a dashed circle.
Discussion
We found that the deamination reaction occurs in an NMR tube during a titration experiment and that the pH value of the solution in the NMR tube increases, probably due to the release of NH3 as a result of the reaction (Harris and Liddament, 2004). The HN and N chemical shift values of some residues were sensitive to the change in pH. In particular, we noticed that the chemical shift values of His residues and residues close to the His residues were sensitive to the pH change. Therefore, at the end of the titration, we adjusted the pH of the protein–DNA complex solution to that of the initial protein alone solution. After the adjustment, the HSQC spectrum of the complex was recorded and the chemical shift values of the complex were obtained. It should be noted that if the adjustment is not carried out, false perturbations originating from the pH change, which do not reflect the interaction with DNA, would be mixed up. For example, although H345 exhibited an apparent chemical shift perturbation of greater than 0.02 p.p.m., the perturbation diminished after adjustment of the pH. Perturbation was reported for H345 of the mutant APOBEC3G (Chen et al, 2008). There is a possibility that this perturbation may have originated just from the pH change. In this sense, our perturbation data are supposed to be more adequate for accurately characterizing the interaction.
It was reported that APOBEC3G binds to any kind of ssDNA of larger than 9 nt with essentially the same affinity even in the absence of a cytidine (Chelico et al, 2006). Therefore, binding of the deaminase domain to 10-mer DNA can be expected even after the complete conversion of cytidines to uridines through deamination in an NMR tube. Thus, the interaction of the deaminase domain with 10-mer DNA can be examined with our method.
Among perturbations observed for various residues, the following are of particular interest. First, intensive perturbations were observed for the following residues that are located close to the catalytic residue, E259; E254, R256, H257, L260, C261, F262 and L263 (Figure 4A). This implies that the substrate cytidine residue is positioned close to the catalytic residue in the APOBEC3G:ssDNA complex. In the crystal structures of Escherichia coli and mouse cytidine deaminases, a substrate cytidine is located close to the zinc-coordinating His residue (Xiang et al, 1997; Teh et al, 2006). On the basis of these structures, an APOBEC3G:ssDNA complex model was proposed in which the substrate cytidine is positioned close to the zinc-coordinating H257 residue (Chen et al, 2008). The perturbation observed for H257 of the wild type is consistent with this model. On the other hand, for the mutant, although perturbation was observed for E259, intensive perturbations for the residues around E259, including H257, were not seen (Figure 4B), which does not fit the model.
The S284-W285-S286 motif is conserved among DNA deaminases including APOBEC3G (Rogozin et al, 2007). Perturbations were observed for T283 and S284 for the wild type (Figure 4A). This implies that the substrate cytidine residue is positioned close to the conserved residues. In the proposed model mentioned above, W285 is located close to the substrate cytidine (Chen et al, 2008). The perturbation data for the wild type are consistent with this model. On the other hand, for the mutant, perturbation was not observed for the residues located close to the conserved residues (Figure 4B), which does not fit the model.
The surface potential of the wild-type deaminase domain is presented at the right of Figure 4C. The residues mentioned above, that is, those positioned close to E259 and S284-W285-S286, are located in a positively charged area, which is suitable for negatively charged ssDNA to interact with these residues. The chemical shift perturbation data and electrostatic features strongly suggest that this area is a key interactive region. A close-up of this area is shown in Figure 4D. The substrate cytidine is supposed to be positioned close to catalytic E259, conserved S284-W285-S286, and zinc-coordinating H257, as indicated by the dashed circle.
The position and polarity of the main chain of ssDNA in the complex with the deaminase domain were proposed on the basis of the results of analysis of the mutant APOBEC3G (Chen et al, 2008), as shown to the right of Figure 4E by a dashed vertical line in black. The model was constructed to have a similar main chain trace to that observed in the adenosine deaminase TadA:RNA complex (Losey et al, 2006). The main chain of ssDNA is located close to the area composed of the C-terminal regions of β3, β4 and β5, and the N-terminal regions of α1 and α2. The 3′ region of the main chain takes off and does not contact the deaminase domain.
