Abstract
Replication protein A (RPA) is involved in many aspects of DNA metabolism including meiotic recombination. Many species possess a single RPA1 gene but Arabidopsis possesses five RPA1 paralogues. This feature has enabled us to gain further insight into the meiotic role of RPA1. Proteomic analysis implicated one of the AtRPA1 family (AtRPA1a) in meiosis. Immunofluorescence studies confirmed that AtRPA1a is associated with meiotic chromosomes from leptotene through to early pachytene. Analysis of an Atrpa1a mutant revealed that AtRPA1a is not essential at early stages in the recombination pathway. DNA double-strand breaks are repaired in Atrpa1a, but the mutant is defective in the formation of crossovers, exhibiting a 60% reduction in chiasma frequency. Consistent with this, localization of recombination proteins AtRAD51 and AtMSH4 appears normal, whereas the numbers of AtMLH1 and AtMLH3 foci at pachytene are significantly reduced. This suggests that the defect in Atrpa1a is manifested at the stage of second-end capture. Analysis of Atrpa1a/Atmsh4 and Atrpa1a/Atmlh3 double mutants indicates that loss of AtRPA1a predominantly affects the formation of class I, interference-dependent crossovers.
Keywords: Arabidopsis , crossover control, meiosis, recombination, replication protein A (RPA)
Introduction
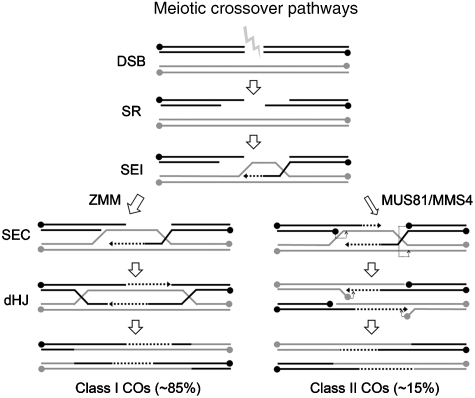
Meiotic recombination is initiated by the formation of DNA double-strand breaks (DSBs) catalyzed by the topoisomerase II-like protein SPO11 (Keeney et al, 1997). The DSBs undergo strand resection to form 3′ single-stranded DNA (ssDNA) tails. Initially, one of these invades the intact homologous duplexes of one of the non-sister chromatids of the other homologue. This displaces the corresponding sequence on the homologue to form a displacement loop (D-loop). DNA synthesis then occurs, extending the invading strand and the D-loop. This allows the capture of the 3′-end of the DNA from the other side of the DSB and following further DNA synthesis, ligation to form two four-way junctions termed double-Holliday junctions (dHjs) that connect the homologues. It was originally proposed that differential resolution of the dHjs resulted in the formation of crossover (CO) or non-crossover products (NCOs), but recent studies have led to a revised model which proposes that the CO/NCO decision occurs in early prophase I, such that most or all dHjs are resolved as COs, whereas NCOs arise through an early branch point in the homologous recombination (HR) pathway (Figure 1) (Allers and Lichten, 2001). In the budding yeast, Saccharomyces cerevisiae and in Arabidopsis this pathway accounts for the majority of meiotic COs. These, so-called class I COs, are dependent on the activities of a set of proteins referred to as the ZMM (Zip1, Zip2, Zip3, Zip4, Msh4, Msh5 and Mer3) complex (Boerner et al, 2004; Higgins et al, 2008b). Furthermore, class I COs are subject to interference, a mechanism which ensures that they do not occur in adjacent chromosomal regions (Jones, 1984). The remaining ZMM-independent (class II) COs do not exhibit interference and a proportion of these arise through a distinct pathway which is dependent on the activity of the Mus81/Mms4 protein complex (Figure 1) (de los Santos et al, 2003; Berchowitz et al, 2007; Higgins et al, 2008a).
Figure 1.
Meiotic crossover formation. Strand-resection (SR) of Spo11-induced DSBs enables Dmc1/Rad51-mediated single-end invasion (SEI) of the homologue followed by second-end capture (SEC). This intermediate is then processed to form either class I or class II COs. Class I, interference-dependent COs arise through a dHJ intermediate and require the ZMM group of proteins. They account for ∼85% of total COs. Remaining, class II COs, are interference-independent and a proportion of them are Mus81/Mms4-dependent whereby the SEC intermediate is cleaved (thin dotted lines) to form a product with 5′ flaps and gaps. It is proposed that an isomerization step follows leading to formation of 3′ flaps, a second-round of Mus81 cleavage then follows prior to filling in of the gaps and ligation to form the CO product scheme based on Hollingsworth and Brill (2004). The formation of non-CO products is not shown.
Replication protein A (RPA) is an ssDNA-binding protein comprised of three subunits of ∼70, ∼32 and 14 kDa (Wold, 1997; Iftode et al, 1999). The RPA heterocomplex is implicated in a wide range of cellular activities associated with DNA metabolism, notably DNA replication, DNA repair and HR. In budding yeast the large RPA subunit is encoded by RFA1. Analysis of mutant alleles of RFA1, rfa1-t11 and rfa1-t48, revealed an overall decrease in meiotic recombination frequency of up to 100-fold thereby confirming the crucial role played by RPA in meiosis (Soustelle et al, 2002). Immunolocalization studies in budding yeast have detected foci corresponding to RPA in nuclei at meiotic S-phase and in early prophase I where they co-localize with the recombination proteins Rad51 and Rad52 (Gasior et al, 1998). RPA is one of the several accessory proteins that participate in co-ordinating the assembly of the strand-exchange proteins Rad51 and Dmc1 on to 3′ ssDNA to form nucleoprotein filaments, which invade homologous duplex DNA. These studies have demonstrated that the strand-exchange activity of Rad51 is significantly enhanced by RPA. Paradoxically, prior saturation of the ssDNA with RPA before Rad51 addition interferes with loading of the recombinase and thus inhibits strand-exchange. This inhibition is overcome by the addition of Rad52 which stimulates Rad51 nucleation and strand-exchange and promotes post-invasion steps (reviewed in Sung et al, 2003).
In mouse, as in budding yeast, RPA1 is essential. Mice carrying a heterozygous point mutation in RPA1 accumulate lymphoid tumours and their litters exhibit early embryonic lethality (Wang et al, 2005). Cells from these mice have a reduced ability to repair DSBs induced by exposure to the replication inhibitor aphidicolin. Studies in mouse spermatocytes using both immunolocalization and immunogold labelling in conjunction with electron microscopy have also detected RPA foci during prophase I (Plug et al, 1997, 1998; Moens et al, 2007; Oliver-Bonet et al, 2007). RPA accumulates during late leptotene/early zygotene and persists through to pachytene, before dissociating from the chromatin at around the time CO-designated recombination intermediates (Marcon and Moens, 2003) acquire the MutL homologue, MLH1. Immunolocalization studies of RPA in human spermatocytes have also revealed that the complex is present throughout prophase I until mid-pachytene (Oliver-Bonet et al, 2007). These observations suggest that RPA may have a role at later stages of prophase I in addition to an earlier role but, because it has not proved possible to generate and analyse RPA-deficient spermatocytes, functional studies have not been conducted.
In contrast to budding yeast and mouse, it has been reported that Arabidopsis has five paralogues of RPA1 (Table I, Supplementary Figure 1) (Shultz et al, 2007). The same authors also report two copies of the ∼32 kDa RPA2 subunit as well as two copies of the 14 kDa RPA3 subunit (Shultz et al, 2007). Paralogues of RPA1 have been reported in a range of other plant species, notably rice, which also contains multiple copies (three) of the gene (Table I) (Ishibashi et al, 2006; Singh et al, 2007).
Table 1.
Sequence homology of AtRPA1a with other RPA1-like proteins in Arabidopsis and other species
| Species/gene identity | Amino acid identity (%) | Amino acid similarity (%) |
|---|---|---|
| A. thaliana At4g19130 | 49 | 68 |
| A. thaliana At5g45400 | 45 | 62 |
| A. thaliana At5g61000 | 32 | 53 |
| A. thaliana At5g08020 | 32 | 52 |
| S. cerevisiae | 30 | 52 |
| Schizosaccharomyces pombe | 30 | 53 |
| Mouse | 33 | 53 |
| Human | 35 | 55 |
| Oryza sativa (rice) | 59 | 75 |
| Pisum sativum (pea) | 68 | 80 |
| Vitis vinifera (grapevine) | 73 | 84 |
| Source: http://www.ncbi.nlm.nih.gov/. | ||
In a survey of the meiotic proteome of Brassica oleracea, which is a close relative of Arabidopsis and provides an effective route to identifying Arabidopsis meiotic genes, we identified peptides corresponding to the predicted product of a member of the AtRPA1 family, At2g06510 (herein referred to as AtRPA1a) suggesting a possible role in meiosis (Sanchez-Moran et al, 2005). We report here the functional analysis of AtRPA1a during meiosis. We present evidence that AtRPA1a is expressed during prophase I of meiosis where it is required to ensure wild-type levels of meiotic COs. The phenotype of meiocytes lacking AtRPA1a indicates that the protein is not required for the repair of meiotic DSBs, but plays an essential role at later stages in the meiotic recombination pathway that is required for the formation of class I COs.
Results
Loss of AtRPA1a results in reduced fertility and meiotic defects
To investigate a potential role for AtRPA1a during meiosis a T-DNA knockout line (SALK_017580) was obtained from the Nottingham Arabidopsis Stock Centre (NASC). Molecular characterization of SALK_017580 confirmed a T-DNA insertion at the splice junction between the first intron and second exon 736 bp downstream from the ATG start codon resulting in a knockout of the AtRPA1a transcript (Supplementary Figure 2A–C). A homozygous line (Atrpa1a) was identified that showed normal vegetative growth, but with reduced fertility (Supplementary Figure 3). Mean silique length was reduced from 15.84 mm (n=50) in wild-type to 10.84 mm (n=50) in Atrpa1a. Mean seed-set per silique in the mutant was 17.72 (n=50) compared with 56.08 (n=50) in wild-type representing a decrease of 68.4%. Pollen viability determined by Alexander's staining revealed a large proportion of non-viable pollen (data not shown) (Alexander, 1969).
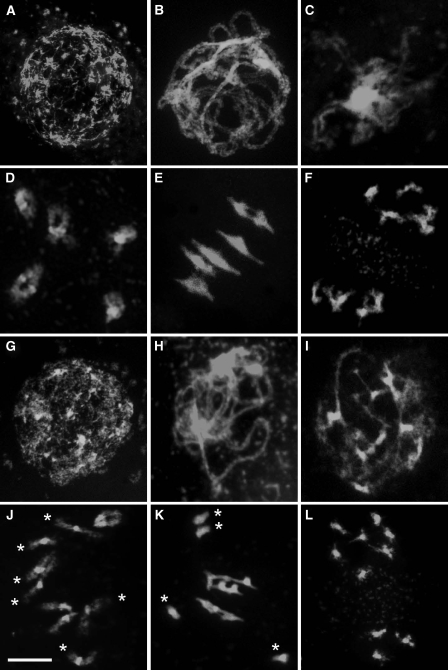
To determine if the reduced fertility phenotype of Atrpa1a was due to a defect in meiosis, chromosome spreads prepared from pollen mother cells (PMCs) were examined using fluorescence microscopy (Figure 2). Early prophase I from G2 through to diplotene was indistinguishable from wild-type, with the chromosomes apparently pairing normally and attaining full synapsis at pachytene (Figure 2A–C, G–I). However, as the chromosomes desynapsed and began to condense during late diplotene/diakinesis, it became apparent that not all the homologous chromosomes pairs were linked by chiasmata, leading to the presence of univalents at metaphase I (Figure 2D, E, J and K). As a result, missegregation occurred at the first meiotic division leading to unbalanced dyads and subsequent aneuploid tetrads following the second meiotic division (Figure 2F and L). These observations demonstrate that AtRPA1a is essential for normal progression through meiosis. Nevertheless, the absence of any evidence of chromosome fragmentation would suggest that meiotic DSB repair per se is not dependent on the activity of AtRPA1a.
Figure 2.
Meiotic stages from wild-type (A–F) and Atrpa1a (G–L) pollen mother cells. (A, G) leptotene. (B, H) pachytene. (C, I) diplotene. (D, J) diakinesis. (E, K) metaphase I. (F, L) metaphase II. Stages to diplotene (A, B, C, G, H, I) are very similar in appearance in wild-type and Atrpa1a. At diakinesis and metaphase I univalents (asterisks) are apparent in Atrpa1a (J, K) which can lead to unbalanced chromosome segregation at the second division (L). Bar, 10 μm.
Subsequent cytological analysis of an independently isolated Atrpa1a allele (GABI_406D08) revealed the same meiotic phenotype (Supplementary Figures 2A and 4).
Complementation of Atrpa1a restores fertility
To confirm that the meiotic phenotype was due to the insertion in AtRPA1a a full-length cDNA of the gene was placed under the control of the AtDMC1 promoter in pPF408 and was transformed into Atrpa1a. Three plants from each of two independently generated Atrpa1a homozygotes were transformed in duplicate. Here, 58 transformants were obtained which on visual inspection were found to have full or partial restoration of fertility. Three lines that showed restoration to full fertility were analysed further. The presence of the transgene and the endogenous knockout gene was confirmed by PCR and cytological examination revealed that normal meiosis had been restored (Supplementary Figure 5). Atrpa1a controls transformed with empty pPF408 did not exhibit any restoration of fertility and meiosis remained aberrant.