Alternative model as to the position of the main chain of ssDNA has been proposed very recently (Holden et al, 2008), as shown to the right of Figure 4E by a dashed kinked horizontal line in green. The model is constructed on the basis of the crystal structure and mutational data. It is suggested that two active centre (AC) loops, AC loops 1 and 3, are important for binding of DNA. AC loop 1 connects α0 and β1, and AC loop 3 does β2′ and α1. For our solution structure, convergence is poor for these loops (Figure 1C). This may be due to either intrinsic flexibility of these loops or accidental lack of sufficient structural constraints for them. It is discussed that N244 and H257 of AC loop 3 are structurally conserved among crystals of Zn deaminases and may be involved in binding of the substrate cytidine (Holden et al, 2008). We observed the perturbations on binding of DNA for C243 and A246, which are located close to N244, and for H257 and neighbouring R256 (Figure 4A), which may be related to their suggestion. It was also discussed that D316 and D317 may be important for positioning the substrate so that the third cytidine of the CCC sequence is most likely to be deaminated. We observed the perturbation on binding of DNA for D316 (Figure 4A), which may be related to their suggestion.
The results of analysis of the wild-type APOBEC3G in solution suggest the third model shown at the left of Figure 4E. For the wild type in solution, large chemical shift perturbations were observed for L271, D272, D274, Q275 and K297, together with medium perturbations for V305 and S306, whereas these were not observed for the mutant (Figure 4A and B). These perturbation data suggest that in the complex with the wild-type deaminase, ssDNA may go along α1 and α2 and that one end of the main chain of ssDNA is located close to the area composed of the N-terminal regions of β3 and β4, and the C-terminal regions of α1 and α2, as shown at the left of Figure 4E. The mutation of L271 to Ala causes the decrease in the activity of APOBEC3G (Supplementary data of Chen et al, 2008), which is consistent with our model in which L271 is supposed to mediate substrate contact. Nonetheless, it seems to us fair to say that at this moment we cannot select one of these possible positions on the basis of currently available information. We do not exclude the possibility that two or even three models are coexisting in solution.
It was found that deamination of ssDNA by APOBEC3G occurs preferentially at the CCCA sequence (Yu et al, 2004; Chelico et al, 2006). The third cytidine residue, underlined, is converted to a uridine residue through deamination. Some deamination of the second cytidine residue was also detected after an extended reaction time, although experimental data for and detailed features of the deamination of the second cytidine residue, such as the reaction rate and yield, were not provided (Chelico et al, 2006). Here, we have succeeded for the first time to monitor the deamination reaction in real-time by means of NMR signals. First, it was revealed by the NMR spectra that the second and third cytidine residues, C5 and C6, of the C4C5C6A7 sequence of 10-mer DNA are fully deaminated and converted to uridine residues, U5 and U6, 24 h after the addition of the wild-type deaminase domain, whereas the first cytidine residue, C4, is not deaminated at all (Figure 5). Second, the time course of the deamination reaction was monitored and chased in real-time using NMR signals. Then, it was revealed that the deamination of the third cytidine residue, C6, and the resultant appearance of the signal of the generated uridine residue, U6, occur at a very early stage of the time course, that is, within 30 min after the start of the reaction (Figure 6B). In contrast, it was found that the deamination of the second cytidine residue, C5, and the resultant appearance of the signal of the generated uridine residue, U5, occur at a very late stage of the time course. The reaction is accomplished between 4.5 and 24 h, probably around 10 h, after the start of the reaction (Figure 6B–E). Virtually, it looks as if the deamination of the C5 residue starts after accomplishment of deamination of the C6 residue. Finally, both the C5 and C6 residues are fully deaminated and converted to uridine residues. Thus, the detailed time course of the deamination reaction of consecutive cytidine residues was visualized by means of NMR signals. The monitoring demonstrated that the deamination reaction occurs in a strict 3′ → 5′ order.
It was reported that when there are two CCCA sequences in one ssDNA, the deamination of the third cytidine residue by APOBEC3G occurs more frequently for the CCCA sequence located closer to the 5′ end. On the basis of this finding, it was concluded that APOBEC3G exerts an effect on multiple CCCA sequences processively with 3′ → 5′ directionality along ssDNA (Chelico et al, 2006). What we found here is the order of the reaction within one CCCA sequence. Therefore, the two findings regard different phenomena but may be linked to each other in the sense of 3′ → 5′ directionality/order.
The method of real-time monitoring of the enzymatic reaction with NMR signals has been established in this work, and its usefulness to address the dynamical aspects of the reaction is demonstrated. This method can be further applied in future to study unique features of APOBEC3G such as 3′ → 5′ directionality and processivity (Chelico et al, 2006). This method could also be applied to other enzymatic systems.