AtRPA1a localizes to meiotic chromosomes during prophase I
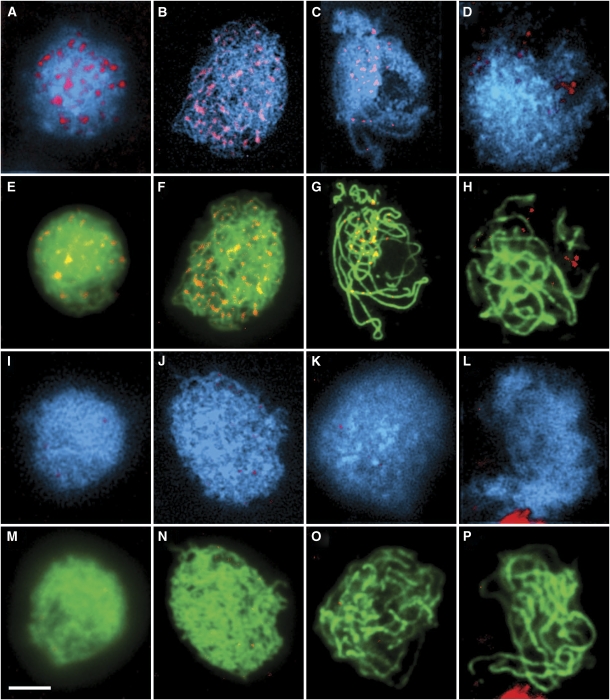
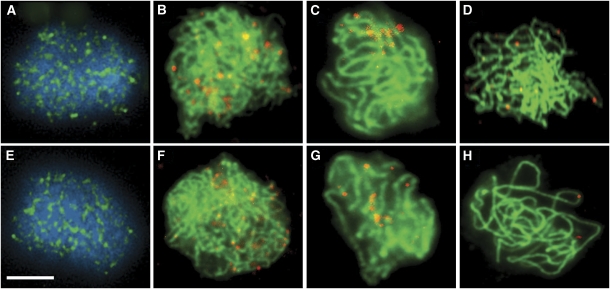
The distribution of AtRPA1a during meiosis was investigated using an antibody (Ab) against a recombinant polypeptide comprising amino acid residues 2–171 of the protein expressed in Escherichia coli (Figure 3). Immunolocalization studies on chromosome spread preparations of wild-type PMCs using the anti-AtRPA1a Ab in conjunction with Abs recognizing the axis-associated protein, ASY1 (Caryl et al, 2000; Armstrong et al, 2002) or the synaptonemal complex (SC) transverse element protein, ZYP1 (Higgins et al, 2005) to aid meiotic staging revealed that AtRPA1a was first detectable as chromatin-associated foci at early leptotene (Figure 3A and E). The number of foci increased to a maximum (175.8±12, n=5) at late leptotene/early zygotene before gradually reducing during pachytene (Figure 3B, C, F and G). By late pachytene the protein was no longer detectable (Figure 3D and H). When the anti-AtRPA1a Ab was applied to spread preparations of Atrpa1a prophase I chromosomes no signal was detectable, thereby confirming the specificity of the Ab (Figure 3I–P) and the RT–PCR analysis which had revealed that the AtRPA1a transcript was absent in the mutant (Supplementary Figure 2C). Identical results were obtained when the studies were conducted with an anti-AtRPA1a Ab based on a peptide comprising residues 116–135 of the protein (Supplementary Figure 6).
Figure 3.
Immunolocalization of AtRPA1a (red) to wildtype (A–H) and Atrpa1a (I–P) prophase I nuclei. In wild-type nuclei AtRPA1a foci first appear during late leptotene/early zygotene (A), reach a maximum number during zygotene (B), reduce in number throughout pachytene (C) before disappearing by late pachytene (D). Corresponding images showing colocalization with AtASY1 (E, F) and AtZYP1 (G, H) (green) show that AtRPA1a foci localize along chromosome axes. Corresponding stages in mutant nuclei show localization of AtASY1 (M, N) and AtZYP1 (O, P) but no AtRPA1 localization (I–P). Nuclei in A–D and I–L are counterstained with DAPI. Bar, 10 μm.
Later stages of meiotic recombination are affected in Atrpa1a
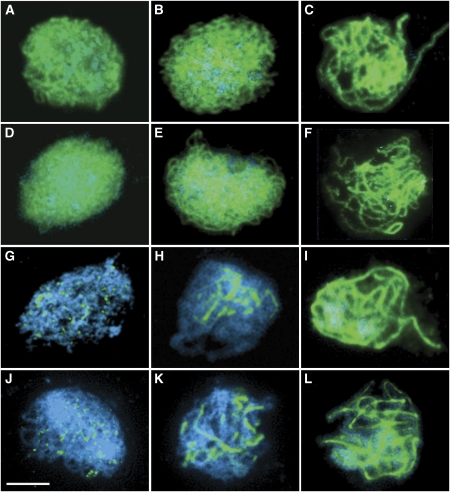
To investigate the meiotic defect in Atrpa1a in more detail, immunolocalization studies were carried out on spread preparations of PMCs from the mutant at various stages throughout prophase I. Localization of ASY1 and ZYP1 in Atrpa1a was indistinguishable from the wild-type control (Figure 4A–L). In both Atrpa1a and wild-type an abundant ASY1 signal was detectable at late leptotene/early zygotene (Figure 4A and D) which by zygotene through pachytene was clearly discernable as a linear axis-associated signal (Figure 4B, C, E and F). ZYP1 was detectable as foci or very short linear signals at late leptotene/early zygotene before progressively polymerizing to form a continuous linear signal through zygotene into pachytene indicative of full synapsis (Figure 4G–L). This indicates that there are no major defects in axis formation and assembly of the SC in Atrpa1a.
Figure 4.
Immunolocalization of AtASY1 (A–F) and AtZYP1 (G–L) proteins (green) to wildtype (A–C, G–I) and Atrpa1a (D–F, J–L) prophase I nuclei. (A, D, G, J) late leptotene/early zygotene. (B, E, H, K) zygotene. (C, F, I, L) pachytene. At each stage wildtype and Atrpa1a are indistinguishable, indicating that there are no major defects in axis assembly and synapsis in Atrpa1a. Bar, 10 μm.
The observation that chromosome synapsis was normal suggested that early stages in the recombination pathway were not substantially perturbed. Immunolocalization with an Ab against the strand-exchange protein AtRAD51 was consistent with this conclusion. This revealed no obvious difference in the number of AtRAD51 foci in Atrpa1a and wild-type chromosome spread preparations at early prophase I (Figure 5A and E). To assess the effect of loss of AtRPA1a at later stages of the recombination pathway, immunolocalization studies were conducted using Abs against the mismatch repair proteins AtMSH4 and AtMLH1 (Higgins et al, 2004, 2008b; Jackson et al, 2006).
Figure 5.
Immunolocalization of strand-exchange and mismatch repair proteins to wild-type (A, B, C, D) and Atrpa1a (E, F, G, H) prophase I nuclei. During early prophase I, wildtype and Atrpa1a show similar patterns of AtRAD51 (green) localization (A, E). Wild-type and Atrpa1a also show similar patterns of AtMSH4 (red) localization; mid-prophase (B, F), late-prophase (C, G). Late-prophase I Atrpa1a nuclei show a marked reduction in numbers of AtMLH1 (red) foci relative to wildtype (H, D respectively). Chromosome axes (green) are labelled using anti-ASY1 Ab in mid-prophase I (B, F); the SC (green) is labelled using anti-ZYP1 Ab in late-prophase I (C, D, G, H). Bar, 10 μm.
The MutS homologue Msh4 functions as a heterodimer with Msh5 and is essential for the formation of class I COs (Ross-Macdonald and Roeder, 1994; Boerner et al, 2004; Higgins et al, 2004). In wild-type Arabidopsis, numerous AtMSH4 foci are detected in chromosome spread preparations at leptotene and persist until early pachytene, but with a gradual reduction in number (Higgins et al, 2004). Comparison of Atrpa1a and wildtype did not reveal any obvious difference in the number of AtMSH4 foci per nucleus at early prophase I (mean Atrpa1=86 versus wildtype=83, n=5), suggesting that the protein associates as normal with the recombination intermediates (Figure 5B and F). Moreover, there was no discernable difference in the progressive reduction in the number of AtMSH4 foci thereafter. Dual-immunolocalization with ZYP1 indicated that by pachytene few AtMSH4 axis-associated foci remained in both Atrpa1a and wild-type nuclei (Figure 5C and G). Hence, it appears that the dynamics of AtMSH4 turnover in the mutant are indistinguishable from wild-type.
The MutL homologue, Mlh1, forms a heterodimer with Mlh3 which co-localizes to CO sites where it is required to ensure resolution of dHjs as COs rather than non-COs (Hunter and Borts, 1997; Wang et al, 1999; Marcon and Moens, 2003; Jackson et al, 2006). Immunolocalization of AtMLH1 in Atrpa1a chromosome spreads at pachytene revealed the presence of foci corresponding to the protein. However, there was a significant reduction in their number compared with wildtype, which typically had 8–10 AtMLH1 foci per nucleus. Although Atrpa1a nuclei with up to five AtMLH1 foci were observed, the majority contained 0–3 foci (Figure 5D and H). When an Ab against AtMLH3 was used in place of the anti-AtMLH1 Ab a corresponding reduction in AtMLH3 foci was observed (Supplementary Figure 7).
Together, these data suggest that recombination is initiated as normal in the absence of AtRPA1a, but that at later stages of prophase I a proportion of the recombination intermediates lose their CO designation and are repaired as non-COs.
Residual chiasmata are randomly distributed in Atrpa1a
Previously, we have shown that mutants lacking AtMSH4 and AtMLH3 are, to different degrees, defective in the formation of chiasmata and that the numerical distribution of residual chiasmata per nucleus is random (Higgins et al, 2004; Jackson et al, 2006). Our initial analysis of Atrpa1a revealed a reduction in chiasma formation. Moreover, the immunolocalization studies indicated that the number of MLH1/MLH3 foci is significantly reduced in Atrpa1a. Hence, to gain further insight into the effect of loss of AtRPA1a in relation to these observations, we conducted a quantitative cytological analysis to determine the frequency and distribution of the residual chiasmata in Atrpa1a at metaphase I (Figure 6). Nuclei containing between 0 and 8 chiasmata were observed giving an overall mean chiasma frequency of 3.98 (n=50) per cell, whereas in wild-type nuclei the number of chiasmata range between 8 and 12 per cell and the overall mean chiasma frequency is 9.86 (Higgins et al, 2004). Moreover, the numerical distribution of chiasmata in Atrpa1a does not significantly deviate from a Poisson distribution (χ2(4)=8.947 P>0.05), indicating that the residual distribution of chiasmata among cells is random. In contrast the distribution of chiasmata in wild-type cells deviates significantly from a Poisson distribution (Higgins et al, 2004).
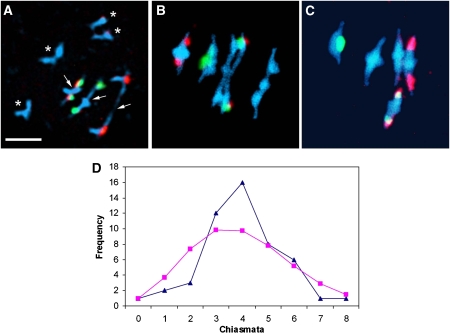
Figure 6.
Metaphase I nuclei of Atrpa1a (A, B) and wild-type (C) pollen mother cells following FISH to label the positions of 5S rDNA (red) and 45S rDNA (green) loci. (A) A nucleus with four univalents (asterisks) and three chiasmata (arrows); a proximal chiasma in chromosome 4 and single distal chiasmata in chromosomes 2 and 5. (B) A nucleus with seven chiasmata; a chiasma in each arm of chromosomes 2 and 5 and single distal chiasmata in chromosomes 1, 3 and 4. (C) A typical wild-type nucleus with 10 chiasmata. (D) Observed (triangles) and Poisson-predicted (squares) distributions of chiasma numbers per cell for Atrpa1a. Bar, 10 μm.
AtRPA1a is required for normal levels of class I crossovers
In principle, the reduction in chiasma formation observed in Atrpa1a could be due to a reduction in type I COs or a general reduction in both classes of CO. Previously, we have shown that formation of class I COs is dependent on the activity of AtMSH4 (Higgins et al, 2004). Loss of the protein results in a dramatic reduction of COs/chiasmata to around 15% of wild-type levels and the residual class II COs exhibit a random numerical distribution consistent with a loss of CO interference. Thus, to determine if AtRPA1a is implicated in one or both CO pathways we constructed an Atrpa1a/Atmsh4 double mutant. This line exhibited a further reduction in fertility compared with Atrpa1a, setting fewer seed (mean=4.85/silique; 8.65% of wild-type) and forming shorter siliques (mean=7.22 mm; 45.58% of wild-type). These figures are consistent with published data for Atmsh4 (Higgins et al, 2004). Cytological analysis of the chiasma frequency in metaphase I chromosome spread preparations revealed a reduction in chiasma frequency to a mean of 1.08 (n=50). Comparison with the residual chiasma frequency (mean=1.2, n=50) and distribution in an Atmsh4 mutant grown under identical conditions revealed that there was no significant difference (P=0.597).
Although class I COs are significantly reduced in Atrpa1a, they are not entirely absent. Simply subtracting the mean chiasma frequencies for Atrpa1a and the Atrpa1a/Atmsh4 double mutant would suggest that about 2–3 class I COs per cell occur in the absence of AtRPA1a. This is consistent with the immunolocalization studies, which revealed the presence of a few AtMLH1 foci in Atrpa1a nuclei at pachytene. In a previous analysis of an Atmlh3 mutant, we demonstrated that loss of the protein results in a significant reduction in CO formation. Evidence suggests that CO imposition is lost such that a majority (∼70%) of class I CO-designated recombination intermediates are resolved as non-COs (Jackson et al, 2006). Based on this, it would be predicted that the chiasma frequency in an Atrpa1a/Atmlh3 double mutant would be intermediate between Atrpa1 and the Atrpa1a/Atmsh4 double mutant. We therefore investigated if this was the case. Chiasma counts from metaphase I chromosome spread preparations from an Atrpa1a/Atmlh3 double mutant gave a mean chiasma frequency of 1.76 (n=50). This figure is significantly lower than the chiasma frequency in either the Atrpa1a or Atmlh3 mutant (mean=3.92, n=50) (P<0.0001; P<0.0001, respectively), but significantly greater than that in Atmsh4 and Atrpa1a/Atmsh4 (P<0.01; P<0.01, respectively). Hence, as predicted, it appears that absence of AtMLH3 activity results in a loss of CO imposition affecting a large proportion, but not all, of the AtMSH4-dependent class I COs that occur in the Atrpa1a mutant.