A C → U conversion through deamination in the minus strand causes a G → A mutation in the plus strand. Deamination of the third cytidine of a CCCA sequence in the minus strand results in the generation of a TAGG sequence in the plus strand, which contains a stop codon, TAG. Deamination of both the second and third cytidines of the CCCA sequence in the minus strand results in the generation of a TAAG sequence in the plus strand, which contains another stop codon, TAA. It is supposed that the anti-HIV activity of APOBEC3G is partially due to the generation of the stop codon through deamination and the resultant premature termination of viral open reading frames (Yu et al, 2004). APOBEC3G searches for the target sequence processively with 3′ → 5′ directionality through sliding and jumping after accomplishment of the deamination at the first target sequence (Chelico et al, 2006). Our results indicate that the deamination of the second cytidine residue of the CCCA sequence occurs at a rather later stage of the reaction time course. When the third cytidine residue of the CCCA sequence is already deaminated, further deamination of the second cytidine residue does not increase the number of stop codons, whereas deamination of the third cytidine residue of the other CCCA sequence does increase the number of stop codons. Therefore, it is reasonable that the deamination of the second cytidine residue of the CCCA sequence is carried out only at a later stage of the reaction time course in terms of the maximization of the anti-HIV activity of APOBEC3G through the generation of the stop codons.
Gag and nef initiation codons are mutated in 60 and 90% of Δvif hu-APOBEC3G+ reverse transcripts, respectively (Yu et al, 2004). The ATG initiation codons of gag and nef are followed by a G, forming a ATGG sequence. The minus strand of ATGG is CCAT. To mutate the initiation codon, deamination should occur at the cytidine residue next to an adenosine residue, not at the other cytidine residue. The phenomenon that the cytidine residue closest to the adenosine residue is preferably deaminated among three cytidine residues of the CCCA sequence may have been selected during evolution to effectively mutate the initiation codon and to maximize the anti-HIV activity of APOBEC3G.
APOBEC3G orthologues from several species are active against a broad range of retroviruses. For example, Vif-defective HIV-1 is blocked by APOPBEC3Gs from human, AGM and mouse (Mariani et al, 2003). Anti-HIV-1 activity of human APOBEC3G is counteracted by Vif of HIV-1 (Conticello et al, 2003; Yu et al, 2003; Mehle et al, 2004; Kobayashi et al, 2005). A far greater degree of specificity is found in the Vif sensitivity. For example, Vif of HIV-1 effectively counteracts human APOBEC3G but not APOBEC3G of AGM. Conversely, Vif of simian immunodeficiency virus (SIV) for AGM counteracts APOBEC3G of AGM but not human APOBEC3G (Mariani et al, 2003). Then, it was found that a single amino acid at the position 128 of human and AGM APOBEC3Gs governs the species-specific sensitivity of these proteins to Vif-mediated inhibition (Mangeat et al, 2004). The amino acid at the position 128 is Asp for human APOBEC3G and Lys for AGM APOBC3G, respectively. It was demonstrated that human APOBEC3GD128K, in which Asp128 of human APOBEC3G was replaced by Lys, exhibited the same Vif sensitivity pattern as AGM APOBEC3G, as it became resistant to Vif of HIV-1, but was effectively blocked by Vif of SIVAGM. Conversely, AGM APOBEC3GK128D, in which Lys128 of AGM APOBEC3G was replaced by Asp, acquired the HIV-1 Vif susceptibility (Mangeat et al, 2004). It was further demonstrated that the phenotype correlates with the ability of Vif to bind APOBEC3G and interfere with its incorporation into virion (Mangeat et al, 2004).
The N-terminal domain of APOBEC3G is responsible for the binding to Vif (Conticello et al, 2003; Harris and Liddament, 2004) and the residue at the position 128 locates in this domain. The structures of the N-terminal domains of human and AGM APOBEC3Gs constructed with homology modelling have given the implication on the mechanism of species-specific sensitivity of APOBEC3G to Vif action. D128 of human APOBEC3G and K128 of AGM APOBEC3G are found to locate at the surface (Figure 7A and B), which would allow them to interact with Vif. It is noted that the surface potential is quite different around at the position 128 between human and AGM APOBEC3Gs, due to intrinsic difference in charge between Asp and Lys residues (Figure 7C and D). It is supposed that this difference in the surface potential is recognized by Vifs of either HIV-1 or SIVAGM, which results in the species-specific sensitivity of APOBEC3G to Vif action. When D128 of human APOBEC3G is replaced by a Lys residue, the surface potential around at the position 128 becomes more similar to that of AGM APOBEC3G (data not shown), which also supports our idea.