Together, the data from the analysis of the double mutant lines are consistent with the proposal that the reduction in COs/chiasmata in Atrpa1a is primarily due to a detrimental effect on the formation of class I COs and that loss of the protein has no obvious effect on the formation of the class II COs.
Discussion
Arabidopsis possesses several RPA1 paralogues
Unlike the situation in budding yeast and mammals, inspection of the Arabidopsis genome reveals five RPA1 paralogues (Shultz et al, 2007). During a survey of the meiotic proteome of B. oleracea, a close relative of Arabidopsis, we identified peptides corresponding to one of these, AtRPA1a (Sanchez-Moran et al, 2005). Subsequent analysis of a homozygous Atrpa1a mutant indicated that the gene was not required for vegetative growth, but was essential for normal levels of fertility. A previous study primarily focusing on RPA1 paralogues in rice had also included a preliminary analysis of Atrpa1a, from which the authors concluded that loss of Atrpa1a was lethal due to a failure to recover Atrpa1a homozygotes. Similarly, they reported that attempts to use RNA interference to knockdown AtRPA1a resulted in lethality (Ishibashi et al, 2005). Although the basis of this discrepancy is uncertain, several factors could account for it. In the earlier study only limited numbers of Atrpa1a progeny were analysed. Also, the Atrpa1a lines were obtained from different sources. The earlier study used seed from ABRC, whereas the line used in this study was obtained later from NASC. Hence, it is likely that our material had been grown through more generations. It is possible that earlier generations contained an additional linked T-DNA insertion that may have affected an essential gene, which combined with a limited analysis may account for the failure to identify homozygous lines. The RNAi studies may have been confounded by the construct downregulating other members of the AtRPA1a family as ‘off-targets'. This phenomenon is reported to be a potential problem (Xu et al, 2006). Regardless of the underlying explanation, our complementation analysis study confirmed that the basis for the meiotic phenotype was due to the loss of AtRPA1a. Moreover, an Atrpa1a homozygote is now available from NASC and our subsequent analysis of an additional independent allele GABI_406D08 confirms the meiotic phenotype.
The finding that Arabidopsis contains several RPA1 paralogues is reminiscent of previous observations with AtSPO11. With the exception of Arabidopsis, all organisms analysed so far possess a single SPO11 gene. In the case of Arabidopsis, three SPO11 paralogues have been identified (Hartung and Puchta, 2000; Grelon et al, 2001). Two of these, AtSPO11-1 and AtSPO11-2, have been shown to work in conjunction to initiate meiotic DSB formation (Grelon et al, 2001; Stacey et al, 2006; Hartung et al, 2007). One general factor that could in part account for these duplications is that Arabidopsis is thought to have had a tetraploid ancestor and that its extant form retains duplicated regions encompassing around 60% of the genome (AGI, 2000).
DNA double-strand break repair is not dependent on AtRPA1a
RPA is implicated in several activities during homologous recombination. In vitro studies using purified RPA from budding yeast have led to the suggestion that in vivo RPA binds to ssDNA prior to RAD51, which then displaces it to form a functional nucleoprotein complex that can mediate strand-exchange. This process is impaired when the RPA complex is prepared using the recombination-deficient rfa1-t11 allele due to the displacement of the mutant form of RPA by RAD51 occurring more slowly than normal (Kantake et al, 2003). Other analyses of the rfa1-t11 and rfa1-t48 mutants have revealed that although DSB formation is normal and strand-resection to form a 3′ single-stranded tail occurs in a timely fashion, the 5′ ends of the DNA at the break sites undergo hyper-resection indicative of a failure in the repair process (Soustelle et al, 2002). More recently a study of repair following HO endonuclease-induced DSB formation at the MAT locus in an rfa1-t11 background indicates that the mutant cells are unable to undergo strand-exchange, despite apparently normal Rad51 loading. It is suggested that RPA may bind to the displaced D-loop, thus stabilizing it and preventing reversal of the strand-exchange reaction (Wang and Haber, 2004). This observation is consistent with conclusions of an earlier biochemical study that RPA stabilized Rad51-mediated strand-exchange intermediates through sequestration of the displaced DNA strand (Eggler et al, 2002).
In Arabidopsis the failure to repair meiotic DSBs is associated with a variety of mutants, such as Atrad51, Atmre11 and Atmnd1, that are defective in early steps of the homologous recombination pathway (Li et al, 2004; Puizina et al, 2004; Kerzendorfer et al, 2006). This is manifested by chromosome fragmentation at metaphase I. In the case of Atrpa1a no evidence of fragmentation was detected. Hence, it is clear that in contrast to the recombination-deficient rfa1 alleles in budding yeast, AtRPA1a is not essential for meiotic DSB repair. This suggests that one or more of the remaining AtRPA1 paralogues present in Arabidopsis may undertake a role during the early stages of the recombination pathway. It is also conceivable that AtRPA1a is also normally active during these early steps, possibly in conjunction with one of the other AtRPA1 proteins, but in its absence this role can be fulfilled by another member of the AtRPA1 family.
Analysis of an Atrpa1a mutant suggests a role in CO formation
Analysis of budding yeast mutants has revealed an important role for RPA1 in the early stages of meiotic recombination. However, immunolocalization studies in mouse have indicated that the protein is present throughout much of prophase I, thereby implying that RPA may play a further role in the later stages of the recombination pathway (Plug et al, 1998; Moens et al, 2007). Consistent with this, recent in vitro biochemical studies of the RPA protein from the budding yeast mutant rfa1-t11 have revealed that the protein is defective in promoting Rad52-mediated strand annealing and second-end capture following D-Loop formation (Sugiyama et al, 2006).
Our studies revealed that the chronology of AtRPA1a localization in Arabidopsis is essentially the same as that of RPA in mouse. The protein is present throughout much of prophase I. Loss of AtRPA1a coincides with the removal of AtMSH4 foci and appearance of AtMLH1, but by the time AtMLH1 forms foci on pachytene chromosomes, AtRPA1a is no longer associated with the chromatin. Thus it seems likely that AtRPA1a is not required to maintain the interaction of the MutL heterocomplex with the CO intermediate.
Studies in chromosome spread preparations of Atrpa1a meiocytes indicated no discernable effect on the localization of AtRAD51 compared with wildtype. Similarly the distribution of AtMSH4 foci was indistinguishable from that in wildtype. This was accompanied by apparently normal elaboration of the chromosome axes and SC based on the localization of AtASY1 and AtZYP1, respectively. Complete polymerization of the SC in Atrpa1a is indicative that the AtRPA1a protein is not essential for early stages in the recombination pathway. This is based on previous studies that Arabidopsis meiotic mutants affecting DSB formation and resection and strand-exchange are asynaptic, whereas mutants deficient in proteins such as AtMSH5 and AtMLH3 which are required later in the recombination pathway undergo full synapsis (Couteau et al, 1999; Jones et al, 2003; Jackson et al, 2006; Higgins et al, 2008b). However, our analysis revealed that although the number and chronology of AtMSH4 foci was normal, a significant deficiency in the number of AtMLH1 foci compared with wildtype was apparent. This implies a defect in the formation of COs and was substantiated by our observation that the chiasma frequency at metaphase I in Atrpa1a is significantly reduced. These data provide direct evidence that AtRPA1a plays a key role at a late stage in the processing of recombination pathway intermediates to form COs.
How might loss of AtRPA1a disrupt CO formation?
Studies have demonstrated that the MutS heterodimer Msh4/Msh5 and the MutL heterodimer Mlh1/Mlh3 are essential for the formation of meiotic COs (Hoffmann and Borts, 2004; Kolas and Cohen, 2004). Biochemical studies of purified human hMSH4/hMSH5 have led to a model whereby the complex forms a sliding clamp structure that encircles the two homologous DNA duplexes in progenitor dHjs to stabilize the formation of the dHj intermediate (Snowden et al, 2004, 2008). It is proposed that the MutL proteins impose resolution of the dHJs as COs, presumably by influencing which DNA strands are cut and religated during resolution, but details of this process have yet to be determined (Wang et al, 1999). Based on the Atrpa1a phenotype, it seems feasible that loss of AtRPA1a compromises either maturation of the unligated progenitor dHj or alternatively, the protein may be required for efficient interaction between the MutL proteins and the dHj intermediate. In both instances this would lead to the reduction in the number of AtMLH1 foci and chiasmata that is observed in the mutant.
In the model for hMSH4/hMSH5 action, it is proposed that the complex binds duplex DNA adjacent to the D-loop, prior to branch migration and second-end capture (Snowden et al, 2004). Taking into account the in vitro studies using the RPA complex from rfa1-t11 (Sugiyama et al, 2006) and assuming that, despite the apparent lack of an obvious RAD52 homologue (Iyer et al, 2002), strand-annealing and second-end capture during meiosis in Arabidopsis are substantially the same as in budding yeast, then one may predict that loss of AtRPA1a could dramatically reduce the efficiency of this step (Figure 7). In Atrpa1a localization of AtMSH4 appears normal, but subsequent localization of AtMLH1 and AtMLH3 is reduced. This could reflect the fact that the heterodimer is compromised in its progression from the initial interaction to the formation of a mature dHj-associated complex. However, if this explanation is correct, then our data indicate that loss of AtRPA1a does not completely block the AtMSH4-dependent class I CO pathway. This is based on the observation that the residual CO/chiasma frequency in Atrpa1a is significantly higher than that in a Atmsh4 mutant. Importantly, it seems that the ‘extra' chiasmata in Atrpa1a are residual class I COs as they are absent in an Atmsh4/Atrpa1a double mutant, which forms the same number of chiasmata as Atmsh4. How might one account for the fact that the residual chiasma frequency in Atrpa1a is greater than in Atmsh4 as it implies that a proportion of the CO-designated intermediates still proceed to dHjs in the absence of AtRPA1a? One possible scenario is that the proposed role for AtRPA1a in second-end capture is partially fulfilled by another of the AtRPA1 family, all of which are transcribed to some extent in stamen tissue (www.genevestigator.ethz.ch/gv/index.jsp). If so, then it would account for the phenotype of the Atmsh4/Atrpa1a double mutant. Despite the capability of forming at least a proportion of the normal complement of progenitor dHJs in the absence of AtMSH4, the mutant would not be able to process them further. Hence, only the class II, AtMSH4-independent, non-interference sensitive COs would remain, just as in the Atmsh4 single mutant. A corollary of this is that it would be predicted that an Atrpa1a/Atmlh3 double mutant would form fewer COs/chiasmata than an Atrpa1a mutant. The rationale for this is that class I AtMSH4-dependent recombination intermediates require the activity of AtMLH3 in conjunction with AtMLH1 to ensure their resolution as COs. In the absence of AtMLH3, COs/chiasmata exhibit a Poisson distribution indicating that normal control is lost such that only a random subset of these CO-designated class I intermediates give rise to COs. Hence, in an Atrpa1a/Atmlh3 mutant the class II COs would be unaffected, but the residual class I-designated COs would resolve at random to either COs or non-COs as in the Atmlh3 mutant. Overall, this would lead to a reduction in the number of chiasmata at metaphase I. This is exactly what was observed in this investigation.
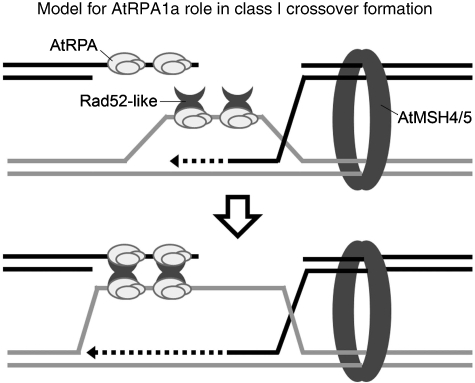
Figure 7.
A model for AtRPA1a during second-end capture. We propose that an RPA complex containing AtRPA1a coats the D-loop and ssDNA tail of the resected second strand from the DSB site to mediate second-end capture through a RAD52-like protein (based on Sugiyama et al, 2006). In the absence of AtRPA1a this step is compromised leading to a reduction in CO formation.
It is possible that the primary effect of loss of AtRPA1a is on the MutL complex rather than the MutS complex. On the balance of evidence this is perhaps less likely, but cannot yet be excluded. Nevertheless, if this is the case, then it is most likely that initial loading of AtMLH1/AtMLH3 is destabilized. This is based on observations in mouse and Arabidopsis which demonstrate that loading of the MutL proteins occurs at the time that the RPA complex is dissociating from the chromatin, such that very few mixed complexes are detected (Moens et al, 2007). However, the fact that Atrpa1a nuclei are observed containing up to 5 AtMLH1 foci reveals that this step cannot be totally dependent upon AtRPA1a.
The finding that there is no significant difference in the chiasma frequency of Atmsh4/Atrpa1a compared with Atmsh4 indicates that the formation of class II interference-independent COs is not dependent on AtRPA1a activity and strongly suggests that the predominant role of AtRPA1a is in class I CO formation. However, we cannot entirely exclude the possibility that AtRPA1a may play a role in class II CO formation. Recent studies indicate that class II COs in Arabidopsis are only partially dependent on Mus81. We estimate that AtMUS81 participates in about a third of class II COs, approximately 5% of the total COs (Higgins et al, 2008a). This suggests that there are at least two pathways leading to class II COs in Arabidopsis. Thus, if AtRPA1a was required for only a proportion of class II COs and accepting that there could be some redundancy with other members of the AtRPA1 family, the overall reduction in CO frequency would be very small and unlikely to be detectable by chiasma counts.
Materials and methods
Plant material and nucleic acid extraction
A. thaliana ecotype Columbia (0) was used for wild-type analysis. T-DNA insertion lines SALK_017580 and GABI_406D08 were obtained from NASC for mutant analysis (Alonso, 2003). Plants were grown, material harvested and nucleic acid extractions were performed as described earlier by Higgins et al (2004).