Materials and methods
Preparation of wild-type APOBEC3G and DNA oligomers
DNA encoding the cytidine deaminase domain of wild-type APOBEC3G (193–384) was cloned into a pET15b expression vector with an N-terminal hexahistidine (His6)-affinity tag and a thrombin cleavage site (Novagen). E. coli BL21(DE3/RIL) cells (Stratagene) were transformed with the vector and grown in 1 l of M9 minimal medium containing 13C-labelled D-glucose and 15NH4Cl as the sole carbon and nitrogen sources, respectively, to an optical density at 600 nm of 0.6. Protein expression was induced with 1 mM IPTG at 37°C, and the cultures were harvested 5 h after induction by centrifugation (3000 g) for 20 min at 4°C. The cells were lysed by sonication in lysis buffer (20 mM Tris–HCl (pH 7.5) and 150 mM NaCl). The insoluble fraction was removed by centrifugation (16 000 g) for 20 min at 4°C. The soluble fraction was loaded onto a Ni Sepharose resin column (GE Healthcare), washed with a solution comprising 20 mM Tris–HCl (pH 7.5), 500 mM NaCl, 30 mM imidazole and eluted with an imidazole gradient of 30–500 mM. The His6 tag was cleaved using 50 U of thrombin protease (GE Healthcare) in 20 mM Tris–HCl (pH 7.5), 500 mM NaCl, 10 μM ZnCl2 and 5 mM DTT at 25°C overnight. After removal of thrombin with benzamidine-Sepharose resin (GE Healthcare), the solution was loaded onto a Ni Sepharose resin column again, washed with a solution comprising 20 mM Tris–HCl (pH 7.5), 500 mM NaCl, 30 mM imidazole, 10 μM ZnCl2 and 5 mM DTT, and eluted with an imidazole gradient of 30–500 mM. The eluate was dialysed against the solution comprising 20 mM Tris–HCl (pH 7.5), 30 mM NaCl, 10 μM ZnCl2 and 5 mM DTT. Finally, Tris was replaced by 2H-labelled Tris using an ultrafiltration cartridge (Millipore). The concentration of wild-type APOBEC3G was 0.1–0.4 mM.
DNA oligomers, synthesized with a DNA synthesizer and purified by reverse-phase HPLC, were purchased (Nippon Seihun).
Gel retardation experiment
32P-labelled DNA (200 nM), d(ATTCCCAATT), was incubated with various concentrations of APOBEC3G (40–80 μM) at 4°C for 1 h in a 10 μl solution comprising 20 mM Tris (pH 7.5), 150 mM NaCl, 10 μM ZnCl2 and 5 mM DTT. The mixtures were run on a 6% polyacrylamide gel containing 45 mM Tris-borate, 150 mM NaCl, 1 mM EDTA, and detected with BAS2000 (Fuji Film).
NMR spectroscopy
NMR spectra were recorded with Bruker DRX800, DRX600 and DRX500 spectrometers equipped with a cryoprobe with a Z-gradient. Sequential assignments of the main chain and side chain 1H, 13C and 15N resonances of wild-type APOBEC3G were made in the standard way using triple-resonance NMR spectra (Clore and Gronenborn, 1994), as reported for other proteins (Miyanoiri et al, 2003; Enokizono et al, 2005). For the chemical shift perturbation experiment, a concentrated DNA solution was added step by step to the APOBEC3G solution up to the APOBEC3G:DNA ratio of 1:10, 1H–15N HSQC spectra being recorded at each step. For real-time monitoring of the cytidine deamination in an NMR tube, a concentrated DNA solution was added at one time to the APOBEC3G solution, the APOBEC3G:DNA ratio being 1:10, and then 1H one-dimensional and 1H–13C HSQC spectra were continuously recorded during the time course of the reaction. The spectra were processed and analysed with XWIN-NMR (Bruker), NMRPipe (Delaglio et al, 1995), Sparky (Goddard and Kneller, 2006) and Kujira (Kobayashi et al, 2007). Combined chemical shift perturbation as to HN and N was defined and calculated as [(ΔδHN)2+(ΔδN/5)2]1/2 where ΔδHN and ΔδN are the chemical shift perturbations for HN and N resonances, respectively. The relative intensity of a 1H–15N correlation peak of the deaminase domain in a free state to that in a complex state was also calculated.