T-DNA insertion site mapping
The T-DNA insertion site of SALK_017580 was amplified with primers LBa1 5′-TGGTTCACGTAGTGGGCCATCG-3′ and RPA-R1 5′-CAACCCTGTTCGGAGGCG-3′. The PCR product was cloned into pDrive (Qiagen) and sequenced. Pairs of primers were used to determine whether the plants were homozygous or heterozygous for the T-DNA insertion. Primers RPA-F1 5′-GAGTATTCGTGGCTATGTATTTGG-3′ and RPA-R1 were used to amplify the wild-type genomic region of SALK_517580 and primers LBa1 and RPA-R1 were used to amplify the region where the T-DNA had inserted.
Construction of the AtRPA1a complementation plasmid
Primers RPA-COMP-F 5′-CCACTAGTAAGCTTCTCCCGCAAAATTCAGC-3′ and RPA-COMP-R 5′-CCACTAGTTAATATAATAGTGTACTAAACTCGAGCTTGC-3′ were used to amplify the entire AtRPA1a coding sequence with flanking 5′ and 3′ UTR regions from cDNA clone pda 11728 (Riken GSC/BRC). The PCR product was cloned into the binary vector pPF408 (Siaud et al, 2004) using SpeI sites incorporated into the primers. The construct was confirmed by sequencing.
Plant transformation
The binary plasmid construct was introduced into Agrobacterium tumefaciens LBA 4404 and plants transformed as described earlier (Higgins et al, 2004). Transformed plants were selected by spraying with glufosinate-ammonium (0.1 g/l), on appearance of the first true leaves, followed by two more sprayings at 8-day intervals.
Antibody production
Primers RPA-SUB-N 5′-CCGCTAGCCCGGTGAGTTTGACTCCGAAC-3′ and RPA-SUB-170 5′-CCCTCGAGAGATGGCCTGAAGCTTGGAGTATTG-3′ were used to amplify a 510 bp fragment encoding a 170aa region comprising residues 2 to 171 of AtRPA1a from cDNA clone pda 11728 (Riken GSC/BRC). The PCR product was cloned into the expression vector pET21b (Novagen) using NheI and Xhol sites incorporated into RPA-SUB-N and RPA-SUB-170, respectively. Recombinant His-tagged protein was isolated from E. coli BL21 (Novagen) under native conditions using Ni-agarose following the manufacturer's protocol (QIAGEN). Polyclonal antiserum against the recombinant protein was raised in rat (ISL, Paignton, UK).
Peptide antiserum was raised in rabbit against the 20aa sequence ETDTEAQKTFSGTGNIPPPN (residues 116–135) conjugated to KLH (Sigma-Genosys Ltd.).
Both antibodies were used at a dilution of 1/500. Specificity was established by application to the Atrpa1a deletion mutant as described in the Results section.
Nucleic acid sequencing
Nucleotide sequencing was carried out by the Functional Genomics Unit, School of Biosciences, University of Birmingham, UK.
Cytological procedures
The cytological methods were carried out as described earlier (Higgins et al, 2004). Besides anti-AtRPA1a (described above), the following antibodies were used in this study: anti-ASY1 (rabbit/rat, 1/500 dilution), anti-MSH4 (rabbit, 1/500 dilution), anti-ZYP1 (rabbit/rat, 1/500 dilution), anti-AtRAD51 (rabbit, 1/500 dilution), anti-AtMLH1 (rabbit, 1/200 dilution), and anti-MLH3 (rabbit/rat, 1/200 dilution) (Mercier et al, 2003; Higgins et al, 2004, 2005). FISH on metaphase I chromosomes was carried out using the 45S rDNA and 5S rDNA probes. Microscopy was conducted using a Nikon Eclipse T300 Microscope (Tokyo, Japan). Image capture and image analysis was done using SmartCapture 2 (Digital Scientific, Cambridge, UK).
Statistical procedures
Observed and Poisson-expected numbers of chiasmata per cell were tested for agreement using a chi-squared (χ2) test. For the purposes of this test, classes were grouped such that none of the expected values fell below five (Lancaster, 1966). The number of degrees of freedom was given by the number of classes (after grouping) minus two, due to the necessity of estimating a parameter, μ, the mean. Analysis of variance was used to compare chiasma frequency between mutant lines.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Acknowledgments
We thank the Biotechnology and Biological Sciences Research Council (UK) for financial support. Valuable technical support was provided by Karen Staples and Steve Price.
References
- AGI (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Alexander MP (1969) Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Allers T, Lichten M (2001) Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57 [DOI] [PubMed] [Google Scholar]
- Alonso JM (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana (vol 301, pg 653, 2003). Science 301: 1849. [DOI] [PubMed] [Google Scholar]
- Armstrong SJ, Caryl AP, Jones GH, Franklin FCH (2002) Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J Cell Science 115: 3645–3655 [DOI] [PubMed] [Google Scholar]
- Berchowitz LE, Francis KE, Bey AL, Copenhaver GP (2007) The role of AtMUS81 in interference-insensitive crossovers in A-thaliana. Plos Genetics 3: 1355–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerner GV, Kleckner N, Hunter N (2004) Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45 [DOI] [PubMed] [Google Scholar]
- Caryl AP, Armstrong SJ, Jones GH, Franklin FCH (2000) A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma 109: 62–71 [DOI] [PubMed] [Google Scholar]
- Couteau F, Belzile F, Horlow C, Grandjean O, Vezon D, Doutriaux MP (1999) Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell 11: 1623–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM (2003) The Mus81/Mms4 endonuclease acts independently of double- holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164: 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggler AL, Inman RB, Cox MM (2002) The Rad51-dependent pairing of long DNA substrates is stabilized by replication protein A. J Biol Chem 277: 39280–39288 [DOI] [PubMed] [Google Scholar]
- Gasior SL, Wong AK, Kora Y, Shinohara A, Bishop DK (1998) Rad52 associates with RPA and functions with Rad55 and Rad57 to assemble meiotic recombination complexes. Genes Dev 12: 2208–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon M, Vezon D, Gendrot G, Pelletier G (2001) AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J 20: 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F, Puchta H (2000) Molecular characterisation of two paralogous SPO11 homologues in Arabidopsis thaliana. Nucleic Acids Res 28: 1548–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F, Wurz-Wildersinn R, Fuchs J, Schubert I, Suer S, Puchta H (2007) The catalytically active tyrosine residues of both SPO11-1 and SPO11-2 are required for meiotic double-strand break induction in Arabidopsis. Plant Cell 19: 3090–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Armstrong SJ, Franklin FCH, Jones GH (2004) The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev 18: 2557–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Buckling EF, Franklin FCH, Jones GH (2008a) Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J 54: 152–162 [DOI] [PubMed] [Google Scholar]
- Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FCH (2005) The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev 19: 2488–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Vignard J, Mercier R, Pugh AH, Franklin FCH, Jones GH (2008b) AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant J 55: 28–39 [DOI] [PubMed] [Google Scholar]
- Hoffmann ER, Borts RH (2004) Meiotic recombination intermediates and mismatch repair proteins. Cytogenet Genome Res 107: 232–248 [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM, Brill SJ (2004) The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev 18: 117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N, Borts RH (1997) Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev 11: 1573–1582 [DOI] [PubMed] [Google Scholar]
- Iftode C, Daniely Y, Borowiec JA (1999) Replication protein A (RPA): The eukaryotic SSB. Crit Rev Biochem Mol Biol 34: 141–180 [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Kimura S, Sakaguchi K (2006) A higher plant has three different types of RPA heterotrimeric complex. J Biochem 139: 99–104 [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Koga A, Yamamoto T, Uchiyama Y, Mori Y, Hashimoto J, Kimura S, Sakaguchi K (2005) Two types of replication protein A in seed plants - Characterization of their functions in vitro and in vivo. FEBS J 272: 3270–3281 [DOI] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Aravind L (2002) Classification and evolutionary history of the single-strand annealing proteins, RecT, Red beta, ERF and RAD52. BMC Genomics 3: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson N, Sanchez-Moran E, Buckling E, Armstrong SJ, Jones GH, Franklin FCH (2006) Reduced meiotic crossovers and delayed prophase I progression At-MLH3-deficient Arabidopsis. EMBO J 25: 1315–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones GH (1984) The control of chiasma distribution. SEB Symposium 38: 293–320 [PubMed] [Google Scholar]
- Jones GH, Armstrong SJ, Caryl AP, Franklin FCH (2003) Meiotic chromosome synapsis and recombination in Arabidopsis thaliana; an integration of cytological and molecular approaches. Chromosome Res 11: 205–215 [DOI] [PubMed] [Google Scholar]
- Kantake N, Sugiyama T, Kolodner RD, Kowalczykowski SC (2003) The recombination-deficient mutant RPA (rfa1-t11) is displaced slowly from single-stranded DNA by Rad51 protein. J Biol Chem 278: 23410–23417 [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384 [DOI] [PubMed] [Google Scholar]
- Kerzendorfer C, Vignard J, Pedrosa-Harand A, Siwiec T, Akimcheva S, Jolivet S, Sablowski R, Armstrong S, Schweizer D, Mercier R, Schlogelhofer P (2006) The Arabidopsis thaliana MND1 homologue plays a key role in meiotic homologous pairing, synapsis and recombination. J Cell Sci 119: 2486–2496 [DOI] [PubMed] [Google Scholar]
- Kolas NK, Cohen PE (2004) Novel and diverse functions of the DNA mismatch repair family in mammalian meiosis and recombination. Cytogenet Genome Res 107: 216–231 [DOI] [PubMed] [Google Scholar]
- Lancaster HO (1966) The Chi-squared distribution. John Wiley and Sons, New York
- Li WX, Chen CB, Markmann-Mulisch U, Timofejeva L, Schmelzer E, Ma H, Reiss B (2004) The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc Natl Acad Sci USA 101: 10596–10601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon E, Moens P (2003) MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics 165: 2283–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Armstrong SJ, Horlow C, Jackson NP, Makaroff CA, Vezon D, Pelletier G, Jones GH, Franklin FCH (2003) The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development 130: 3309–3318 [DOI] [PubMed] [Google Scholar]
- Moens PB, Marcon E, Shore JS, Kochakpour N, Spyropoulos B (2007) Initiation and resolution of interhomolog connections: crossover and non-crossover sites along mouse synaptonemal complexes. J Cell Sci 120: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Oliver-Bonet M, Campillo M, Turek PJ, Ko E, Martin RH (2007) Analysis of replication protein A (RPA) in human spermatogenesis. Mol Hum Reprod 13: 837–844 [DOI] [PubMed] [Google Scholar]
- Plug AW, Peters A, Keegan KS, Hoekstra MF, de Boer P, Ashley T (1998) Changes in protein composition of meiotic nodules during mammalian meiosis. J Cell Sci 111: 413–423 [DOI] [PubMed] [Google Scholar]
- Plug AW, Peters A, Xu Y, Keegan KS, Hoekstra MF, Baltimore D, deBoer P, Ashley T (1997) ATM and RPA in meiotic chromosome synapsis and recombination. Nat Genet 17: 457–461 [DOI] [PubMed] [Google Scholar]
- Puizina J, Siroky J, Mokros P, Schweizer D, Riha K (2004) Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 16: 1968–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald P, Roeder GS (1994) Mutation of a meiosis-specific MutS homolog decreases crossing-over but not mismatch correction. Cell 79: 1069–1080 [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran E, Mercier R, Higgins JD, Armstrong SJ, Jones GH, Franklin FCH (2005) A strategy to investigate the plant meiotic proteome. Cytogenet Genome Res 109: 181–189 [DOI] [PubMed] [Google Scholar]
- Shultz RW, Tatineni VM, Hanley-Bowdoin L, Thompson WF (2007) Genome-wide analysis of the core DNA replication machinery in the higher plants Arabidopsis and rice(1[W][OA]). Plant Physiol 144: 1697–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaud N, Dray E, Gy I, Gerard E, Takvorian N, Doutriaux MP (2004) Brca2 is involved in meiosis in Arabidopsis thaliana as suggested by its interaction with Dmc1. EMBO J 23: 1392–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DK, Islam MN, Choudhury NR, Karjee S, Mukherjee SK (2007) The 32 kDa subunit of replication protein A (RPA) participates in the DNA replication of Mung bean yellow mosaic India virus (MYMIV) by interacting with the viral Rep protein. Nucleic Acids Res 35: 755–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden T, Acharya S, Butz C, Berardini M, Fishel R (2004) hMSH4-hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell 15: 437–451 [DOI] [PubMed] [Google Scholar]
- Snowden T, Shim KS, Schmutte C, Acharya S, Fishel R (2008) hMSH4-hMSH5 adenosine nucleotide processing and interactions with homologous recombination machinery. J Biol Chem 283: 145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soustelle C, Vedel M, Kolodner R, Nicolas A (2002) Replication protein A is required for meiotic recombination in Saccharomyces cerevisiae. Genetics 161: 535–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey NJ, Kuromori T, Azumi Y, Roberts G, Breuer C, Wada T, Maxwell A, Roberts K, Sugimoto-Shirasu K (2006) Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J 48: 206–216 [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Kantake N, Wu Y, Kowalczykowski SC (2006) Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. EMBO J 25: 5539–5548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P, Krejci L, Van Komen S, Sehorn MG (2003) Rad51 recombinase and recombination mediators. J Biol Chem 278: 42729–42732 [DOI] [PubMed] [Google Scholar]
- Wang TF, Kleckner N, Hunter N (1999) Functional specificity of MutL homologs in yeast: Evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc Natl Acad Sci USA 96: 13914–13919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Haber JE (2004) Role of Saccharomyces single-stranded DNA-binding protein RPA in the strand invasion step of double-strand break repair. Plos Biol 2: 104–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YX, Putnam CD, Kane MF, Zhang WJ, Edelmann L, Russell R, Carrion DV, Chin L, Kucherlapati R, Kolodner RD, Edelmann W (2005) Mutation in Rpa1 results in defective DNA double-strand break repair, chromosomal instability and cancer in mice. Nat Genet 37: 750–755 [DOI] [PubMed] [Google Scholar]
- Wold MS (1997) Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem 66: 61–92 [DOI] [PubMed] [Google Scholar]
- Xu P, Zhang YJ, Kang L, Roossinck MJ, Mysore KS (2006) Computational estimation and experimental verification of off-target silencing during posttranscriptional gene silencing in plants. Plant Physiol 142: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7