Structure determination
In total, 2380 distance constraints (1117 intra-residue, 581 sequential, 311 medium-range and 371 long-range ones) were obtained from three-dimensional 13C- and 15N-NOESY spectra. In total, 262 backbone dihedral constraints were obtained from TALOS (Cornilescu et al, 1999) based on 13Cα, 13Cβ and 13C′ chemical shifts. Additionally, in the later stage of the calculations, 112 distance constraints for 56 hydrogen bonds were included for slowly exchanging amide protons within the identified secondary structure elements. Also, in the later stage, a Zn2+ molecule was involved and constrained with respect to the H257, C288 and C291 residues (Prochnow et al, 2007; Chen et al, 2008) in the structure calculations.
Structure calculations were carried out using distance and dihedral angle constraints (2754 constraints in total) with the simulated annealing protocol supplied with Xplor-NIH v. 2.18 (Schwieters et al, 2003). A final set of 10 structures was selected from the 200 calculations on the basis of the criteria of the smallest residual energy values. The quality of the structure was evaluated with PROCHECK (Laskowski et al, 1996). Ramachandran plot analysis of the final structures for residues 221–379 showed that 70.6, 25.7, 3.1 and 0.6% were in the most favourable, additionally allowed, generously allowed and disallowed regions, respectively. Coordinates have been deposited in the Protein Data Bank (PDB) with the accession code 2kbo.
Acknowledgments
MK was supported by a Grant-in-Aid for Scientific Research (no. 19036026), PRESTO of JST, and the Strategic Research Project of YCU (no. K20008). TN was supported by a Grant-in-Aid for Scientific Research (no. 2057011) and Strategic Research Project of YCU (no. W20011). This study was supported by a Grant-in-Aid for High Technology Research (HTR) from the Ministry of Education, Science, Sports, and Culture, Japan, research grants from the Human Science Foundation (HIV-K-14719), a Grant-in-Aid for AIDS research from the Ministry of Health, Labour, and Welfare, Japan (H17-AIDS-002), the RIKEN Structural Genomics/Proteomics Initiative (RSGI), the National Project on Protein Structural and Functional Analyses, Ministry of Education, Science, Sports, and Culture, Japan.
References
- Beale RC, Petersen-Mahrt SK, Watt IN, Harris RS, Rada C, Neuberger MS (2004) Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J Mol Biol 337: 585–596 [DOI] [PubMed] [Google Scholar]
- Cen S, Guo F, Niu M, Saadatmand J, Deflassieux J, Kleiman L (2004) The interaction between HIV-1 Gag and APOBEC3G. J Biol Chem 279: 33177–33184 [DOI] [PubMed] [Google Scholar]
- Chelico L, Pham P, Calabrese P, Goodman MF (2006) APOBEC3G DNA deaminase acts processively 3′ → 5′ on single-stranded DNA. Nat Struct Mol Biol 13: 392–399 [DOI] [PubMed] [Google Scholar]
- Chen KM, Harjes E, Gross PJ, Fahmy A, Lu Y, Shindo K, Harris RS, Matsuo H (2008) Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature 452: 116–119 [DOI] [PubMed] [Google Scholar]
- Clore GM, Gronenborn AM (1994) Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods Enzymol 239: 349–363 [DOI] [PubMed] [Google Scholar]
- Conticello SG, Harris RS, Neuberger MS (2003) The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr Biol 13: 2009–2013 [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13: 289–302 [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Enokizono Y, Konishi Y, Nagata K, Ouhashi K, Uesugi S, Ishikawa F, Katahira M (2005) Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D. J Biol Chem 280: 18862–18870 [DOI] [PubMed] [Google Scholar]
- Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O (2005) APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433: 430–433 [DOI] [PubMed] [Google Scholar]
- Greene WC (2004) The brightening future of HIV therapeutics. Nat Immunol 5: 867–871 [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG (2006) SPARKY 3. San Francisco: University of California [Google Scholar]
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH (2003) DNA deamination mediates innate immunity to retroviral infection. Cell 113: 803–809 [DOI] [PubMed] [Google Scholar]
- Harris RS, Liddament MT (2004) Retroviral restriction by APOBEC proteins. Nat Rev Immunol 4: 868–877 [DOI] [PubMed] [Google Scholar]
- Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, Stevens RC, Goodman MF, Chen XS (2008) Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature 456: 121–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Takaori-Kondo A, Miyauchi Y, Iwai K, Uchiyama T (2005) Ubiquitination of APOBEC3G by an HIV-1 Vif–Cullin5–Elongin B–Elongin C complex is essential for Vif function. J Biol Chem 280: 18573–18578 [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Iwahara J, Koshiba S, Tomizawa T, Tochio N, Guntert P, Kigawa T, Yokoyama S (2007) KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high-throughput NMR structure studies. J Biomol NMR 39: 31–52 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8: 477–486 [DOI] [PubMed] [Google Scholar]
- Lecossier D, Bouchonnet F, Clavel F, Hance AJ (2003) Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300: 1112. [DOI] [PubMed] [Google Scholar]
- Losey HC, Ruthenburg AJ, Verdine GL (2006) Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat Struct Mol Biol 13: 153–159 [DOI] [PubMed] [Google Scholar]
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D (2003) Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424: 99–103 [DOI] [PubMed] [Google Scholar]
- Mangeat B, Turelli P, Liao S, Trono D (2004) A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J Biol Chem 279: 14481–14483 [DOI] [PubMed] [Google Scholar]
- Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, Bollman B, Munk C, Nymark-McMahon H, Landau NR (2003) Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114: 21–31 [DOI] [PubMed] [Google Scholar]
- Marin M, Rose KM, Kozak SL, Kabat D (2003) HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med 9: 1398–1403 [DOI] [PubMed] [Google Scholar]
- Marti-Renom MA, Stuar AC, Fiser A, Sanchez R, Melo F, Sali A (2000) Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct 29: 291–325 [DOI] [PubMed] [Google Scholar]
- Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D (2004) Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J Biol Chem 279: 7792–7798 [DOI] [PubMed] [Google Scholar]
- Miyanoiri Y, Kobayashi H, Imai T, Watanabe M, Nagata T, Uesugi S, Okano H, Katahira M (2003) Origin of higher affinity to RNA of the N-terminal RNA-binding domain than that of the C-terminal one of a mouse neural protein, musashi1, as revealed by comparison of their structures, modes of interaction, surface electrostatic potentials, and backbone dynamics. J Biol Chem 278: 41309–41315 [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T (1999) Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem 274: 18470–18476 [DOI] [PubMed] [Google Scholar]
- Navaratnam N, Morrison JR, Bhattacharya S, Patel D, Funahashi T, Giannoni F, Teng BB, Davidson NO, Scott J (1993) The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J Biol Chem 268: 20709–20712 [PubMed] [Google Scholar]
- Navarro F, Bollman B, Chen H, Konig R, Yu Q, Chiles K, Landau NR (2005) Complementary function of the two catalytic domains of APOBEC3G. Virology 333: 374–386 [DOI] [PubMed] [Google Scholar]
- Prochnow C, Bransteitter R, Klein MG, Goodman MF, Chen XS (2007) The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature 445: 447–451 [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Iyer LM, Liang L, Glazko GV, Liston VG, Pavlov YI, Aravind L, Pancer Z (2007) Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol 8: 647–656 [DOI] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM (2003) The Xplor-NIH NMR molecular structure determination package. J Magn Reson 160: 65–73 [DOI] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH (2002) Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418: 646–650 [DOI] [PubMed] [Google Scholar]
- Shindo K, Takaori-Kondo A, Kobayashi M, Abudu A, Fukunaga K, Uchiyama T (2003) The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J Biol Chem 278: 44412–44416 [DOI] [PubMed] [Google Scholar]
- Suspene R, Sommer P, Henry M, Ferris S, Guetard D, Pochet S, Chester A, Navaratnam N, Wain-Hobson S, Vartanian JP (2004) APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res 32: 2421–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh AH, Kimura M, Yamamoto M, Tanaka N, Yamaguchi I, Kumasaka T (2006) The 1.48 A resolution crystal structure of the homotetrameric cytidine deaminase from mouse. Biochemistry 45: 7825–7833 [DOI] [PubMed] [Google Scholar]
- Teng B, Burant CF, Davidson NO (1993) Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science 260: 1816–1819 [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD (1994) The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR 4: 171–180 [DOI] [PubMed] [Google Scholar]
- Xiang S, Short SA, Wolfenden R, Carter CW Jr (1997) The structure of the cytidine deaminase-product complex provides evidence for efficient proton transfer and ground-state destabilization. Biochemistry 36: 4768–4774 [DOI] [PubMed] [Google Scholar]
- Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR (2004) Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol 11: 435–442 [DOI] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF (2003) Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302: 1056–1060 [DOI] [PubMed] [Google Scholar]
- Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L (2003) The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424: 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]